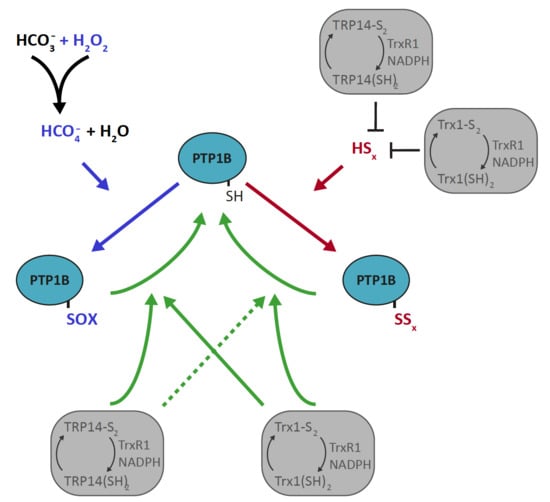
Qualitative Differences in Protection of PTP1B Activity by the Reductive Trx1 or TRP14 Enzyme Systems upon Oxidative Challenges with Polysulfides or H2O2 Together with Bicarbonate
Abstract
1. Introduction
2. Materials and Methods
2.1. Recombinant Proteins
2.2. Inactivation of Recombinant PTP1B Using Polysulfide or HCO3−/H2O2
2.3. PTP Activity Assay
3. Results
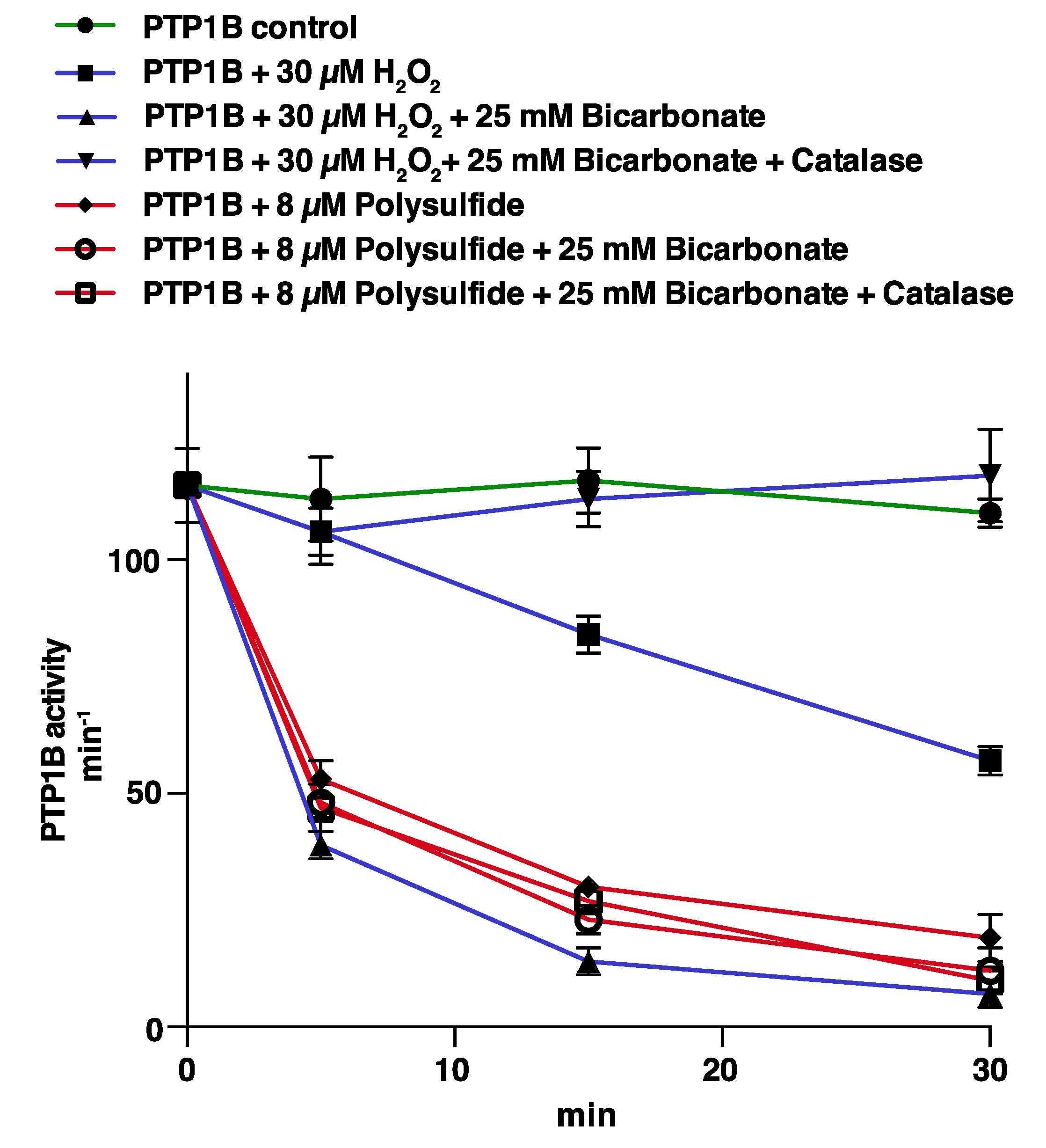
3.1. PTP1B Inactivation by H2O2 is Potentiated by HCO3− but that of Polysulfides is Not
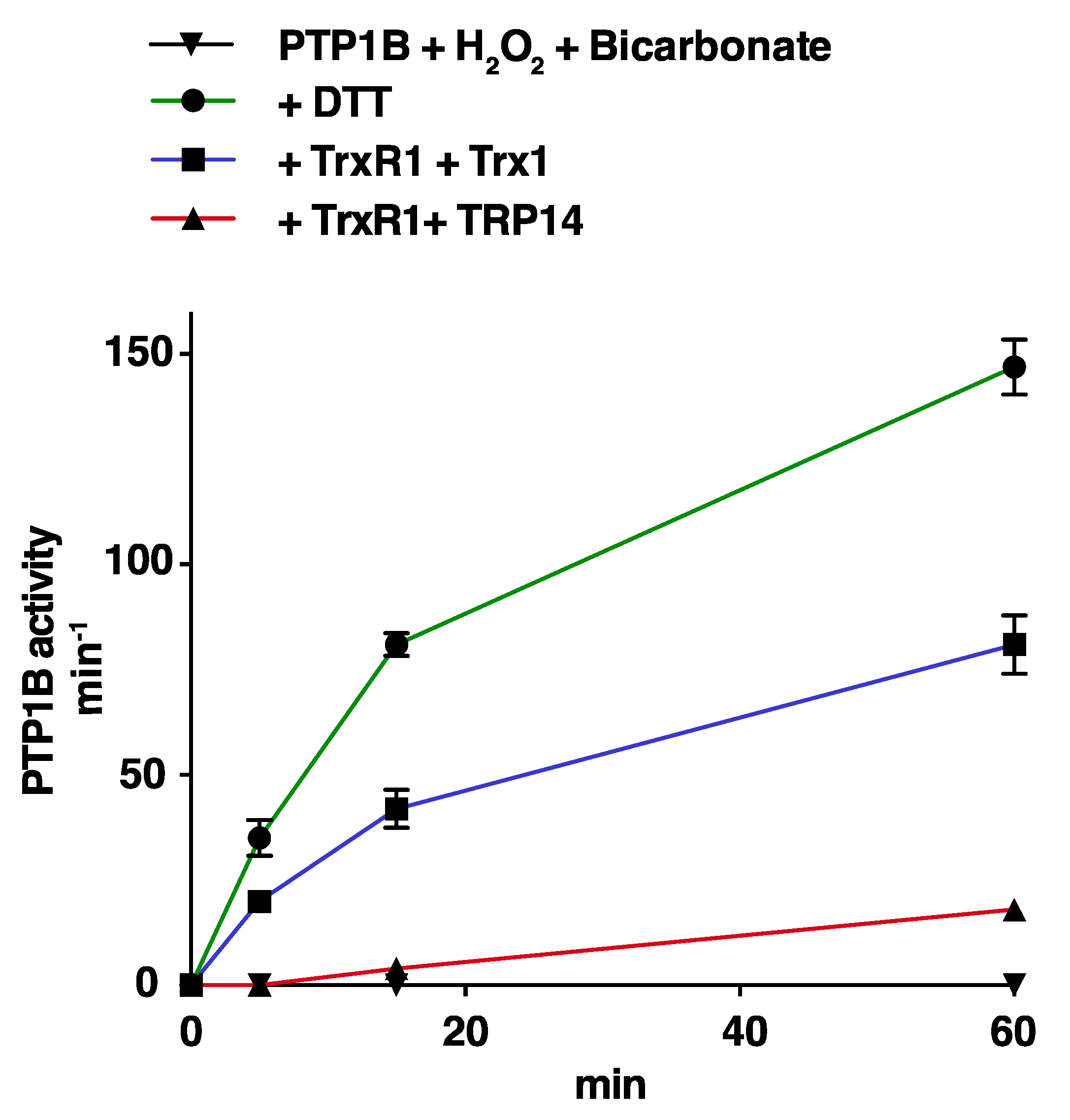
3.2. Trx1 as well as TRP14 Can Potently Protect PTP1B from Inactivation by Polysulfides but Not from H2O2 in Combination with HCO3−
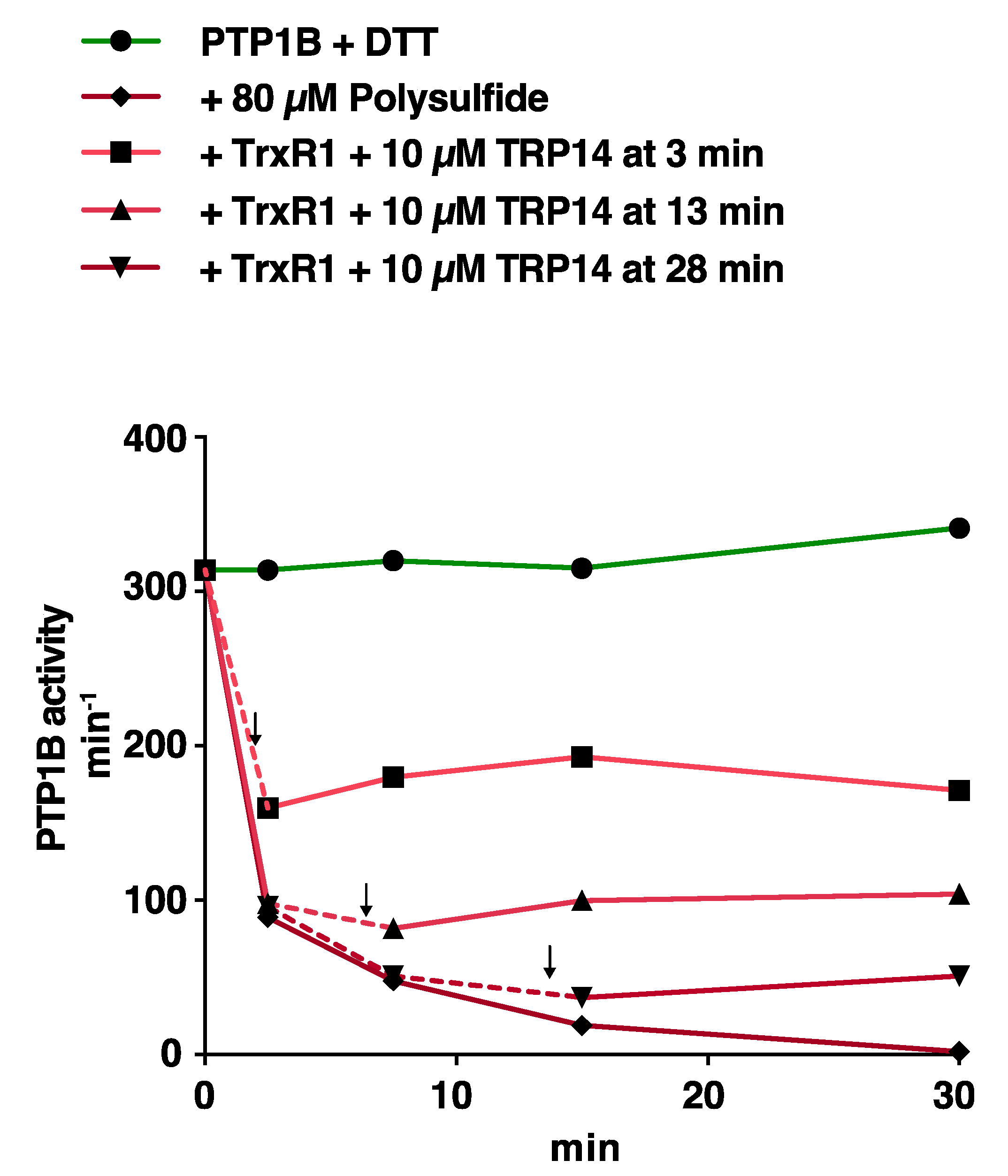
3.3. Reactivation of Oxidized PTP1B is More Efficient with Trx1 Than with TRP14 and More EFficient for H2O2- and HCO3−-Inactivated PTP1B Than for Polysulfide-Inactivated PTP1B
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell. Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.N.; Mortensen, O.H.; Peters, G.H.; Drake, P.G.; Iversen, L.F.; Olsen, O.H.; Jansen, P.G.; Andersen, H.S.; Tonks, N.K.; Moller, N.P. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 2001, 21, 7117–7136. [Google Scholar] [CrossRef] [PubMed]
- Frijhoff, J.; Dagnell, M.; Godfrey, R.; Ostman, A. Regulation of protein tyrosine phosphatase oxidation in cell adhesion and migration. Antioxid. Redox. Signal. 2014, 20, 1994–2010. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T. Redox-dependent signal transduction. FEBS Lett. 2000, 476, 52–54. [Google Scholar] [CrossRef]
- Tanner, J.J.; Parsons, Z.D.; Cummings, A.H.; Zhou, H.; Gates, K.S. Redox regulation of protein tyrosine phosphatases: Structural and chemical aspects. Antioxid. Redox. Signal 2011, 15, 77–97. [Google Scholar] [CrossRef]
- Bertolotti, M.; Farinelli, G.; Galli, M.; Aiuti, A.; Sitia, R. AQP8 transports NOX2-generated H2O2 across the plasma membrane to promote signaling in B cells. J. Leukoc. Biol. 2016, 100, 1071–1079. [Google Scholar] [CrossRef]
- Medrano-Fernandez, I.; Bestetti, S.; Bertolotti, M.; Bienert, G.P.; Bottino, C.; Laforenza, U.; Rubartelli, A.; Sitia, R. Stress Regulates Aquaporin-8 Permeability to Impact Cell Growth and Survival. Antioxid. Redox. Signal 2016, 24, 1031–1044. [Google Scholar] [CrossRef]
- Frijhoff, J.; Dagnell, M.; Augsten, M.; Beltrami, E.; Giorgio, M.; Ostman, A. The mitochondrial reactive oxygen species regulator p66Shc controls PDGF-induced signaling and migration through protein tyrosine phosphatase oxidation. Free Radic. Biol. Med. 2014, 68, 268–277. [Google Scholar] [CrossRef]
- Dagnell, M.; Cheng, Q.; Rizvi, S.H.M.; Pace, P.E.; Boivin, B.; Winterbourn, C.C.; Arner, E.S.J. Bicarbonate is essential for protein-tyrosine phosphatase 1B (PTP1B) oxidation and cellular signaling through EGF-triggered phosphorylation cascades. J. Biol. Chem. 2019, 294, 12330–12338. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Singh, H.; Parsons, Z.D.; Lewis, S.M.; Bhattacharya, S.; Seiner, D.R.; LaButti, J.N.; Reilly, T.J.; Tanner, J.J.; Gates, K.S. The biological buffer bicarbonate/CO2 potentiates H2O2-mediated inactivation of protein tyrosine phosphatases. J. Am. Chem Soc. 2011, 133, 15803–15805. [Google Scholar] [CrossRef]
- Bakhmutova-Albert, E.V.; Yao, H.; Denevan, D.E.; Richardson, D.E. Kinetics and mechanism of peroxymonocarbonate formation. Inorg. Chem. 2010, 49, 11287–11296. [Google Scholar] [CrossRef] [PubMed]
- Trindade, D.F.; Cerchiaro, G.; Augusto, O. A role for peroxymonocarbonate in the stimulation of biothiol peroxidation by the bicarbonate/carbon dioxide pair. Chem. Res. Toxicol. 2006, 19, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Kwon, K.S.; Kim, S.R.; Rhee, S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998, 273, 15366–15372. [Google Scholar] [CrossRef] [PubMed]
- van Montfort, R.L.; Congreve, M.; Tisi, D.; Carr, R.; Jhoti, H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 2003, 423, 773–777. [Google Scholar] [CrossRef]
- Salmeen, A.; Andersen, J.N.; Myers, M.P.; Meng, T.C.; Hinks, J.A.; Tonks, N.K.; Barford, D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 2003, 423, 769–773. [Google Scholar] [CrossRef]
- Sivaramakrishnan, S.; Cummings, A.H.; Gates, K.S. Protection of a single-cysteine redox switch from oxidative destruction: On the functional role of sulfenyl amide formation in the redox-regulated enzyme PTP1B. Bioorg. Med. Chem. Lett. 2010, 20, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Rinna, A.; Torres, M.; Forman, H.J. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic. Biol. Med. 2006, 41, 86–91. [Google Scholar] [CrossRef]
- Barrett, W.C.; DeGnore, J.P.; Konig, S.; Fales, H.M.; Keng, Y.F.; Zhang, Z.Y.; Yim, M.B.; Chock, P.B. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry 1999, 38, 6699–6705. [Google Scholar] [CrossRef]
- Krishnan, N.; Fu, C.; Pappin, D.J.; Tonks, N.K. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci. Signal. 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Doka, E.; Ida, T.; Dagnell, M.; Abiko, Y.; Luong, N.C.; Balog, N.; Takata, T.; Espinosa, B.; Nishimura, A.; Cheng, Q.; et al. Control of protein function through oxidation and reduction of persulfidated states. Sci. Adv. 2020, 6, eaax8358. [Google Scholar] [CrossRef] [PubMed]
- Dagnell, M.; Frijhoff, J.; Pader, I.; Augsten, M.; Boivin, B.; Xu, J.; Mandal, P.K.; Tonks, N.K.; Hellberg, C.; Conrad, M.; et al. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-beta receptor tyrosine kinase signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Schwertassek, U.; Haque, A.; Krishnan, N.; Greiner, R.; Weingarten, L.; Dick, T.P.; Tonks, N.K. Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. FEBS J. 2014, 281, 3545–3558. [Google Scholar] [CrossRef]
- Parsons, Z.D.; Gates, K.S. Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry 2013, 52, 6412–6423. [Google Scholar] [CrossRef]
- Dagnell, M.; Pace, P.E.; Cheng, Q.; Frijhoff, J.; Ostman, A.; Arner, E.S.J.; Hampton, M.B.; Winterbourn, C.C. Thioredoxin reductase 1 and NADPH directly protect protein tyrosine phosphatase 1B from inactivation during H2O2 exposure. J. Biol. Chem. 2017, 292, 14371–14380. [Google Scholar] [CrossRef]
- Xu, J.; Eriksson, S.E.; Cebula, M.; Sandalova, T.; Hedstrom, E.; Pader, I.; Cheng, Q.; Myers, C.R.; Antholine, W.E.; Nagy, P.; et al. The conserved Trp114 residue of thioredoxin reductase 1 has a redox sensor-like function triggering oligomerization and crosslinking upon oxidative stress related to cell death. Cell. Death Dis. 2015, 6, e1616. [Google Scholar] [CrossRef]
- Pader, I.; Sengupta, R.; Cebula, M.; Xu, J.; Lundberg, J.O.; Holmgren, A.; Johansson, K.; Arner, E.S. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc. Natl. Acad. Sci. USA 2014, 111, 6964–6969. [Google Scholar] [CrossRef]
- Cheng, Q.; Arner, E.S. Selenocysteine insertion at a predefined UAG codon in a release factor 1 (RF1) depleted Escherichia coli host strain bypasses species barriers in recombinant selenoprotein translation. J. Biol. Chem. 2017, 292, 5476–5487. [Google Scholar] [CrossRef]
- Montalibet, J.; Skorey, K.I.; Kennedy, B.P. Protein tyrosine phosphatase: Enzymatic assays. Methods 2005, 35, 2–8. [Google Scholar] [CrossRef]
- Meng, T.C.; Buckley, D.A.; Galic, S.; Tiganis, T.; Tonks, N.K. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J. Biol. Chem. 2004, 279, 37716–37725. [Google Scholar] [CrossRef] [PubMed]
- Cuevasanta, E.; Moller, M.N.; Alvarez, B. Biological chemistry of hydrogen sulfide and persulfides. Arch. Biochem. Biophys. 2017, 617, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Greiner, R.; Dick, T.P. Real-time assays for monitoring the influence of sulfide and sulfane sulfur species on protein thiol redox states. Methods Enzymol. 2015, 555, 57–77. [Google Scholar] [PubMed]
- Doka, E.; Pader, I.; Biro, A.; Johansson, K.; Cheng, Q.; Ballago, K.; Prigge, J.R.; Pastor-Flores, D.; Dick, T.P.; Schmidt, E.E.; et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016, 2, e1500968. [Google Scholar] [CrossRef] [PubMed]
- Peskin, A.V.; Pace, P.E.; Winterbourn, C.C. Enhanced hyperoxidation of peroxiredoxin 2 and peroxiredoxin 3 in the presence of bicarbonate/CO2. Free Radic. Biol. Med. 2019, 145, 1–7. [Google Scholar] [CrossRef]
- Truzzi, D.R.; Coelho, F.R.; Paviani, V.; Alves, S.V.; Netto, L.E.S.; Augusto, O. The bicarbonate/carbon dioxide pair increases hydrogen peroxide-mediated hyperoxidation of human peroxiredoxin 1. J. Biol. Chem. 2019, 294, 14055–14067. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dagnell, M.; Cheng, Q.; Arnér, E.S.J. Qualitative Differences in Protection of PTP1B Activity by the Reductive Trx1 or TRP14 Enzyme Systems upon Oxidative Challenges with Polysulfides or H2O2 Together with Bicarbonate. Antioxidants 2021, 10, 111. https://doi.org/10.3390/antiox10010111
Dagnell M, Cheng Q, Arnér ESJ. Qualitative Differences in Protection of PTP1B Activity by the Reductive Trx1 or TRP14 Enzyme Systems upon Oxidative Challenges with Polysulfides or H2O2 Together with Bicarbonate. Antioxidants. 2021; 10(1):111. https://doi.org/10.3390/antiox10010111
Chicago/Turabian StyleDagnell, Markus, Qing Cheng, and Elias S.J. Arnér. 2021. "Qualitative Differences in Protection of PTP1B Activity by the Reductive Trx1 or TRP14 Enzyme Systems upon Oxidative Challenges with Polysulfides or H2O2 Together with Bicarbonate" Antioxidants 10, no. 1: 111. https://doi.org/10.3390/antiox10010111
APA StyleDagnell, M., Cheng, Q., & Arnér, E. S. J. (2021). Qualitative Differences in Protection of PTP1B Activity by the Reductive Trx1 or TRP14 Enzyme Systems upon Oxidative Challenges with Polysulfides or H2O2 Together with Bicarbonate. Antioxidants, 10(1), 111. https://doi.org/10.3390/antiox10010111