Machine Learning Predicts Phenoconversion from Polysomnography in Isolated REM Sleep Behavior Disorder
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Recordings
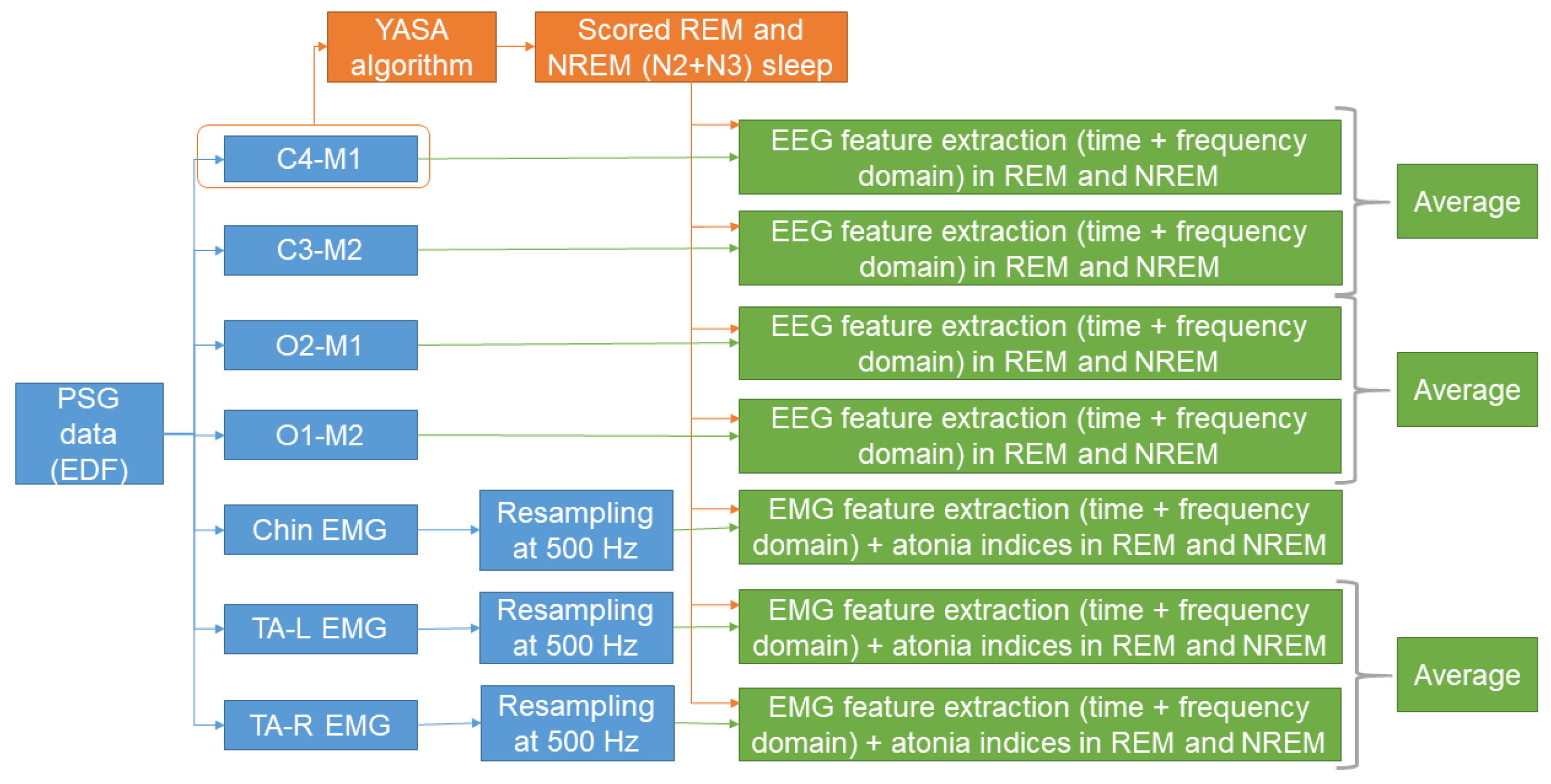
2.2. Automatic Sleep Stage Scoring
2.3. Feature Extraction
2.3.1. EEG Features
2.3.2. EMG Features
2.4. Machine Learning Model Training and Testing
2.5. Code
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AHI | apnea-hypopnea index |
| AUC | area under the receiver operating characteristic curve |
| DLB | dementia with Lewy bodies |
| EEG | electroencephalography |
| EEGREM | EEG features calculated in REM sleep |
| EEGNREM | EEG features calculated in NREM sleep |
| EMG | electromyography |
| EMGREM | EMG features calculated in REM sleep |
| EMGNREM | EMG features calculated in NREM sleep |
| κ | Cohen’s kappa |
| iRBD | isolated REM sleep behavior disorder |
| mbest | best number of features |
| MSA | multiple system atrophy |
| N1 | NREM stage 1 |
| N2 | NREM stage 2 |
| N3 | NREM stage 3 |
| NREM | non-REM |
| REM | rapid eye movement |
| RWA | REM sleep without atonia |
| PD | Parkinson’s disease |
| PLMS | periodic leg movements in sleep |
| PSG | polysomnography |
| TA | tibialis anterior |
| v-PSG | video-PSG |
| W | wakefulness |
References
- Calabresi, P.; Mechelli, A.; Natale, G.; Volpicelli-Daley, L.; Di Lazzaro, G.; Ghiglieri, V. Alpha-Synuclein in Parkinson’s Disease and Other Synucleinopathies: From Overt Neurodegeneration Back to Early Synaptic Dysfunction. Cell Death Dis. 2023, 14, 176. [Google Scholar] [CrossRef]
- Savica, R.; Boeve, B.F.; Mielke, M.M. When Do α-Synucleinopathies Start? An Epidemiological Timeline: A Review. JAMA Neurol. 2018, 75, 503–509. [Google Scholar] [CrossRef]
- Mahlknecht, P.; Marini, K.; Werkmann, M.; Poewe, W.; Seppi, K. Prodromal Parkinson’s Disease: Hype or Hope for Disease-Modification Trials? Transl. Neurodegener. 2022, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- American Academy of Sleep Medicine. International Classification of Sleep Disorders, 3rd, text revision ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2023. [Google Scholar]
- Cesari, M.; Heidbreder, A.; St Louis, E.K.; Sixel-Döring, F.; Bliwise, D.L.; Baldelli, L.; Bes, F.; Fantini, M.L.; Iranzo, A.; Knudsen-Heier, S.; et al. Video-Polysomnography Procedures for Diagnosis of Rapid Eye Movement Sleep Behavior Disorder (RBD) and the Identification of Its Prodromal Stages: Guidelines from the International RBD Study Group. Sleep 2022, 45, zsab257. [Google Scholar] [CrossRef] [PubMed]
- Miglis, M.G.; Adler, C.H.; Antelmi, E.; Arnaldi, D.; Baldelli, L.; Boeve, B.F.; Cesari, M.; Dall’Antonia, I.; Diederich, N.J.; Doppler, K.; et al. Biomarkers of Conversion to α-Synucleinopathy in Isolated Rapid-Eye-Movement Sleep Behaviour Disorder. Lancet Neurol. 2021, 20, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, A.; Verga, L.; Giora, E.; Zucconi, M.; Ferini-Strambi, L. The Risk of Neurodegeneration in REM Sleep Behavior Disorder: A Systematic Review and Meta-Analysis of Longitudinal Studies. Sleep Med. Rev. 2019, 43, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Videnovic, A.; Ju, Y.-E.S.; Arnulf, I.; Cochen-De Cock, V.; Högl, B.; Kunz, D.; Provini, F.; Ratti, P.-L.; Schiess, M.C.; Schenck, C.H.; et al. Clinical Trials in REM Sleep Behavioural Disorder: Challenges and Opportunities. J. Neurol. Neurosurg. Psychiatry 2020, 91, 740–749. [Google Scholar] [CrossRef]
- Högl, B.; Stefani, A.; Videnovic, A. Idiopathic REM Sleep Behaviour Disorder and Neurodegeneration—An Update. Nat. Rev. Neurol. 2018, 14, 40–55. [Google Scholar] [CrossRef]
- Figorilli, M.; Lanza, G.; Congiu, P.; Lecca, R.; Casaglia, E.; Mogavero, M.P.; Puligheddu, M.; Ferri, R. Neurophysiological Aspects of REM Sleep Behavior Disorder (RBD): A Narrative Review. Brain Sci. 2021, 11, 1588. [Google Scholar] [CrossRef]
- Christensen, J.A.E.; Kempfner, J.; Zoetmulder, M.; Leonthin, H.L.; Arvastson, L.; Christensen, S.R.; Sorensen, H.B.D.; Jennum, P. Decreased Sleep Spindle Density in Patients with Idiopathic REM Sleep Behavior Disorder and Patients with Parkinson’s Disease. Clin. Neurophysiol. 2014, 125, 512–519. [Google Scholar] [CrossRef]
- Christensen, J.A.E.; Cesari, M.; Pizza, F.; Antelmi, E.; Frandsen, R.A.V.; Plazzi, G.; Jennum, P. Nocturnal Eye Movements in Patients with Idiopathic Rapid Eye Movement Sleep Behaviour Disorder and Patients with Parkinson’s Disease. J. Sleep Res. 2021, 30, e13125. [Google Scholar] [CrossRef]
- Christensen, J.A.E.; Zoetmulder, M.; Koch, H.; Frandsen, R.; Arvastson, L.; Christensen, S.R.; Jennum, P.; Sorensen, H.B.D. Data-Driven Modeling of Sleep EEG and EOG Reveals Characteristics Indicative of Pre-Parkinson’s and Parkinson’s Disease. J. Neurosci. Methods 2014, 235, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R.B.; Gagnon, J.F.; Rompré, S.; Montplaisir, J.Y. Severity of REM Atonia Loss in Idiopathic REM Sleep Behavior Disorder Predicts Parkinson Disease. Neurology 2010, 74, 239–244. [Google Scholar] [CrossRef] [PubMed]
- McCarter, S.J.; Sandness, D.J.; McCarter, A.R.; Feemster, J.C.; Teigen, L.N.; Timm, P.C.; Boeve, B.F.; Silber, M.H.; St Louis, E.K. REM Sleep Muscle Activity in Idiopathic REM Sleep Behavior Disorder Predicts Phenoconversion. Neurology 2019, 93, e1171–e1179. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Lam, S.P.; Yu, M.W.M.; Li, S.X.; Zhou, J.; Chan, N.Y.; Wang, J.; Feng, H.; Chan, A.; et al. Electromyography Activity Level in Rapid Eye Movement Sleep Predicts Neurodegenerative Diseases in Idiopathic Rapid Eye Movement Sleep Behavior Disorder: A 5-Year Longitudinal Study. Sleep Med. 2019, 56, 128–134. [Google Scholar] [CrossRef]
- Singh, A.; Williams, S.; Calabrese, A.; Riha, R. Tonic REM Sleep Muscle Activity Is the Strongest Predictor of Phenoconversion Risk to Neurodegenerative Disease in Isolated REM Sleep Behaviour Disorder. J. Sleep Res. 2023, 32, e13792. [Google Scholar] [CrossRef] [PubMed]
- Melpignano, A.; Parrino, L.; Santamaria, J.; Gaig, C.; Trippi, I.; Serradell, M.; Mutti, C.; Riccò, M.; Iranzo, A. Isolated Rapid Eye Movement Sleep Behavior Disorder and Cyclic Alternating Pattern: Is Sleep Microstructure a Predictive Parameter of Neurodegeneration? Sleep 2019, 42, zsz142. [Google Scholar] [CrossRef]
- Iranzo, A.; Isetta, V.; Molinuevo, J.L.; Serradell, M.; Navajas, D.; Farre, R.; Santamaria, J. Electroencephalographic Slowing Heralds Mild Cognitive Impairment in Idiopathic REM Sleep Behavior Disorder. Sleep Med. 2010, 11, 534–539. [Google Scholar] [CrossRef]
- Gong, S.-Y.; Shen, Y.; Gu, H.-Y.; Zhuang, S.; Fu, X.; Wang, Q.-J.; Mao, C.-J.; Hu, H.; Dai, Y.-P.; Liu, C.-F. Generalized EEG Slowing Across Phasic REM Sleep, Not Subjective RBD Severity, Predicts Neurodegeneration in Idiopathic RBD. Nat. Sci. Sleep 2022, 14, 407–418. [Google Scholar] [CrossRef]
- Rodrigues Brazète, J.; Gagnon, J.-F.; Postuma, R.B.; Bertrand, J.-A.; Petit, D.; Montplaisir, J. Electroencephalogram Slowing Predicts Neurodegeneration in Rapid Eye Movement Sleep Behavior Disorder. Neurobiol. Aging 2016, 37, 74–81. [Google Scholar] [CrossRef]
- Ruffini, G.; Ibañez, D.; Castellano, M.; Dubreuil-Vall, L.; Soria-Frisch, A.; Postuma, R.; Gagnon, J.-F.; Montplaisir, J. Deep Learning With EEG Spectrograms in Rapid Eye Movement Behavior Disorder. Front. Neurol. 2019, 10, 806. [Google Scholar] [CrossRef]
- Arnaldi, D.; Mattioli, P.; Famà, F.; Girtler, N.; Brugnolo, A.; Pardini, M.; Donniaquio, A.; Massa, F.; Orso, B.; Raffa, S.; et al. Stratification Tools for Disease-Modifying Trials in Prodromal Synucleinopathy. Mov. Disord. 2022, 37, 52–61. [Google Scholar] [CrossRef]
- Jeong, E.; Woo Shin, Y.; Byun, J.-I.; Sunwoo, J.-S.; Roascio, M.; Mattioli, P.; Giorgetti, L.; Famà, F.; Arnulfo, G.; Arnaldi, D.; et al. EEG-Based Machine Learning Models for the Prediction of Phenoconversion Time and Subtype in Isolated Rapid Eye Movement Sleep Behavior Disorder. Sleep 2024, 47, zsae031. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.; Lina, J.-M.; Dubé, J.; Lafrenière, A.; Gagnon, J.-F.; Montplaisir, J.-Y.; Postuma, R.B.; Carrier, J. EEG Rhythmic and Arrhythmic Spectral Components and Functional Connectivity at Resting State May Predict the Development of Synucleinopathies in Idiopathic REM Sleep Behavior Disorder. Sleep 2024, zsae074. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Christensen, J.A.E.; Sixel-Döring, F.; Trenkwalder, C.; Mayer, G.; Oertel, W.H.; Jennum, P.; Sorensen, H.B.D. Validation of a New Data-Driven Automated Algorithm for Muscular Activity Detection in REM Sleep Behavior Disorder. J. Neurosci. Methods 2019, 312, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Cooray, N.; Andreotti, F.; Lo, C.; Symmonds, M.; Hu, M.T.M.; De Vos, M. Detection of REM Sleep Behaviour Disorder by Automated Polysomnography Analysis. Clin. Neurophysiol. 2019, 130, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Gunter, K.M.; Brink-Kjaer, A.; Mignot, E.; Sorensen, H.B.D.; During, E.; Jennum, P. SViT: A Spectral Vision Transformer for the Detection of REM Sleep Behavior Disorder. IEEE J. Biomed. Health Inform. 2023, 27, 4285–4292. [Google Scholar] [CrossRef]
- Rechichi, I.; Iadarola, A.; Zibetti, M.; Cicolin, A.; Olmo, G. Assessing REM Sleep Behaviour Disorder: From Machine Learning Classification to the Definition of a Continuous Dissociation Index. Int. J. Environ. Res. Public Health 2021, 19, 248. [Google Scholar] [CrossRef]
- Rechtschaffen, A.; Kales, A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects; Public Health Service, US Government Printing Office: Washington, DC, USA, 1968.
- Iber, C.; Ancoli-Israel, S.; Chesson, A.J.; Quan, S. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, 1st ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2007. [Google Scholar]
- Berry, R.; Brooks, R.; Gamaldo, C.; Harding, S.; Marcus, C.; Vaughn, B.; Tangredi, M. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, 2nd ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2012. [Google Scholar]
- Stefani, A.; Heidbreder, A.; Hackner, H.; Hogl, B. Validation of a Leg Movements Count and Periodic Leg Movements Analysis in a Custom Polysomnography System. BMC Neurol. 2017, 17, 42. [Google Scholar] [CrossRef]
- American Academy of Sleep Medicine. International Classification of Sleep Disorders: Diagnostic and Coding Manual, Revised, 1st ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2001. [Google Scholar]
- American Academy of Sleep Medicine. International Classification of Sleep Disorders: Diagnostic and Coding Manual, 2nd ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2005. [Google Scholar]
- American Academy of Sleep Medicine. The International Classification of Sleep Disorders (ICSD-3), 3rd ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2014. [Google Scholar]
- Cesari, M.; Stefani, A.; Penzel, T.; Ibrahim, A.; Hackner, H.; Heidbreder, A.; Szentkirályi, A.; Stubbe, B.; Völzke, H.; Berger, K.; et al. Interrater Sleep Stage Scoring Reliability between Manual Scoring from Two European Sleep Centers and Automatic Scoring Performed by the Artificial Intelligence-Based Stanford-STAGES Algorithm. J. Clin. Sleep Med. 2021, 17, 1237–1247. [Google Scholar] [CrossRef]
- Vallat, R.; Walker, M.P. An Open-Source, High-Performance Tool for Automated Sleep Staging. eLife 2021, 10, e70092. [Google Scholar] [CrossRef] [PubMed]
- Troester, M.; Quan, S.; Berry, R.; Plante, D.; Abreu, A.; Alzoubaidi, M.; Bandyopadhyay, A.; DelRosso, L.; Ebben, M.; Kwon, Y.; et al. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, 3rd ed.; American Academy of Sleep Medicine: Darien, IL, USA, 2023. [Google Scholar]
- Hjorth, B. EEG Analysis Based on Time Domain Properties. Electroencephalogr. Clin. Neurophysiol. 1970, 29, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Vidaurre, C.; Krämer, N.; Blankertz, B.; Schlögl, A. Time Domain Parameters as a Feature for EEG-Based Brain–Computer Interfaces. Neural Netw. 2009, 22, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J.F. On a Simple Algorithm to Calculate the “energy” of a Signal. In Proceedings of the International Conference on Acoustics, Speech, and Signal Processing, Albuquerque, NM, USA, 3–6 April 1990; Volume 1, pp. 381–384. [Google Scholar]
- Bandt, C.; Pompe, B. Permutation Entropy: A Natural Complexity Measure for Time Series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Ferri, R.; Manconi, M.; Plazzi, G.; Bruni, O.; Vandi, S.; Montagna, P.; Ferini-Strambi, L.; Zucconi, M. A Quantitative Statistical Analysis of the Submentalis Muscle EMG Amplitude during Sleep in Normal Controls and Patients with REM Sleep Behavior Disorder. J. Sleep Res. 2008, 17, 89–100. [Google Scholar] [CrossRef]
- Ferri, R.; Rundo, F.; Manconi, M.; Plazzi, G.; Bruni, O.; Oldani, A.; Ferini-Strambi, L.; Zucconi, M. Improved Computation of the Atonia Index in Normal Controls and Patients with REM Sleep Behavior Disorder. Sleep Med. 2010, 11, 947–949. [Google Scholar] [CrossRef]
- Larracy, R.; Phinyomark, A.; Scheme, E. Machine Learning Model Validation for Early Stage Studies with Small Sample Sizes. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Mexico, 1–5 November 2021; pp. 2314–2319. [Google Scholar] [CrossRef]
- Uno, H.; Cai, T.; Pencina, M.J.; D’Agostino, R.B.; Wei, L.J. On the C-Statistics for Evaluating Overall Adequacy of Risk Prediction Procedures with Censored Survival Data. Stat. Med. 2011, 30, 1105–1117. [Google Scholar] [CrossRef]
- Grambsch, P.M.; Therneau, T.M. Proportional Hazards Tests and Diagnostics Based on Weighted Residuals. Biometrika 1994, 81, 515–526. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus Machine Learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef]
- Coelho, B.F.O.; Massaranduba, A.B.R.; dos Santos Souza, C.A.; Viana, G.G.; Brys, I.; Ramos, R.P. Parkinson’s Disease Effective Biomarkers Based on Hjorth Features Improved by Machine Learning. Expert Syst. Appl. 2023, 212, 118772. [Google Scholar] [CrossRef]
- Lee, S.-B.; Kim, Y.-J.; Hwang, S.; Son, H.; Lee, S.K.; Park, K.-I.; Kim, Y.-G. Predicting Parkinson’s Disease Using Gradient Boosting Decision Tree Models with Electroencephalography Signals. Park. Relat. Disord. 2022, 95, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Christensen, J.A.E.; Sixel-Doring, F.; Muntean, M.-L.; Mollenhauer, B.; Trenkwalder, C.; Jennum, P.; Sorensen, H.B.D. A Clinically Applicable Interactive Micro and Macro-Sleep Staging Algorithm for Elderly and Patients with Neurodegeneration. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 3649–3652. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Christensen, J.A.E.; Muntean, M.-L.; Mollenhauer, B.; Sixel-Döring, F.; Sorensen, H.B.D.; Trenkwalder, C.; Jennum, P. A Data-Driven System to Identify REM Sleep Behavior Disorder and to Predict Its Progression from the Prodromal Stage in Parkinson’s Disease. Sleep Med. 2021, 77, 238–248. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Non-Converters | Converters | p-Value |
|---|---|---|---|
| Number | 48 | 18 | - |
| Males (%) | 95.8 | 83.3 | 0.087 |
| Age (years) | 68 [63–73] | 72.5 [66–76] | 0.160 |
| Time in bed (min) | 485 [473–492.5] | 485.5 [476–509] | 0.324 |
| Total sleep time (min) | 382.5 ± 57.8 | 399.9 ± 77.8 | 0.327 |
| Sleep period time (min) | 463.5 [443.5–472.5] | 467 [415–478] | 0.560 |
| Sleep efficiency (%) | 80.45 [69.45–88.4] | 84.95 [79.3–87.1] | 0.476 |
| Sleep latency (min) | 14 [8.55–25.3] | 12.5 [4.3–23.8] | 0.293 |
| REM latency (min) | 97.5 [64.5–191.75] | 97.5 [75–121.5] | 0.703 |
| Wake (%SPT) | 13.7 [8.7–21.7] | 13.2 [10.0–15.77] | 0.757 |
| N1 (%SPT) | 12.5 [3.1–19.6] | 14.0 [9.6–18.8] | 0.658 |
| N2 (%SPT) | 47.4 [37.2–56.2] | 44.9 [39.6–54.7] | 0.713 |
| N3 (%SPT) | 2.3 [0.0–8.2] | 5.6 [0.0–8.9] | 0.667 |
| REM (%SPT) | 14.8 ± 7.4 | 16.2 ± 7.9 | 0.523 |
| AHI (/h) | 2.9 [1.5–5.3] | 2.9 [0.3–8.8] | 0.857 |
| AHI in REM (/h) | 2.5 [0.0–8.1] | 2.1 [0.0–6.5] | 0.724 |
| PLMS index (/h) | 20.3 [6.7–57.6] | 24.0 [11.4–47.0] | 0.812 |
| Sleep-related breathing disorder (%) | 89.6 | 77.8 | 0.213 |
| Restless legs syndrome (%) | 22.9 | 27.8 | 0.681 |
| Antidepressants (%) | 43.8 | 22.2 | 0.108 |
| Benzodiazepines (%) | 8.3 | 16.7 | 0.327 |
| Antipsychotics (%) | 14.6 | 5.6 | 0.316 |
| Beta-blockers (%) | 14.6 | 5.6 | 0.316 |
| Antiepileptics (%) | 10.4 | 11.1 | 0.934 |
| Dopamine agonists (%) | 2.1 | 5.6 | 0.463 |
| Clonazepam (%) | 6.3 | 16.7 | 0.189 |
| Feature Name | Type | Description |
|---|---|---|
| Zero-crossing rate | Time domain | The number of zero-crossings, normalized by the window length |
| Hjorth parameters | Time domain | The three Hjorth parameters (activity, mobility, and complexity) [40] |
| Time domain properties | Time domain | Time domain properties derived using the log-power and the log-amplitude of each derivative, up to the 10th derivative [41] |
| Percent differential | Time domain | Difference between the 75th and 25th percentile of signal amplitude |
| Coastline | Time domain | The sum of the rectified sample derivative of the signal |
| Root mean square | Time domain | The root mean square of the signal |
| Variance | Time domain | The variance of the signal |
| Peak-to-peak | Time domain | The difference between the maximum and minimum peak of the signal |
| Crest factor | Time domain | The ratio between the absolute peak of the signal and the root mean square |
| Form factor | Time domain | The ratio between the root mean square and the average of the rectified signal |
| Pulse indicator | Time domain | The ratio between the absolute peak of the signal and the average of the rectified signal |
| Teager–Kaiser energy operator | Time domain | The energy of the signal, calculated according to [42] |
| Permutation entropy | Time domain | A non-linear measure that characterizes the complexity of the signal, calculated to the 10th order [43] |
| Shannon entropy | Time domain | The normalized Shannon entropy [44], calculated with the number of bins equal to the square root of the signal samples |
| Peak-power frequency | Frequency domain | The frequency at which the maximum in the power spectrum is achieved |
| Spectral edge frequency | Frequency domain | The frequency below which 95% of the signal power is contained |
| Relative power spectra | Frequency domain | Relative power calculated in the delta (0.5–4 Hz), theta (4–8 Hz), alpha (8–13 Hz), and beta (13–35 Hz) bands. |
| Slow-to-fast ratio | Frequency domain | The ratio between the relative powers in delta and theta bands with respect to the relative powers in alpha and beta bands |
| Feature Name | Type | Description |
|---|---|---|
| Zero-crossing rate | Time domain | The number of zero-crossings, normalized by the window length |
| Root mean square | Time domain | The root mean square of the signal |
| Variance | Time domain | The variance of the signal |
| Peak-to-peak | Time domain | The difference between the maximum and minimum peak of the signal |
| Crest factor | Time domain | The ratio between the absolute peak of the signal and the root mean square |
| Form factor | Time domain | The ratio between the root mean square and the average of the rectified signal |
| Pulse indicator | Time domain | The ratio between the absolute peak of the signal and the average of the rectified signal |
| Teager–Kaiser energy operator | Time domain | The energy of the signal, calculated according to [42] |
| Permutation entropy | Time domain | A non-linear measure that characterizes the complexity of the signal, calculated to the 10th order [43] |
| Shannon entropy | Time domain | The normalized Shannon entropy [44], calculated with the number of bins equal to the square root of the signal samples |
| Shannon entropy of rectified signal | Time domain | The normalized Shannon entropy of the rectified signal [44], calculated with the number of bins equal to the square root of the signal samples |
| Normalized Wilson amplitude | Time domain | The number of samples with absolute sample derivative over three times a threshold 1, normalized by the signal length |
| Myopulse indicator | Time domain | The percentage of samples with amplitude over three times a threshold 1 |
| Normalized integral | Time domain | The sum of the rectified signal, normalized by its length |
| Normalized wavelength | Time domain | The sum of the rectified sample derivative of the signal, normalized by its length |
| Energy | Time domain | The sum of the signal samples squared |
| 75th percentile | Time domain | The 75th percentile of the rectified signal |
| Fractal exponent | Frequency domain | The negative slope of the spectral density using a logarithmic on both the frequency and power. |
| Gamma power | Frequency domain | The absolute power in the frequency 30–45 Hz |
| Peak-power frequency | Frequency domain | The frequency at which the maximum in the power spectrum is achieved |
| Spectral entropy | Frequency domain | A measure of the random process uncertainty from the frequency distribution. |
| Spectral edge frequency | Frequency domain | The frequency below which 95% of the signal power is contained |
| All Patients | Non-Converters | Converters | |
|---|---|---|---|
| κ (overall) | 0.56 ± 0.14 | 0.55 ± 0.13 | 0.52 ± 0.08 |
| κ (W) | 0.61 ± 0.17 | 0.61 ± 0.18 | 0.61 ± 0.14 |
| κ (N1) | 0.17 ± 0.11 | 0.18 ± 0.11 | 0.17 ± 0.10 |
| κ (N2) | 0.60 ± 0.15 | 0.61 ± 0.16 | 0.58 ± 0.12 |
| κ (N3) | 0.53 ± 0.25 | 0.54 ± 0.24 | 0.48 ± 0.26 |
| κ (REM) | 0.56 ± 0.26 | 0.57 ± 0.26 | 0.52 ± 0.26 |
| Experiment | mbest | Harrel’s C-Index | Uno’s C-Index | Integrated Brier Score | AUC |
|---|---|---|---|---|---|
| EMGREM | 15 | 0.539 ± 0.135 | 0.548 ± 0.125 | 0.226 ± 0.07 | 0.619 ± 0.160 |
| EMGNREM | 15 | 0.542 ± 0.124 | 0.538 ± 0.125 | 0.243 ± 0.082 | 0.560 ± 0.165 |
| EEGREM | 5 | 0.723 ± 0.113 | 0.741 ± 0.110 | 0.174 ± 0.06 | 0.780 ± 0.145 |
| EEGNREM | 10 | 0.558 ± 0.109 | 0.559 ± 0.118 | 0.24 ± 0.085 | 0.602 ± 0.162 |
| EMGREM + EMGNREM | 20 | 0.545 ± 0.143 | 0.553 ± 0.141 | 0.24 ± 0.087 | 0.584 ± 0.157 |
| EEGREM + EEGNREM | 15 | 0.700 ± 0.139 | 0.710 ± 0.132 | 0.194 ± 0.061 | 0.746 ± 0.152 |
| EMGREM + EEGREM | 25 | 0.649 ± 0.145 | 0.653 ± 0.13 | 0.214 ± 0.081 | 0.701 ± 0.178 |
| EMGNREM + EEGNREM | 25 | 0.601 ± 0.111 | 0.594 ± 0.121 | 0.233 ± 0.084 | 0.616 ± 0.161 |
| All | 50 | 0.634 ± 0.141 | 0.639 ± 0.140 | 0.171± 0.088 | 0.688 ± 0.171 |
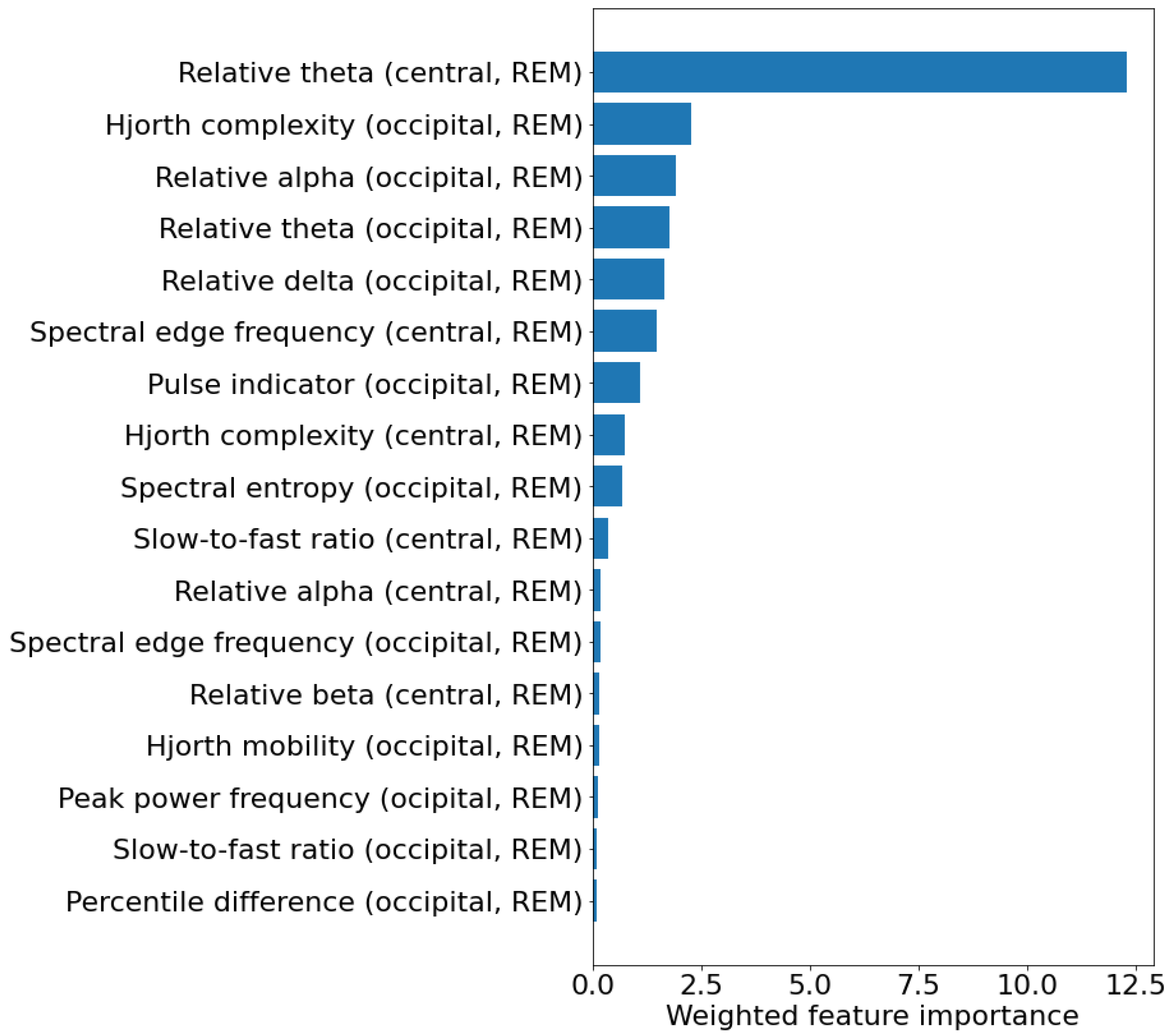
| Feature | Non-Converters (N = 48) | Converters (N = 18) | Hazard Ratio | p-Value |
|---|---|---|---|---|
| Relative theta (central, REM) [%] | 16.6 ± 4.5 | 18.4 ± 5.4 | 1.101 [1.007–1.203] | 0.033 |
| Hjorth complexity (occipital, REM) [u.a.] | 2.59 ± 0.39 | 2.73 ± 0.77 | 1.845 [0.834–4.079] | 0.131 |
| Relative alpha (occipital, REM) [%] | 13.87 ± 4.07 | 11.70 ± 4.04 | 0.878 [0.774–0.995] | 0.042 |
| Relative theta (occipital, REM) [%] | 18.90 ± 4.20 | 20.39 ± 5.84 | 1.086 [0.990–1.192] | 0.081 |
| Relative delta (occipital, REM) [%] | 39.94 ± 6.44 | 40.00 ± 11.90 | 1.009 [0.952–1.070] | 0.756 |
| Spectral edge frequency (central, REM) [Hz] | 18.12 ± 2.30 | 17.91 ± 3.79 | 0.890 [0.723–1.094] | 0.268 |
| Pulse indicator (occipital, REM) [-] | 4.87 ± 0.70 | 5.10 ± 1.06 | 1.262 [0.711–2.242] | 0.427 |
| Hjorth complexity (central, REM) [u.a.] | 2.59 ± 0.39 | 2.73 ± 0.77 | 1.845 [0.834–4.079] | 0.131 |
| Spectral entropy (occipital, REM) [u.a.] | 2.64 ± 0.163 | 2.64 ± 0.292 | 0.522 [0.048–5.630] | 0.592 |
| Slow-to-fast ratio (central, REM) [-] | 3.81 ± 1.36 | 4.59 ± 3.26 | 1.307 [1.073–1.590] | 0.008 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesari, M.; Portscher, A.; Stefani, A.; Angerbauer, R.; Ibrahim, A.; Brandauer, E.; Feuerstein, S.; Egger, K.; Högl, B.; Rodriguez-Sanchez, A. Machine Learning Predicts Phenoconversion from Polysomnography in Isolated REM Sleep Behavior Disorder. Brain Sci. 2024, 14, 871. https://doi.org/10.3390/brainsci14090871
Cesari M, Portscher A, Stefani A, Angerbauer R, Ibrahim A, Brandauer E, Feuerstein S, Egger K, Högl B, Rodriguez-Sanchez A. Machine Learning Predicts Phenoconversion from Polysomnography in Isolated REM Sleep Behavior Disorder. Brain Sciences. 2024; 14(9):871. https://doi.org/10.3390/brainsci14090871
Chicago/Turabian StyleCesari, Matteo, Andrea Portscher, Ambra Stefani, Raphael Angerbauer, Abubaker Ibrahim, Elisabeth Brandauer, Simon Feuerstein, Kristin Egger, Birgit Högl, and Antonio Rodriguez-Sanchez. 2024. "Machine Learning Predicts Phenoconversion from Polysomnography in Isolated REM Sleep Behavior Disorder" Brain Sciences 14, no. 9: 871. https://doi.org/10.3390/brainsci14090871
APA StyleCesari, M., Portscher, A., Stefani, A., Angerbauer, R., Ibrahim, A., Brandauer, E., Feuerstein, S., Egger, K., Högl, B., & Rodriguez-Sanchez, A. (2024). Machine Learning Predicts Phenoconversion from Polysomnography in Isolated REM Sleep Behavior Disorder. Brain Sciences, 14(9), 871. https://doi.org/10.3390/brainsci14090871








