Co-Occurring Methylenetetrahydrofolate Reductase (MTHFR) rs1801133 and rs1801131 Genotypes as Associative Genetic Modifiers of Clinical Severity in Rett Syndrome
Abstract
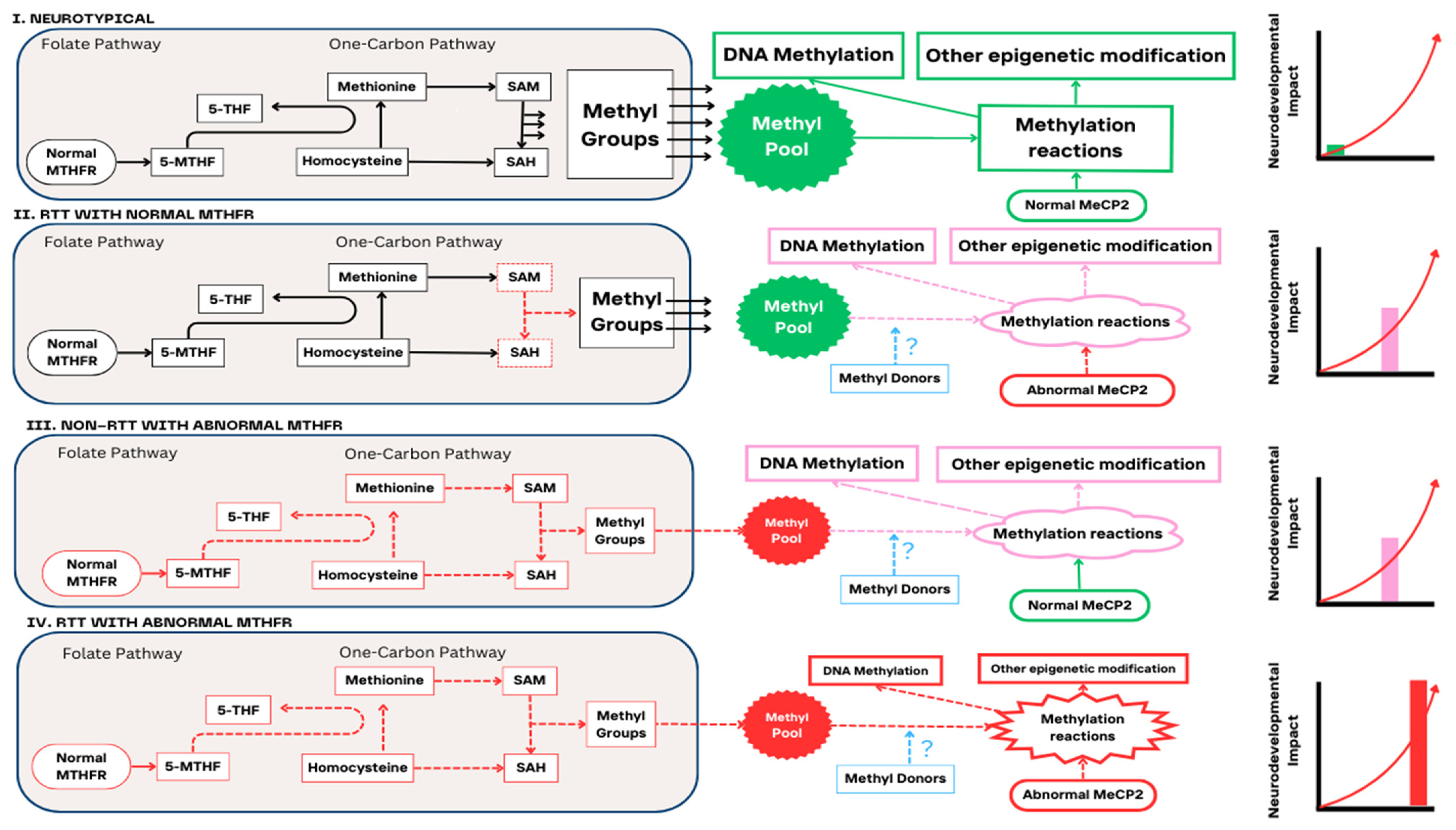
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Determination of MTHFR Variant Frequency
2.2.1. Sample Collection
2.2.2. PGx Test and MTHFR Reporting
PGx Test
MTHFR Reporting
- 1.
- Homozygous designation (n = 17)
- 2.
- Non-homozygous designation (n = 48)
2.3. Assessment of Clinical Severity
2.4. Assessment of AEDs
2.5. Data Extraction and Statistical Analyses
2.6. Informed Consent and Ethics
3. Results
3.1. Study Characteristics
3.2. Frequency of MTHFR rs1801133 and rs1801131 Genotypes in the Sample
3.3. Clinical Severity and MTHFR rs1801133 and rs1801131 Genotypes
3.4. AED Use and MTHFR Genotypes in RTT Patients
3.5. RTT Diagnoses Categorised by MTHFR rs1801133 and rs1801131 Genotypes
4. Discussion
5. Conclusions
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, A.; Miller, M.T.; Li, K.; Sur, M.; Kaufmann, W.E. Towards a better diagnosis and treatment of Rett syndrome: A model synaptic disorder. Brain 2019, 142, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Petriti, U.; Dudman, D.C.; Scosyrev, E.; Lopez-Leon, S. Global prevalence of Rett syndrome: Systematic review and meta-analysis. Syst. Rev. 2023, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Gabel, H.W.; Kinde, B.; Stroud, H.; Gilbert, C.S.; Harmin, D.A.; Kastan, N.R.; Hemberg, M.; Ebert, D.H.; Greenberg, M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Nettles, S.A.; Ikeuchi, Y.; Lefton, K.B.; Abbasi, L.; Erickson, A.; Agwu, C.; Papouin, T.; Bonni, A.; Gabel, H.W. MeCP2 represses the activity of topoisomerase IIβ in long neuronal genes. Cell Rep. 2023, 42, 113538. [Google Scholar] [CrossRef] [PubMed]
- USFDA. Available online: https://www.fda.gov/drugs/news-events-human-drugs/fda-approves-first-treatment-rett-syndrome (accessed on 30 December 2023).
- Neul, J.L.; Percy, A.K.; Benke, T.A.; Berry-Kravis, E.M.; Glaze, D.G.; Marsh, E.D.; Lin, T.; Stankovic, S.; Bishop, K.M.; Youakim, J.M. Trofinetide for the treatment of Rett syndrome: A randomized phase 3 study. Nat. Med. 2023, 29, 1468–1475. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M.; James, S.; Meakin, S.; Green, C.; Scott, A.K.; Walley, T.J.; Farrar, K.; Park, B.K.; Breckenridge, A.M. Adverse drug reactions as cause of admission to hospital: Prospective analysis of 18 820 patients. BMJ 2004, 329, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Osanlou, R.; Walker, L.; Hughes, D.A.; Burnside, G.; Pirmohamed, M. Adverse drug reactions, multimorbidity and polypharmacy: A prospective analysis of 1 month of medical admissions. BMJ Open 2022, 12, e055551. [Google Scholar] [CrossRef] [PubMed]
- Kimpton, J.E.; Carey, I.M.; Threapleton, C.J.D.; Robinson, A.; Harris, T.; Cook, D.G.; DeWilde, S.; Baker, E.H. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: An analysis to inform design of pre-emptive pharmacogenomic testing. Br. J. Clin. Pharmacol. 2019, 85, 2734–2746. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Li, Y.; Zhang, Z.; Sun, Z.; He, Y.; Li, R. Methylenetetrahydrofolate reductase and psychiatric diseases. Transl. Psychiatry 2018, 8, 242. [Google Scholar] [CrossRef]
- Mallhi, T.H.; Shahid, M.; Rehman, K.; Khan, Y.H.; Alanazi, A.S.; Alotaibi, N.H.; Akash, M.S.H.; Butt, M.H. Biochemical Association of MTHFR C677T Polymorphism with Myocardial Infarction in the Presence of Diabetes Mellitus as a Risk Factor. Metabolites 2023, 13, 251. [Google Scholar] [CrossRef]
- Barretta, F.; Uomo, F.; Fecarotta, S.; Albano, L.; Crisci, D.; Verde, A.; Fisco, M.G.; Gallo, G.; Stagna, D.D.; Pricolo, M.R.; et al. Contribution of Genetic Test to Early Diagnosis of Methylenetetrahydrofolate Reductase (MTHFR) Deficiency: The Experience of a Reference Center in Southern Italy. Genes 2023, 14, 980. [Google Scholar] [CrossRef]
- El-Khawaga, O.Y.; Al-Azzawy, M.F.; El-Dawa, A.N.; ElSaid, A.M.; Mustafa, W.; Saad, M. Association study between genetic polymorphisms in MTHFR and stroke susceptibility in Egyptian population: A case–control study. Sci. Rep. 2024, 14, 114. [Google Scholar] [CrossRef]
- van der Pol, K.H.; Nijenhuis, M.; Soree, B.; de Boer-Veger, N.J.; Buunk, A.M.; Guchelaar, H.J.; Risselada, A.; van Schaik, R.H.; Swen, J.J.; Touw, D.; et al. Dutch pharmacogenetics working group guideline for the gene-drug interaction of ABCG2, HLA-B and Allopurinol, and MTHFR, folic acid and methotrexate. Eur. J. Hum. Genet. 2024, 32, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Tulsyan, S.; Agarwal, G.; Lal, P.; Agrawal, S.; Mittal, R.D.; Mittal, B. Relationship of MTHFR and NQO1 Pharmacogenetics and Chemotherapy Clinical Outcomes in Breast Cancer Patients. Biochem. Genet. 2015, 53, 211–222. [Google Scholar] [CrossRef]
- D’Angelo, V.; Ramaglia, M.; Iannotta, A.; Addeo, R. Pharmacogenetics of methotrexate in pediatric hematological neoplasm treatment: Does it need a personalized regimen based on MTHFR polymorphisms? Expert Rev. Hematol. 2014, 7, 517–519. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salazar, J.; Altés, A.; del Río, E.; Estella, J.; Rives, S.; Tasso, M.; Navajas, A.; Molina, J.; Villa, M.; Vivanco, J.L.; et al. Methotrexate consolidation treatment according to pharmacogenetics of MTHFR ameliorates event-free survival in childhood acute lymphoblastic leukaemia. Pharmacogenom. J. 2012, 12, 379–385. [Google Scholar] [CrossRef]
- Miranda-Vilela, A.L. Role of polymorphisms in factor V (FV Leiden), prothrombin, plasminogen activator inhibitor type-1 (PAI-1), methylenetetrahydrofolate reductase (MTHFR) and cystathionine β-synthase (CBS) genes as risk factors for thrombophilias. Mini Rev. Med. Chem. 2012, 12, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- González-Mercado, M.G.; Rivas, F.; Gallegos-Arreola, M.P.; Morán-Moguel, M.C.; Salazar-Páramo, M.; González-López, L.; Gámez-Nava, J.I.; Munoz-Valle, J.F.; Medina-Coss y León, R.; González-Mercado, A.; et al. MTRR A66G, RFC1 G80A, and MTHFR C677T and A1298C Polymorphisms and Disease Activity in Mexicans with Rheumatoid Arthritis Treated with Methotrexate. Genet. Test. Mol. Biomark. 2017, 21, 698–704. [Google Scholar] [CrossRef]
- Lv, S.; Fan, H.; Li, J.; Yang, H.; Huang, J.; Shu, X.; Zhang, L.; Xu, Y.; Li, X.; Zuo, J.; et al. Genetic Polymorphisms of TYMS, MTHFR, ATIC, MTR, and MTRR Are Related to the Outcome of Methotrexate Therapy for Rheumatoid Arthritis in a Chinese Population. Front. Pharmacol. 2018, 9, 1390. [Google Scholar] [CrossRef]
- Nishimoto, E.; Ito, Y.; Sakakibara, T.; Nishikubo, T. Early treatment using betaine and methionine for a neonate with MTHFR deficiency. Pediatr. Int. 2019, 61, 1265–1266. [Google Scholar] [CrossRef]
- Bottiglieri, T.; Laundy, M.; Crellin, R.; Toone, B.K.; Carney, M.W.P.; Reynolds, E.H. Homocysteine, folate, methylation, and monoamine metabolism in depression. J. Neurol. Neurosurg. Psychiatry 2000, 69, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Friso, S.; Choi, S.-W.; Girelli, D.; Mason, J.B.; Dolnikowski, G.G.; Bagley, P.J.; Olivieri, O.; Jacques, P.F.; Rosenberg, I.H.; Corrocher, R.; et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 2002, 99, 5606–5611. [Google Scholar] [CrossRef]
- Froese, D.S.; Huemer, M.; Suormala, T.; Burda, P.; Coelho, D.; Guéant, J.L.; Landolt, M.A.; Kožich, V.; Fowler, B.; Baumgartner, M.R. Mutation Update and Review of Severe Methylenetetrahydro-folate Reductase Deficiency. Hum. Mutat. 2016, 37, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Mulder-Bleile, R.; Burda, P.; Froese, D.S.; Suormala, T.; Zeev, B.B.; Chinnery, P.F.; Dionisi-Vici, C.; Dobbelaere, D.; Gökcay, G.; et al. Clinical pattern, mutations and in vitro residual activity in 33 patients with severe 5, 10 methylenetetrahydrofolate reductase (MTHFR) deficiency. J. Inherit. Metab. Dis. 2016, 39, 115–124. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Yang, H.-Y.; Ding, X.-X.; Zhao, X.; Chen, J.; Bi, P.; Sun, Y.-H. Association between methylenetetrahydrofolate reductase C677T polymorphism and epilepsy susceptibility: A meta-analysis. Seizure 2014, 23, 411–416. [Google Scholar] [CrossRef]
- Gales, A.; Masingue, M.; Millecamps, S.; Giraudier, S.; Grosliere, L.; Adam, C.; Salim, C.; Navarro, V.; Nadjar, Y. Adolescence/adult onset MTHFR deficiency may manifest as isolated and treatable distinct neuro-psychiatric syndromes. Orphanet J. Rare Dis. 2018, 13, 29. [Google Scholar] [CrossRef]
- D’Aco, K.E.; Bearden, D.; Watkins, D.; Hyland, K.; Rosenblatt, D.S.; Ficicioglu, C. Severe 5,10-methylenetetrahydrofolate reductase deficiency and two MTHFR variants in an adolescent with progressive myoclonic epilepsy. Pediatr. Neurol. 2014, 51, 266–270. [Google Scholar] [CrossRef]
- Shimura, M.; Yamada, H.; Takahashi, H.; Yamada, N.; Go, S.; Yamanaka, G.; Kawashima, H. Antiepileptic drug-induced psychosis associated with MTHFRC677T: A case report. J. Med. Case Rep. 2019, 13, 250. [Google Scholar] [CrossRef] [PubMed]
- McInnes, G.; Lavertu, A.; Sangkuhl, K.; Klein, T.E.; Whirl-Carrillo, M.; Altman, R.B. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin. Pharmacol. Ther. 2020, 109, 1528–1537. [Google Scholar] [CrossRef]
- Swen, J.J.; van der Wouden, C.H.; Manson, L.E.; Abdullah-Koolmees, H.; Blagec, K.; Blagus, T.; Böhringer, S.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.-C.; et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: An open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet 2023, 401, 347–356. [Google Scholar] [CrossRef]
- Lavery, L.A.; Zoghbi, H.Y. The distinct methylation landscape of maturing neurons and its role in Rett syndrome pathogenesis. Curr. Opin. Neurobiol. 2019, 59, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Gorgone, G.; Caccamo, D.; Pisani, L.R.; Currò, M.; Parisi, G.; Oteri, G.; Ientile, R.; Rossini, P.M.; Pisani, F. Hyperhomocysteinemia in patients with epilepsy: Does it play a role in the pathogenesis of brain atrophy? A preliminary report. Epilepsia 2009, 50, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Neul, J.L.; Kaufmann, W.E.; Glaze, D.G.; Christodoulou, J.; Clarke, A.J.; Bahi-Buisson, N.; Leonard, H.; Bailey, M.E.S.; Schanen, N.C.; Zappella, M.; et al. Rett syndrome: Revised diagnostic criteria and nomenclature. Ann. Neurol. 2010, 68, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Ameenpur, S.; Ahmed, R.; Basheer, S.; Chishti, S.; Lawrence, R.; Fiori, F.; Santosh, P. An Observational Study of Heart Rate Variability Using Wearable Sensors Provides a Target for Therapeutic Monitoring of Autonomic Dysregulation in Patients with Rett Syndrome. Biomedicines 2022, 10, 1684. [Google Scholar] [CrossRef] [PubMed]
- Neul, J.L.; Glaze, D.G.; Percy, A.K.; Feyma, T.; Beisang, A.; Dinh, T.; Suter, B.; Anagnostou, E.; Snape, M.; Horrigan, J.; et al. Improving Treatment Trial Outcomes for Rett Syndrome: The Development of Rett-specific Anchors for the Clinical Global Impression Scale. J. Child Neurol. 2015, 30, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Suormala, T.; Heuberger, D.; Schäfer, A.; Fowler, B.; Froese, D.S.; Baumgartner, M.R. Functional characterization of missense mutations in severe methylenetetrahydrofolate reductase deficiency using a human expression system. J. Inherit. Metab. Dis. 2017, 40, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Das, S.K.; Sharma, P.; Karthikeyan, G.; Ramakrishnan, L.; Sengupta, S. Homocysteine levels are associated with MTHFR A1298C polymorphism in Indian population. J. Hum. Genet. 2005, 50, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Graydon, J.S.; Claudio, K.; Baker, S.; Kocherla, M.; Ferreira, M.; Roche-Lima, A.; Rodríguez-Maldonado, J.; Duconge, J.; Ruaño, G. Ethnogeographic prevalence and implications of the 677C>T and 1298A>C MTHFR polymorphisms in US primary care populations. Biomark. Med. 2019, 13, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.P.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M. 1000 Genomes Project Consortium; A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Chango, A.; Boisson, F.; Barbé, F.; Quilliot, D.; Droesch, S.; Pfister, M.; Fillon-Emery, N.; Lambert, D.; Frémont, S.; Rosenblatt, D.S.; et al. The effect of 677C-->T and 1298A-->C mutations on plasma homocysteine and 5,10-methylenetetrahydrofolate reductase activity in healthy subjects. Br. J. Nutr. 2000, 83, 593–596. [Google Scholar] [CrossRef]
- Belcastro, V.; Striano, P.; Gorgone, G.; Costa, C.; Ciampa, C.; Caccamo, D.; Pisani, L.R.; Oteri, G.; Marciani, M.G.; Aguglia, U.; et al. Hyperhomocysteinemia in epileptic patients on new antiepileptic drugs. Epilepsia 2010, 51, 274–279. [Google Scholar] [CrossRef]
- Ramaekers, V.; Hansen, S.; Holm, J.; Opladen, T.; Senderek, J.; Häusler, M.; Heimann, G.; Fowler, B.; Maiwald, R.; Blau, N. Reduced folate transport to the CNS in female Rett patients. Neurology 2003, 61, 506–515. [Google Scholar] [CrossRef]
- Ramaekers, V.T.; Sequeira, J.M.; Artuch, R.; Blau, N.; Temudo, T.; Ormazabal, A.; Pineda, M.; Aracil, A.; Roelens, F.; Laccone, F.; et al. Folate receptor autoantibodies and spinal fluid 5-methyltetrahydrofolate deficiency in Rett syndrome. Neuropediatrics 2007, 38, 179–183. [Google Scholar] [CrossRef]
- Ormazabal, A.; Artuch, R.; Vilaseca, M.A.; Aracil, A.; Pineda, M. Cerebrospinal fluid concentrations of folate, biogenic amines and pterins in Rett syndrome: Treatment with folinic acid. Neuropediatrics 2005, 36, 380–385. [Google Scholar] [CrossRef]
- Neul, J.L.; Maricich, S.M.; Islam, M.; Barrish, J.; Smith, E.O.; Bottiglieri, T.; Hyland, K.; Humphreys, P.; Percy, A.; Glaze, D. Spinal fluid 5-methyltetrahydrofolate levels are normal in Rett syndrome. Neurology 2005, 64, 2151–2152. [Google Scholar] [CrossRef]
- Diekman, E.F.; de Koning, T.J.; Verhoeven-Duif, N.M.; Rovers, M.M.; van Hasselt, P.M. Survival and psychomotor development with early betaine treatment in patients with severe methylenetetrahydrofolate reductase deficiency. JAMA Neurol. 2014, 71, 188–194. [Google Scholar] [CrossRef]
- Lam, N.S.K.; Long, X.X.; Li, X.; Saad, M.; Lim, F.; Doery, J.C.; Griffin, R.C.; Galletly, C. The potential use of folate and its derivatives in treating psychiatric disorders: A systematic review. Biomed. Pharmacother. 2022, 146, 112541. [Google Scholar] [CrossRef]
- Glaze, D.G.; Percy, A.K.; Motil, K.J.; Lane, J.B.; Isaacs, J.S.; Schultz, R.J.; Barrish, J.O.; Neul, J.L.; O’Brien, W.E.; Smith, E.O. A study of the treatment of Rett syndrome with folate and betaine. J. Child Neurol. 2009, 24, 551–556. [Google Scholar] [CrossRef]
- Hagebeuk, E.E.O.; Duran, M.; Koelman, J.H.T.M.; Abeling, N.G.G.M.; Vyth, A.; Poll-The, B.-T. Folinic acid supplementation in Rett syndrome patients does not influence the course of the disease: A randomized study. J. Child Neurol. 2012, 27, 304–309. [Google Scholar] [CrossRef]
- Temudo, T.; Rios, M.; Prior, C.; Carrilho, I.; Santos, M.; Maciel, P.; Sequeiros, J.; Fonseca, M.; Monteiro, J.; Cabral, P.; et al. Evaluation of CSF neurotransmitters and folate in 25 patients with Rett disorder and effects of treatment. Brain Dev. 2009, 31, 46–51. [Google Scholar] [CrossRef]
- Hagebeuk, E.E.; Duran, M.; Abeling, N.G.; Vyth, A.; Poll-The, B.T. S-adenosylmethionine and S-adenosylhomocysteine in plasma and cerebrospinal fluid in Rett syndrome and the effect of folinic acid supplementation. J. Inherit. Metab. Dis. 2013, 36, 967–972. [Google Scholar] [CrossRef]
- Huemer, M.; Diodato, D.; Schwahn, B.; Schiff, M.; Bandeira, A.; Benoist, J.F.; Burlina, A.; Cerone, R.; Couce, M.L.; Garcia-Cazorla, A.; et al. Guidelines for diagnosis and management of the cobalamin-related remethylation disorders cblC, cblD, cblE, cblF, cblG, cblJ and MTHFR deficiency. J. Inherit. Metab. Dis. 2017, 40, 21–48. [Google Scholar] [CrossRef]
- Huemer, M.; Diodato, D.; Martinelli, D.; Olivieri, G.; Blom, H.; Gleich, F.; Kölker, S.; Kožich, V.; Morris, A.A.; Seifert, B.; et al. Phenotype, treatment practice and outcome in the cobalamin-dependent remethylation disorders and MTHFR deficiency: Data from the E-HOD registry. J. Inherit. Metab. Dis. 2019, 42, 333–352. [Google Scholar] [CrossRef]
- Yverneau, M.; Leroux, S.; Imbard, A.; Gleich, F.; Arion, A.; Moreau, C.; Nassogne, M.; Szymanowski, M.; Tardieu, M.; Touati, G.; et al. Influence of early identification and therapy on long-term outcomes in early-onset MTHFR deficiency. J. Inherit. Metab. Dis. 2022, 45, 848–861. [Google Scholar] [CrossRef]
- Cuddapah, V.A.; Pillai, R.B.; Shekar, K.V.; Lane, J.B.; Motil, K.J.; Skinner, S.A.; Tarquinio, D.C.; Glaze, D.G.; McGwin, G.; Kaufmann, W.E.; et al. Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J. Med. Genet. 2014, 51, 152–158. [Google Scholar] [CrossRef]
- Ham, A.L.; Kumar, A.; Deeter, R.; Schanen, N.C. Does genotype predict phenotype in Rett syndrome? J. Child Neurol. 2005, 20, 768–778. [Google Scholar] [CrossRef]
- Halbach, N.S.; Smeets, E.E.; Braak, N.v.D.; van Roozendaal, K.E.; Blok, R.M.; Schrander-Stumpel, C.T.; Frijns, J.; Maaskant, M.A.; Curfs, L.M. Genotype-phenotype relationships as prognosticators in Rett syndrome should be handled with care in clinical practice. Am. J. Med. Genet. Part A 2011, 158A, 340–350. [Google Scholar] [CrossRef]
- Fu, C.; Armstrong, D.; Marsh, E.; Lieberman, D.; Motil, K.; Witt, R.; Standridge, S.; Nues, P.; Lane, J.; Dinkel, T.; et al. Consensus guidelines on managing Rett syndrome across the lifespan. BMJ Paediatr. Open 2020, 4, e000717. [Google Scholar] [CrossRef]
- Aka, I.; Bernal, C.J.; Carroll, R.; Maxwell-Horn, A.; Oshikoya, K.A.; Van Driest, S.L. Clinical Pharmacogenetics of Cytochrome P450-Associated Drugs in Children. J. Pers. Med. 2017, 7, 14. [Google Scholar] [CrossRef]
- Roberts, T.A.; Wagner, J.A.; Sandritter, T.; Black, B.T.; Gaedigk, A.; Stancil, S.L. Retrospective Review of Pharmacogenetic Testing at an Academic Children’s Hospital. Clin. Transl. Sci. 2020, 14, 412–421. [Google Scholar] [CrossRef]
- Bousman, C.A.; Al Maruf, A.; Marques, D.F.; Brown, L.C.; Müller, D.J. The emergence, implementation, and future growth of pharmacogenomics in psychiatry: A narrative review. Psychol. Med. 2023, 53, 7983–7993. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M. Pharmacogenomics: Current status and future perspectives. Nat. Rev. Genet. 2023, 24, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Skol, A.D.; Perera, M.A. The limits of genome-wide methods for pharmacogenomic testing. Pharmacogenetics Genom. 2012, 22, 261–272. [Google Scholar] [CrossRef]
- Johnson, D.; Wilke, M.A.; Lyle, S.M.; Kowalec, K.; Jorgensen, A.; Wright, G.E.; Drögemöller, B.I. A Systematic Review and Analysis of the Use of Polygenic Scores in Pharmacogenomics. Clin. Pharmacol. Ther. 2021, 111, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Micale, V.; Di Bartolomeo, M.; Di Martino, S.; Stark, T.; Dell’Osso, B.; Drago, F.; D’Addario, C. Are the epigenetic changes predictive of therapeutic efficacy for psychiatric disorders? A translational approach towards novel drug targets. Pharmacol. Ther. 2023, 241, 108279. [Google Scholar] [CrossRef]
- Marques, D.F.; Ota, V.K.; Santoro, M.L.; Talarico, F.; Costa, G.O.; Spindola, L.M.; Cogo-Moreira, H.; Carvalho, C.M.; Xavier, G.; Cavalcante, D.A.; et al. LINE-1 hypomethylation is associated with poor risperidone response in a first episode of psychosis cohort. Epigenomics 2020, 12, 1041–1051. [Google Scholar] [CrossRef]
- McInnes, G.; Yee, S.W.; Pershad, Y.; Altman, R.B. Genomewide Association Studies in Pharmacogenomics. Clin. Pharmacol. Ther. 2021, 110, 637–648. [Google Scholar] [CrossRef]
- Popejoy, A.B. Diversity in Precision Medicine and Pharmacogenetics: Methodological and Conceptual Considerations for Broadening Participation. Pharmacogenomics Pers. Med. 2019, 12, 257–271. [Google Scholar] [CrossRef]
| (A) | ||||
| Patients with a PGx Test (n = 65) | ||||
| Diagnoses | ||||
| Rett Syndrome | 60 | |||
| Atypical Rett Syndrome | 5 | |||
| Ethnicity | ||||
| White: English, Welsh, Scottish, Northern Irish or British | 53 | |||
| Mixed/Multiple ethnic groups | 8 | |||
| Black/African/Caribbean/Black British | 1 | |||
| Asian/Asian British | 2 | |||
| Other ethnic groups | 1 | |||
| Age (years) | ||||
| Mean (min. max) | 18.7 (2.5; 47.5) | |||
| Standard deviation | ±12.1 | |||
| (B) | ||||
| MTHFR Genotypes | Phenotype (MTHFR Activity) * | n | ||
| rs1801133 (c.665 C>T) | rs1801131 (c.1286 A>C) | |||
| AG | GT | Mild | 10 | |
| AG | TT | Partial | 15 | |
| GA | GT | Partial | 1 | |
| GA | TT | Partial | 2 | |
| GG | GT | Partial | 13 | |
| TT | Yes (reduced) | 1 | ||
| AA | TT | Yes (reduced) | 5 | |
| GG | GG | Yes (reduced) | 11 | |
| CC | Normal | 1 | ||
| GG | TT | Normal | 6 | |
| (A) | |||||||
| CLINICAL DOMAINS | CGI-S: 1 | CGI-S: 2 | CGI-S: 3 | CGI-S: 4 | CGI-S: 5 | CGI-S: 6 | CGI-S: 7 |
| Language/ Communication | (CSS = 0) | (CSS < 5) | (CSS 5–10) | (CSS 10–20) | (CSS 20–25) | (CSS 25–35) | (CSS 35–40) |
| Normal | Appropriate. May have unusual features such as perseveration/echolalia. Reading disability/dyslexia | Phrase-sentences. May have conversations or echolalia | Words (<5). Babbles. Makes choices. 25–50% | No words. Babbles. Makes choices ≤25% | Vocalizations. Occasionally screams. Makes no choices or only rarely makes choices | No words. No vocalizations. Screams. No choices | |
| Homozygous n = 16 | 0 (0%) | 0 (0%) | 0 (0%) | 5 (31.2%) | 5 (31.2%) | 6 (37.5%) | 0 (0%) |
| Heterozygous n = 43 | 0 (0%) | 0 (0%) | 2 (4.65%) | 11 (25.5%) | 13 (30.2%) | 15 (34.8%) | 2 (4.6%) |
| Ambulation | No impairment | Normal, may have slight evidence of dystonia/ataxia/dyspraxia upon careful examination | Walks, able to use stairs/run. May ride tricycle or climb | Walks independently, unable to use stairs or run | Walks with assistance | Stands with support or independently. May walk with support. Sits independently or with support | Cannot sit. Does not stand or walk |
| Homozygous n = 15 | 0 (0%) | 0 (0%) | 0 (0%) | 2 (13.3%) | 2 (13.3%) | 9 (60%) | 2 (13.3%) |
| Heterozygous n = 44 | 0 (0%) | 1 (2.27%) | 6 (13.6%) | 7 (15.9%) | 12 (27.2%) | 14 (31.8%) | 4 (9.1%) |
| Hand Use | Completely normal, no impairment | Normal, may have slight fine motor issues | Bilateral pincer grasp. May use pen to write but has some fine motor issues like tremor | Reaches for objects, raking grasp or unilateral pincer. May use utensils/cup | Reaches. No grasps. | Rarely occasionally reaches out. No grasp | None |
| Homozygous n = 17 | 0 (0%) | 0 (0%) | 0 (0%) | 5 (29.4%) | 5 (29.4%) | 6 (35.2%) | 1 (5.88%) |
| Heterozygous n = 44 | 1 (2.27%) | 0 (0%) | 2 (4.55%) | 20 (45.4%) | 15 (34.1%) | 5 (11.3%) | 1 (2.27%) |
| Social (Eye Contact) | Normal | Occasional eye gaze avoidance | Appropriate eye contact, >30 s | Eye contact <20 s | Eye contact <10 s | Eye contact, inconsistent 5 s | No eye contact |
| Homozygous n = 14 | 0 (0%) | 0 (0%) | 1 (7.14%) | 7 (50%) | 6 (42.8%) | 0 (0%) | 0 (0%) |
| Heterozygous n = 39 | 0 (0%) | 5 (12.8%) | 9 (23.1% | 12 (30.7%) | 10 (25.6%) | 3 (7.69%) | 0 (0%) |
| Autonomic | None | Minimal | No or minimal breathing abnormalities (<5% of times observed) and warm, pink extremities | Breathing dysrhythmia <50%. No cyanosis. Cool UE and LE pink | Breathing dysrhythmia 50%. No cyanosis. Cool UE and LE pink | Breathing dysrhythmia, 50–100%, maybe with cyanosis. Cold LE or UE, may be blue | Breathing dysrhythmia, constantly with cyanosis. Cold UE and LE. Mottled/blue |
| Homozygous n = 17 | 0 (0%) | 1 5.88%) | 4 (23.5%) | 3 (17.6%) | 4 (23.5%) | 5 (29.4%) | 0 (0%) |
| Heterozygous n = 47 | 0 (0%) | 0 (0%) | 6 (12.7%) | 22 (46.8%) | 12 (25.5%) | 7 (14.8%) | 0 (0%) |
| Seizures | None | None or controlled | None, with or without meds | Monthly– Weekly | Weekly | Weekly–Daily | Daily |
| Homozygous n= 17 | 1 (5.8%) | 3 (17.6%) | 1 (5.88%) | 4 (23.5%) | 0 (0%) | 6 (35.2%) | 2 (11.7%) |
| Heterozygous n = 47 | 10 (21.2%) | 8 (17.0%) | 5 (10.6%) | 9 (19.1%) | 5 (10.6%) | 6 (12.7%) | 4 (8.51%) |
| Attentiveness | Entirely normal | Occasional inattention | Attentive to conversation and follows commands | 50–100% of the time | 50% of the time | Less than 50% of the time | 0% |
| Homozygous n = 17 | 0 (0%) | 0 (0%) | 2 (11.7%) | 4 (23.5%) | 7 (41.1%) | 4 (23.5%) | 0 (0%) |
| Heterozygous n = 42 | 0 (0%) | 0 (0%) | 8 (19.0%) | 11 (26.1%) | 11 (26.1%) | 12 (28.5%) | 0 (0%) |
| (B) | |||||||
| RTT CGI-S Clinical Domains | MTHFR rs1801133 and rs1801131 Genotypes | ||||||
| Non-Homozygous (n = 47 *) | Homozygous (n = 17) | ||||||
| Normal * (1) | 0 | 0 | |||||
| Borderline impaired (2) | 0 | 0 | |||||
| Mildly impaired (3) | 4 | 0 | |||||
| Moderately impaired (4) | 21 | 3 | |||||
| Markedly impaired (5) | 16 | 10 | |||||
| Severely impaired (6) | 6 | 4 | |||||
| Extremely impaired (7) | 0 | 0 | |||||
| Number of AEDs * | MTHFR rs1801133 and rs1801131 Genotypes | |||
|---|---|---|---|---|
| Homozygous (n = 17) | Homozygous (%) | Non-Homozygous (n = 48) | Non-Homozygous (%) | |
| None | 5 | 29% | 15 | 31% |
| 1 | 7 | 41% | 24 | 50% |
| 2 | 4 | 24% | 7 | 15% |
| 3 | 1 | 6% | 1 | 2% |
| 4 | 0 | 0% | 0 | 0% |
| 5 | 0 | 0% | 1 | 2% |
| Average number of AEDs used | 1.1 ± 0.9 | 100% | 1.0 ± 0.9 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, J.; Wilkins, G.; Goodman-Vincent, E.; Chishti, S.; Bonilla Guerrero, R.; McFadden, L.; Zahavi, Z.; Santosh, P. Co-Occurring Methylenetetrahydrofolate Reductase (MTHFR) rs1801133 and rs1801131 Genotypes as Associative Genetic Modifiers of Clinical Severity in Rett Syndrome. Brain Sci. 2024, 14, 624. https://doi.org/10.3390/brainsci14070624
Singh J, Wilkins G, Goodman-Vincent E, Chishti S, Bonilla Guerrero R, McFadden L, Zahavi Z, Santosh P. Co-Occurring Methylenetetrahydrofolate Reductase (MTHFR) rs1801133 and rs1801131 Genotypes as Associative Genetic Modifiers of Clinical Severity in Rett Syndrome. Brain Sciences. 2024; 14(7):624. https://doi.org/10.3390/brainsci14070624
Chicago/Turabian StyleSingh, Jatinder, Georgina Wilkins, Ella Goodman-Vincent, Samiya Chishti, Ruben Bonilla Guerrero, Leighton McFadden, Zvi Zahavi, and Paramala Santosh. 2024. "Co-Occurring Methylenetetrahydrofolate Reductase (MTHFR) rs1801133 and rs1801131 Genotypes as Associative Genetic Modifiers of Clinical Severity in Rett Syndrome" Brain Sciences 14, no. 7: 624. https://doi.org/10.3390/brainsci14070624
APA StyleSingh, J., Wilkins, G., Goodman-Vincent, E., Chishti, S., Bonilla Guerrero, R., McFadden, L., Zahavi, Z., & Santosh, P. (2024). Co-Occurring Methylenetetrahydrofolate Reductase (MTHFR) rs1801133 and rs1801131 Genotypes as Associative Genetic Modifiers of Clinical Severity in Rett Syndrome. Brain Sciences, 14(7), 624. https://doi.org/10.3390/brainsci14070624






