Abstract
Elemental sulfur and sulfite have been used to inhibit the growth of yeasts, but thiosulfate has not been reported to be toxic to yeasts. We observed that thiosulfate was more inhibitory than sulfite to Saccharomyces cerevisiae growing in a common yeast medium. At pH < 4, thiosulfate was a source of elemental sulfur and sulfurous acid, and both were highly toxic to the yeast. At pH 6, thiosulfate directly inhibited the electron transport chain in yeast mitochondria, leading to reductions in oxygen consumption, mitochondrial membrane potential and cellular ATP. Although thiosulfate was converted to sulfite and H2S by the mitochondrial rhodanese Rdl1, its toxicity was not due to H2S as the rdl1-deletion mutant that produced significantly less H2S was more sensitive to thiosulfate than the wild type. Evidence suggests that thiosulfate inhibits cytochrome c oxidase of the electron transport chain in yeast mitochondria. Thus, thiosulfate is a potential agent against yeasts.
1. Introduction
Thiosulfate is spontaneously produced by reacting sulfite with elemental sulfur [1], and it is a key intermediate in the biogeochemical cycle of sulfur [2]. Many heterotrophic bacteria oxidize H2S to thiosulfate as a detoxification mechanism [3,4,5], and other bacteria oxidize thiosulfate to sulfate to gain energy for growth [6,7,8]. Further, bacteria and yeast readily use thiosulfate as a sulfur source for growth [9,10,11,12]. We recently characterized the pathway of thiosulfate assimilation in Saccharomyces cerevisiae [10]. Yeast uses sulfate transporters to uptake thiosulfate and then uses a mitochondrial rhodanese Rdl1 to transfer a zero-valence sulfur from thiosulfate to glutathione (GSH), producing glutathione persulfide (GSSH). GSSH spontaneously reacts with another GSH to release H2S and glutathione disulfide (GSSG) [13]. The produced H2S is used for the production of cysteine and methionine [10,14].
Under acidic conditions, thiosulfate spontaneously breaks into sulfurous acid (H2SO3) and elemental sulfur (S0) [15]. Both sulfite and S0 are toxic to yeast. The inhibitory effect of sulfite on yeast has been extensively investigated, as sulfite is widely used in fruit and vegetable preservation [16,17] and in ethanol fermentation to inhibit the growth of wild yeasts [18,19]. Undissociated H2SO3 at low pH is the most effective species as it kills yeasts [20]. At neutral pH, sulfite is also inhibitory because it decreases ATP production by inhibiting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and alcohol dehydrogenase in S. cerevisiae [21,22]. Sulfite does not inhibit the electron transport chain in yeast [23]. Further, sulfite can be oxidized by reactive oxygen species to generate sulfur trioxide free radicals (HSO3− and SO3−) [24,25], which may damage DNA and destroy tryptophan [26,27]. S0 is a common fungicide [28,29]. S0 is believed to be transported into the cell as hydrogen polysulfide (H2Sn) [30,31], which reacts with cellular thiols, including protein thiols and GSH, to form organic persulfide and polysulfide (RSnH, n ≥ 2). GSH spontaneously reacts with RSnH to produce H2S and GSSG, and S0 toxicity could be due to the combination of thiol modification and H2S production [32]. In addition, RSnH may inhibit enzymes with metal ions or heme in their active centers [33].
Thiosulfate is a relatively benign sulfur species, and it is commonly used as a remedy to treat cyanide poisoning [34]. Its toxicity to microorganisms has not been reported to date. Here, we report that thiosulfate is more inhibitory than sulfite to the yeast. At low pH, thiosulfate is converted to S0 and H2SO3, both of which are toxic; at neutral pH, thiosulfate is inhibitory by itself.
2. Materials and Methods
2.1. Materials and Reagents
Sodium thiosulfate, sodium sulfite, sulfur powder (S0), glyceraldehyde 3-phosphate, N, N, N′, N′-Tetramethyl-p-phenylenediamine dihydrochloride (TMPD), ascorbate and nicotinamide adenine dinucleotide (NAD+) were purchased from Sigma Chemical (St. Louis, MO, USA). The enzymes used for DNA manipulations were obtained from Thermo Fisher (Waltham, MA, USA). PCR enzymes were purchased from Toyobo (Osaka, Japan). The RNA extract kit (R6834-01) was purchased from Omega (Norcross, GA, USA). Excess sulfur powder was added to acetone to obtain saturated sulfur in acetone (S0, 20 mM).
2.2. Strains, Mutants, and Plasmids
General cloning and site-directed mutagenesis were performed by using previously reported methods [35,36]. Sequencing and PCR were carried out according to standard procedures. The yeast strain used in this study was S. cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). The Escherichia coli strain DH5α served as the host strain for all plasmid constructions, and it was grown in lysogeny broth (LB) medium (0.5% yeast extract, 1% peptone, and 1% NaCl). When required, 50 μg/mL ampicillin was added to LB medium. S. cerevisiae strains were grown at 30 °C in yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic defined (SD) medium (0.17% YNB, 5% (NH4)2SO4, 2% glucose, amino acid mixtures) supplemented with auxotrophic requirements. The pH of the SD medium (liquid and plate) was routinely adjusted to 6 by adding NaOH. One-step PCR-mediated gene disruption was carried out to delete RDL1 (NP_014928.1) and RDL2 (NP_014929.3) in BY4742 [37]. The recombinant plasmids were constructed using a previously reported method [10]. The strains and plasmids used in this study are listed in Table 1. All primers are listed in Table 2.

Table 1.
The strains and plasmids used in the study.

Table 2.
The primers used in the study.
2.3. Measurement of Growth Curves of S. cerevisiae in the Presence of Thiosulfate or Sulfite
Fresh cells of S. cerevisiae were inoculated in 5 mL of SD medium and grown overnight at 30 °C with shaking at 200 rpm. Cells were then centrifuged at 10,000× g for 5 min, and the pellet was resuspended in SD medium. Equal amounts of cells (OD600nm = 0.1) were cultured in 400 μL of SD medium containing additional sulfite, thiosulfate or S0 at 30 °C with shaking; the growth was measured at OD600nm by using a microplate reader (BioTek, Synergy H1). The Vmax of yeast cells growth in SD medium with different amount of thiosulfate was calculated during the middle log phase, which was used for analyzing the IC50 values of thiosulfate.
Thiosulfate and sulfite tolerance assays were performed on SD agar plates (SD medium with 1.5% agar). The cells of overnight culture in SD medium were serially diluted with fresh SD medium, and 5 µL of each dilution was spotted on the SD agar plates containing thiosulfate or sulfite. The plates were incubated at 30 °C for 48 h before scanning with a scanner (Fluor Chem Q Alpha, Santa Clara, CA, USA).
The pH of SD medium (liquid and plate) was adjusted from 6 to 4 and 5 by adding HCl when needed.
2.4. Detection of Killing Effect of Thiosulfate at Low pH
Fresh cells of the S. cerevisiae strain were inoculated in 5 mL of SD medium and grown overnight at 30 °C with shaking at 200 rpm. Cells were collected by centrifugation (10,000× g, 5 min) and suspended in water. Equal amounts of cells (OD600nm = 1) were suspended in citric acid–sodium phosphate dibasic buffer (CPBS, pH 3.4, 4, 5, 6) with or without 10 mM thiosulfate and incubated at 30 °C for 1 h. The suspensions were then diluted 1:10,000 times with sterile water. Dilutions (100 μL) were added to the YPD plate and incubated at 30 °C for 2 days. Colony-forming units (CFUs) were counted.
The yeast cells incubated in CPBS buffer (pH 3.4) were centrifuged (10,000× g, 5 min), and the pellet was suspended in HEPES buffer (50 mM, pH 7.0). The yeast cells were observed under an Olympus microscope (IX83, Olympus, Tokyo, Japan).
2.5. Rhodanese Assay
The standard rhodanese assay was conducted in the same manner as previously reported [3]. Briefly, fresh yeast cells were collected, washed twice with water, an amount corresponding to an OD600nm of 2 was suspended in ice-cold 50 mM Tris-HCl (pH 8) and disrupted using a pressure cell homogenizer (Stansted Fluid Power LTD SPCH-18, Harlow, UK). The suspension was centrifuged at 12,500× g for 10 min to remove cell debris, and the protein concentration in cell lysate was measured by using a microspectrophotometer (Bio future, K5500). An assay volume of 1 mL included 50 mM Tris-HCl (pH 8), 5 mM sodium thiosulfate, 10 mM KCN and cell lysate. Reactions were initiated by the addition of KCN and terminated after 10 min by boiling for 3 min. Subsequently, 0.1 mL of ferric nitrate reagent was added and centrifuged at 12,500× g for 5 min; the absorbance was measured at 460 nm and compared with a standard curve of thiocyanate.
2.6. Determination of Cellular Thiosulfate Concentration
The sulfur-starved yeast cells were prepared and resuspended at an OD600nm of 10 in sterile phosphate buffered solution (PBS) buffer containing 2% glucose, as previously reported [10]. A final concentration of 200 μM of thiosulfate was added, and the cells were incubated at 30 °C for 60 min. Cells were centrifuged (13,000× g, 5 min), washed twice with water, resuspended in ice-cold 50 mM Tris-HCl (pH 8) to an OD600nm of 5 and disrupted using a pressure cell homogenizer. The cell lysate was again centrifuged at 12,500× g for 10 min to remove cell debris and thiosulfate in the supernatant was derivatized with monobromobimane (mBBr) and detected as previously described [10]. Thiosulfate, sulfite and sulfide react with mBBr to produce derivatives that can be separated by HPLC and detected with a fluorescence detector [38]. The cellular concentration of thiosulfate was calculated using a reported haploid cell volume of 50 fL [39,40].
2.7. Measurements of Mitochondrial Membrane Potential and Cellular ATP Concentration
Overnight yeast cells were collected and suspended in SD medium at an OD600nm of 1 with or without 10 mM thiosulfate, and the samples were incubated at 30 °C for 1 h. Yeast cells were collected, washed twice with ice-cold HEPES buffer (50 mM, pH 7.0) and suspended in the same buffer to an OD600nm of 2. The cells were used to measure mitochondrial membrane potential by using a fluorescent probe 5,5’,6,6’-tetrachloro-l,l’,3,3’-tetraethylbenzimidazolo-carbocyanine iodide (JC-1) (Beyotime Biotech, Shanghai, China) [41], following the manufacturers’ instructions. Briefly, 1 mL of the yeast cell suspension was mixed with an equal volume of the purchased JC-1 staining solution (5 μg/mL), incubated at 30 °C for 20 min, rinsed twice with ice-cold HEPES buffer (50 mM, pH 7.0) and suspended in the ice-cold HEPES buffer at OD600nm of 0.5. The fluorescence (red fluorescence: Ex = 556 nm and Em = 590 nm; green fluorescence: Ex = 490nm and Em = 530 nm) was measured by using a microplate reader (BioTek, Synergy H1). The thiosulfate-treated and untreated yeast cells were tested. In theory, JC-1 exists as monomers that emit green fluorescence in cells with low mitochondrial membrane potential, and it forms aggregates that emit red fluorescence in cells with high mitochondrial membrane potential. A decrease in the ratio of red to green fluorescence indicates a reduction in the mitochondrial membrane potential [42].
For ATP determination, the cells were disrupted, and the lysate was centrifuged at 12,500× g for 10 min to remove cell debris. The protein concentration in the supernatant was measured by using a microspectrophotometer (Bio future, K5500). The ATP concentration was detected using firefly luciferase, and the produced chemiluminescence was measured by using a plate reader (BioTek, Synergy H1) [43].
2.8. Assaying GAPDH Activity
The assay of GAPDH activity was carried out according to a reporter method with minor modification [44]. Fresh yeast cells were collected by centrifugation, washed twice with distilled water, suspended in ice-cold lysis buffer (40 mM triethanolamine, 50 mM Na2HPO4, 5 mM EDTA; pH 8.6) to an OD600nm of 2, and disrupted using a pressure cell homogenizer. The lysate was centrifuged at 12,500× g for 10 min to remove cell debris, and the protein concentration was measured by using a microspectrophotometer (Bio future, K5500). The lysate was incubated with 10 mM thiosulfate or 10 mM sulfite at 30 °C for 30 min. A control without thiosulfate and sulfite was also incubated. Then, the treated lysate containing 0.5 mg of protein was incubated with 1.5 mM glyceraldehyde 3-phosphate and 1 mM NAD+ in 1 mL of the lysis buffer. The generation of NADH was analyzed by recording the absorbance increase at 340 nm for 30 s. The rate of NADH production was calculated.
2.9. Measuring Oxygen Consumption
The sulfur-starved yeast cells were prepared and resuspended in sterile PBS buffer containing 2% glucose at an OD600nm of 10, as previously reported [10]. Thiosulfate was added to a final concentration of 10 mM, and oxygen was monitored by using an Orion RDO meter (Thermo Scientific Inc., Waltham, WA, USA). The RDO meter was calibrated with air-saturated water according to the manufacturers’ instructions. Yeast cells without thiosulfate addition were used as the control.
The sulfur-starved yeast cells were resuspended in sterile PBS buffer containing 12.5 mM ascorbate and 1.4 mM N, N, N′, N′-tetramethyl-p-phenylenediamine (TMPD) at an OD600nm of 10. Thiosulfate or sulfite was added to a final concentration of 10 mM, and oxygen was monitored by using the Orion RDO meter (Thermo Scientific Inc., Waltham, WA, USA). Yeast cells without thiosulfate addition were used as the control.
3. Results
3.1. Thiosulfate Inhibited the Growth of S. cerevisiae at pH 6 and Killed It at pH 3.4
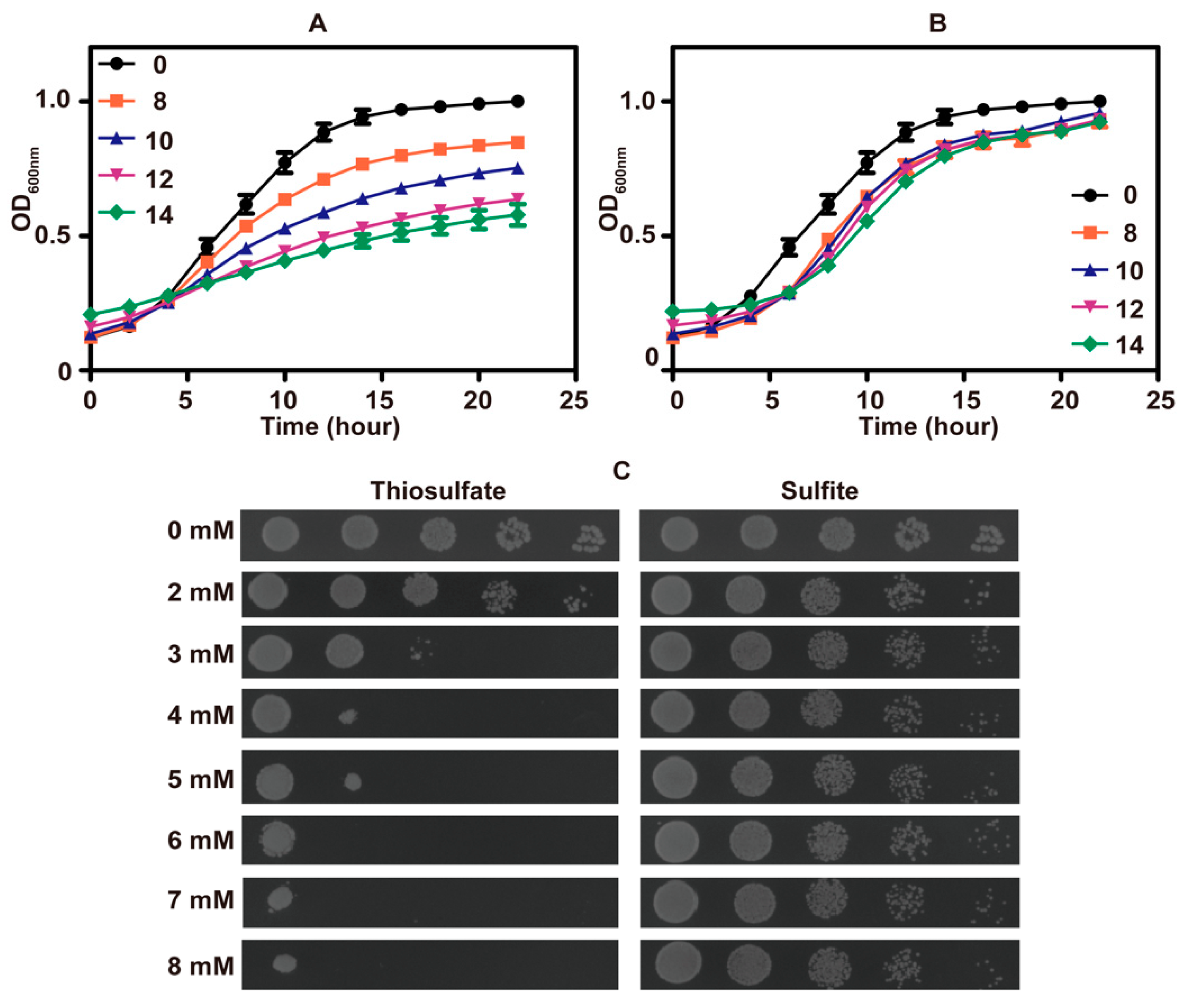
The growth of S. cerevisiae in SD medium was inhibited by high concentrations of thiosulfate (Figure 1A) and sulfite (Figure 1B). Surprisingly, thiosulfate was more inhibitory than sulfite (Figure 1C). The pH of fresh SD medium was approximately 6.0, and the pH decreased to about 2.1 after cultivating the yeast.
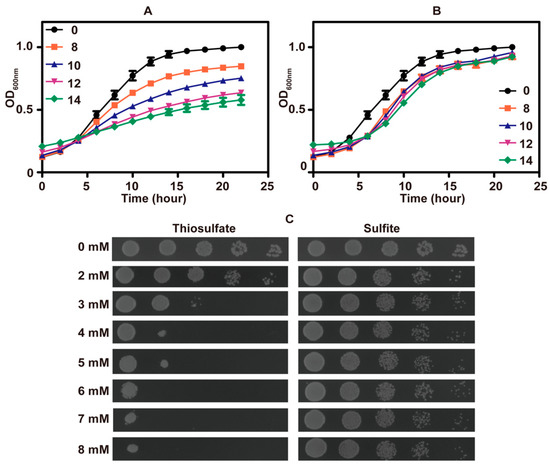
Figure 1.
The phenotype analysis of S. cerevisiae in SD medium. The growth curves of S. cerevisiae in SD medium (pH 6) with different amounts of thiosulfate (A) or sulfite (B); the concentrations of thiosulfate or sulfite are in mM. The data are averages with standard deviations from three replicates. (C) The thiosulfate or sulfite tolerance assay on SD agar (pH 6) plates.
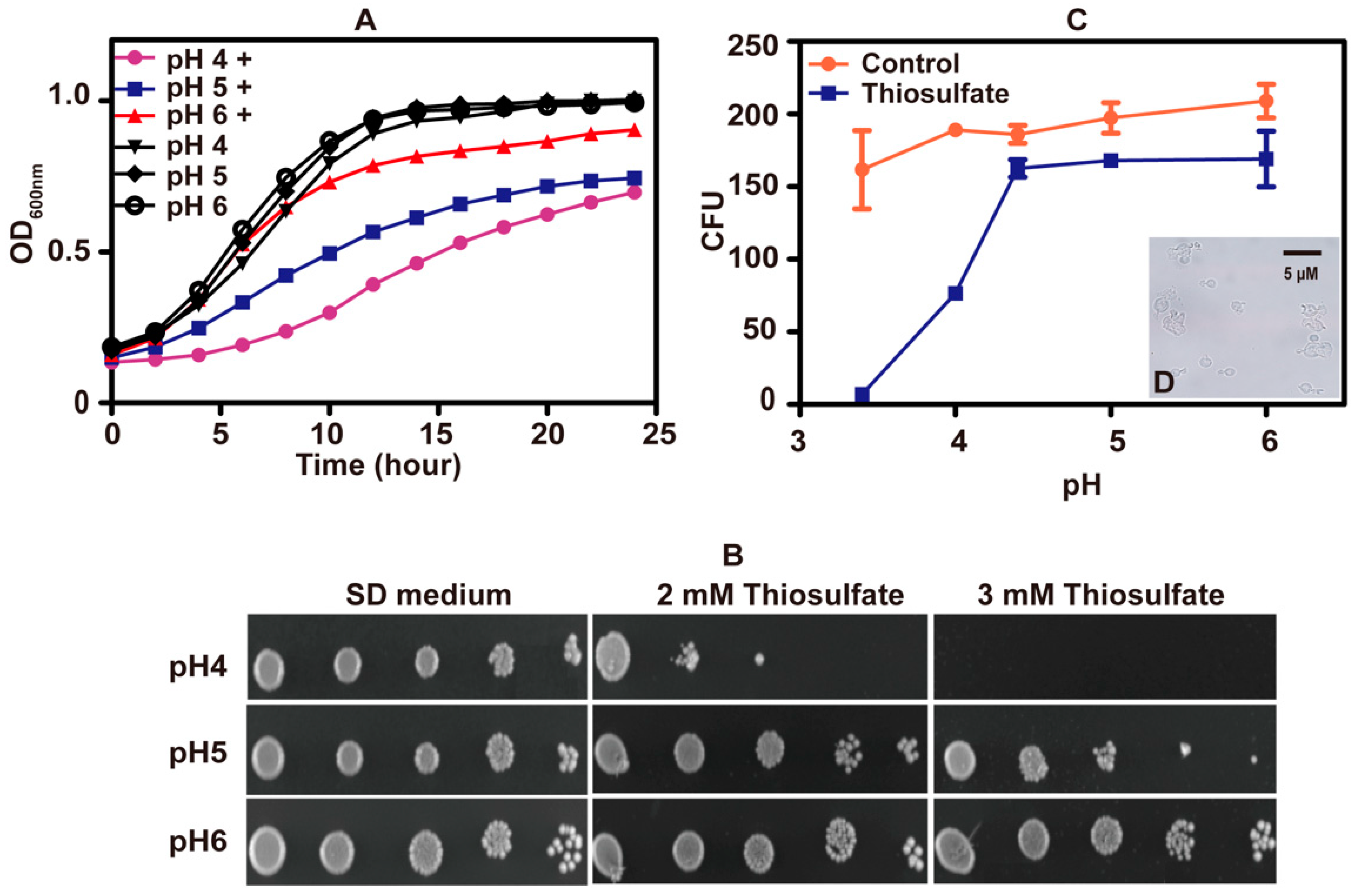
To test whether pH affected the tolerance of S. cerevisiae to thiosulfate, a series of SD mediums with different starting pHs were used. Yeast cells grew well in SD mediums adjusted to various pH values. When 10 mM thiosulfate was added to the medium, S. cerevisiae displayed reduced growth as the pH dropped, suggesting that thiosulfate has pH-dependent effects on S. cerevisiae (Figure 2A). The pH-dependent inhibitions also occurred on agar plates (Figure 2B).
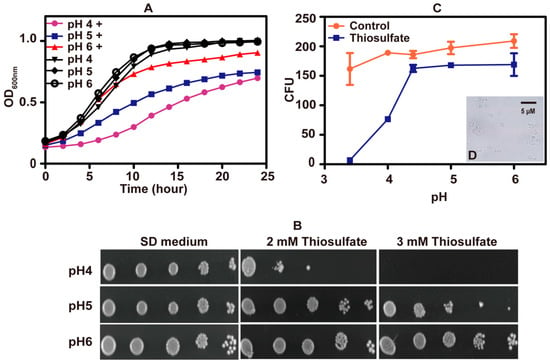
Figure 2.
The toxicity effect of thiosulfate changed with pH. (A) The growth curves of S. cerevisiae in SD medium at different pH. “+” label indicates “with 10 mM thiosulfate”. (B) The tolerance of S. cerevisiae to thiosulfate on SD agar plates of different pH. (C) CFUs of S. cerevisiae after incubating for 1 h in citric acid–sodium phosphate dibasic buffer of different pH (3.4, 4, 4.4, 5 and 6) with 10 mM thiosulfate or without thiosulfate (control). The growth curves are averages of three samples in the 48-well plate (Figure 2A); the data in Figure 2C are averages of three samples with standard deviations. (D) (Figure 2C insert) The image of lysed S. cerevisiae cells after incubating in citric acid–sodium phosphate dibasic buffer (pH 3.4) with 10 mM thiosulfate for 1 h.
Colony forming units (CFUs) were measured after incubation with 10 mM thiosulfate for 60 min in citric acid–sodium phosphate dibasic buffer at different pH values. CFUs did not decrease at pH 4.4 to 6.0, but CFUs sharply decreased at pH < 4 (Figure 2C). At pH 3.4, thiosulfate killed almost all S. cerevisiae cells with erupted cells observed under microscopy (Figure 2D). The results indicate that thiosulfate mainly inhibits the yeast growth at slightly acidic pHs but kills the yeast under acidic conditions.
HPLC analysis showed that approximately 8% of 10 mM thiosulfate decomposed at pH 3.4 in 1 h, likely producing 0.8 mM sulfite and 0.8 mM S0 as previously reported [45]. When yeast cells (OD600nm = 1) in 1 mL of citric acid–sodium phosphate dibasic buffer were incubated with sulfite, S0 or thiosulfate at pH 3.4 and 30 °C for 1 h, sulfite killed almost all the yeast cells and 0.8 mM sulfur powder killed approximately 2/3 of the yeast cells, but 0.8 mM thiosulfate did not have any apparent lethal effect on the yeast cells (Figure S1). At pH 6, 0.8 sulfite and thiosulfate did not reduce CFUs, but 0.8 mM S0 slightly reduced CFUs (Figure S1). These results indicate that at acidic pHs thiosulfate is slowly decomposed to S0 and H2SO3 that kill yeast cells.
3.2. Thiosulfate Is Actively Transported into the Cells for Its Inhibition
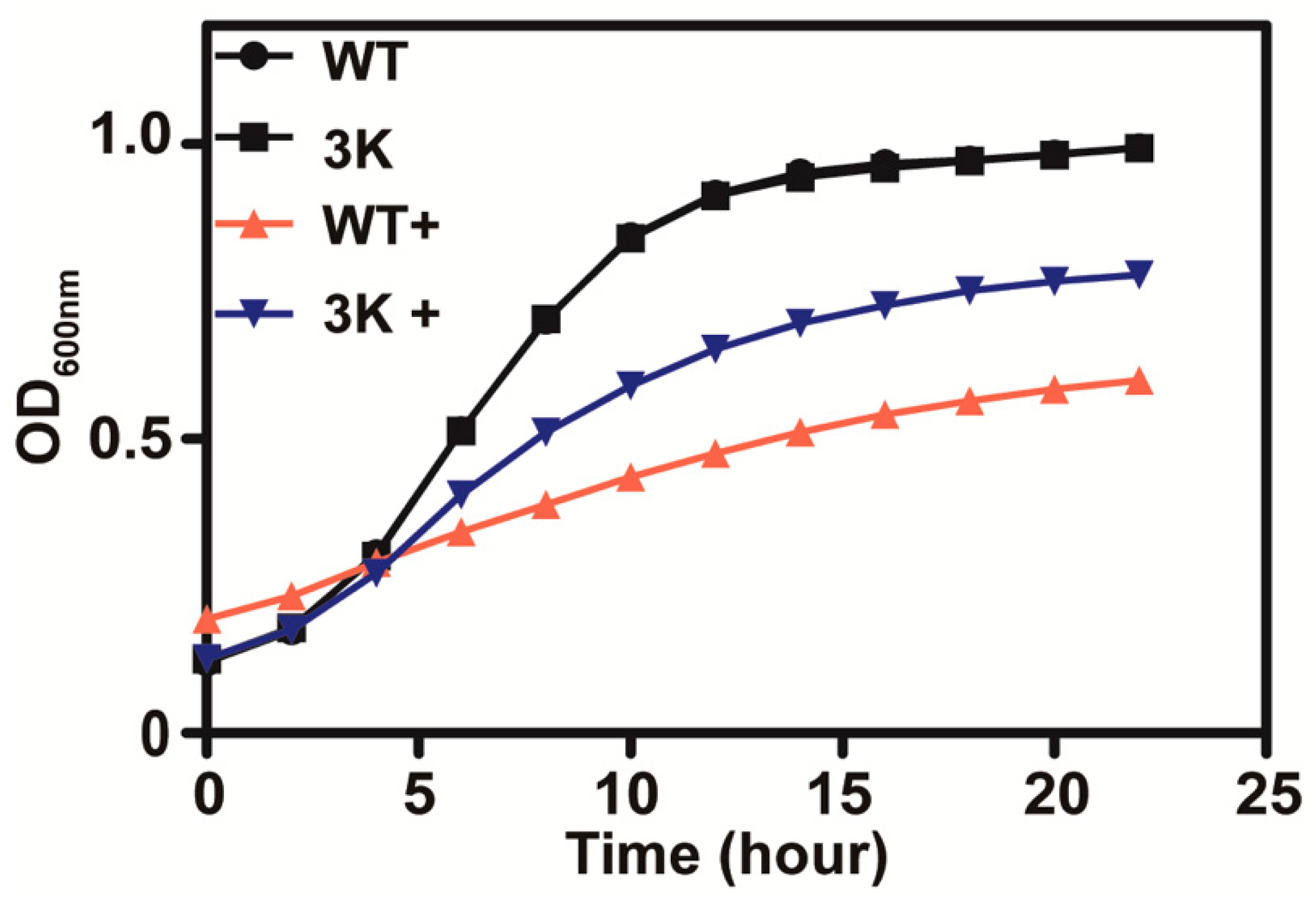
Sul1, Sul2 and Soa1 are the three main permeases responsible for sulfate and thiosulfate uptake in S. cerevisiae, and a mutant with the three genes deleted (triple mutant) was found to be deficient in thiosulfate uptake [10]. The triple mutant yeast grew better than the wild type in SD medium with 10 mM thiosulfate (Figure 3), suggesting that thiosulfate is actively transported into yeast cells where it exhibits its toxicity.
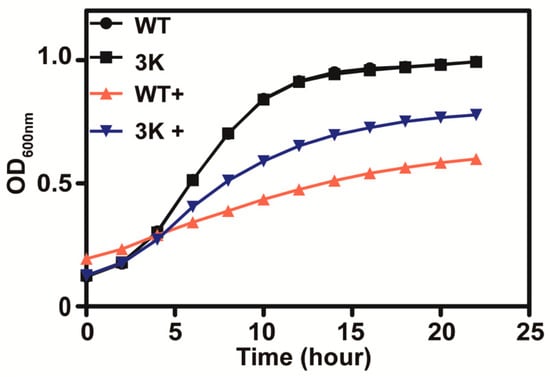
Figure 3.
The thiosulfate transporter mutant (Δsul1 Δsul2 Δsoa1) is less sensitive to thiosulfate in SD medium. The presence of 10 mM thiosulfate is indicated with “+”. The curves of the wild type and the 3K mutant without thiosulfate overlap. The growth curves are averages of three samples in 48-well plates.
3.3. Thiosulfate Itself Is Inhibitory to the Yeast at pH 6
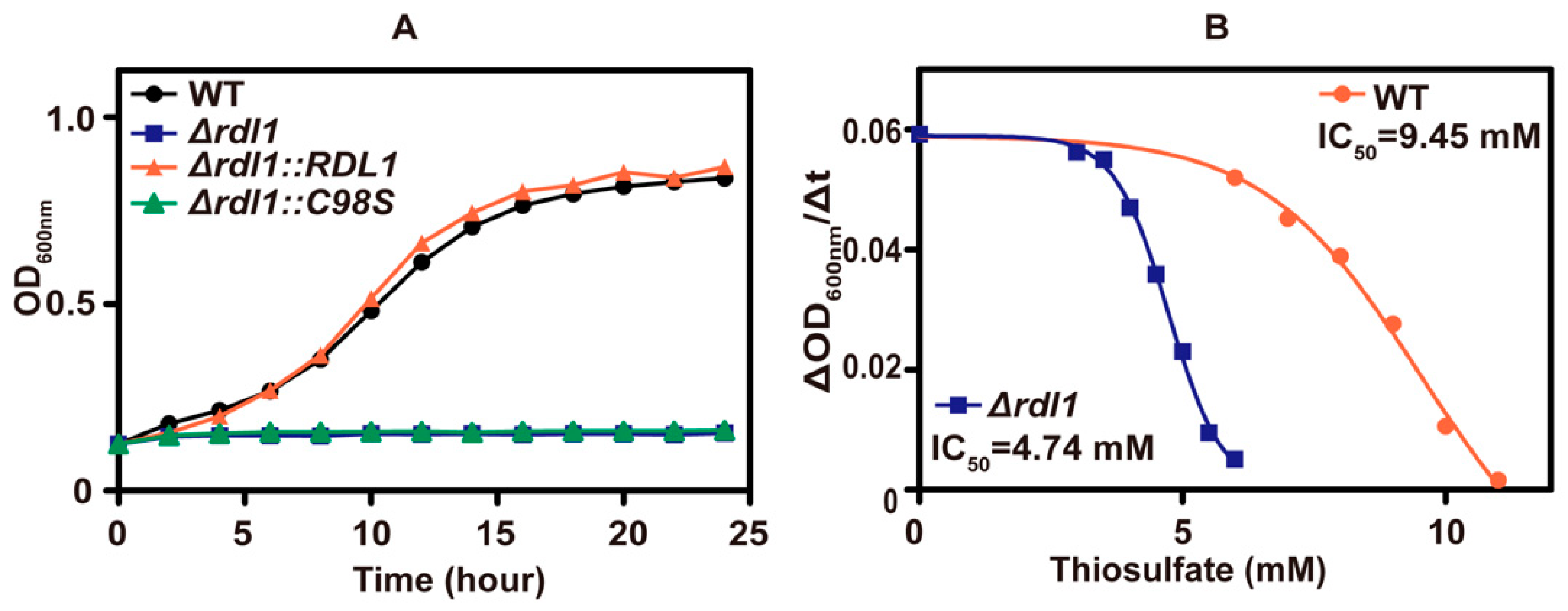
Inside the yeast, the mitochondrial rhodanese, Rdl1, is mainly responsible for converting thiosulfate to H2S [10], and its deletion reduced the whole-cell rhodanese activity by half (Table 3). The RDL1 mutant was more sensitive to thiosulfate than the wild type (Figure 4A). Complementation of Rdl1 to the Δrdl1 mutant restored the relative resistance, but complementing the mutant protein, Rdl1-C98S, without rhodanese activity to the Δrdl1 mutant did not alleviate thiosulfate inhibition (Figure 4A). The IC50 values of thiosulfate were 9.45 mM and 4.74 mM for the wild type and Δrdl1 mutant, respectively (Figure 4B). When sulfur-starved yeast cells were incubated with 0.2 mM thiosulfate in 50 mM potassium phosphate (pH 6) containing glucose for 1 h, the wild type and Δrdl1 mutant accumulated ~0.9 mM and ~1.6 mM thiosulfate inside the cell, respectively. During the incubation, the wild type released large amounts of H2S, whereas the Δrdl1 mutant did not, and the released H2S was detected using a lead acetate paper strip affixed in the gas phase of the tube (Figure S2). When another rhodanese, Rdl2, was deleted, the Δrdl2 mutant and the wild type showed similar resistance to thiosulfate (Figure S3); however, the Δrdl1 Δrdl2 double mutant showed slightly more sensitive to thiosulfate than the Δrdl1 mutant. Overexpression of RDL2 in the Δrdl1 strain did not restore the relative tolerance of the Δrdl1 strain to thiosulfate (Figure S3), perhaps due to variations in subcellular location. These results collectively show that thiosulfate is inhibitory to the yeast growth at pH 6 and Rdl1 alleviates thiosulfate toxicity by actively converting thiosulfate into H2S and sulfite. Further, the produced H2S is not inhibitory to the yeast.

Table 3.
Total rhodanese activities in wild type and rhodanese-deletion strains.
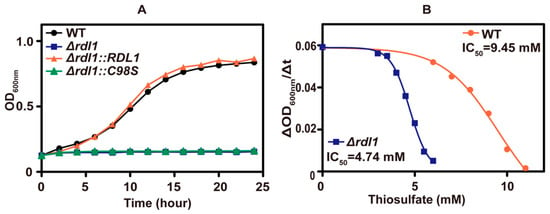
Figure 4.
The tolerance of different S. cerevisiae strains to thiosulfate. (A) The growth curves of different S. cerevisiae strains in SD medium containing 8 mM thiosulfate. (B) The IC50 of thiosulfate in yeast cells. The Vmax of yeast cells growth was calculated in the middle log phase and used for analyzing IC50 of thiosulfate. The growth curves are averages of three replicates in 48-well plates. The P values (by t test) for the Δrdl1 versus wild type were all <0.01.
3.4. Thiosulfate Perturbs the Mitochondrial Bioenergetics in S. cerevisiae
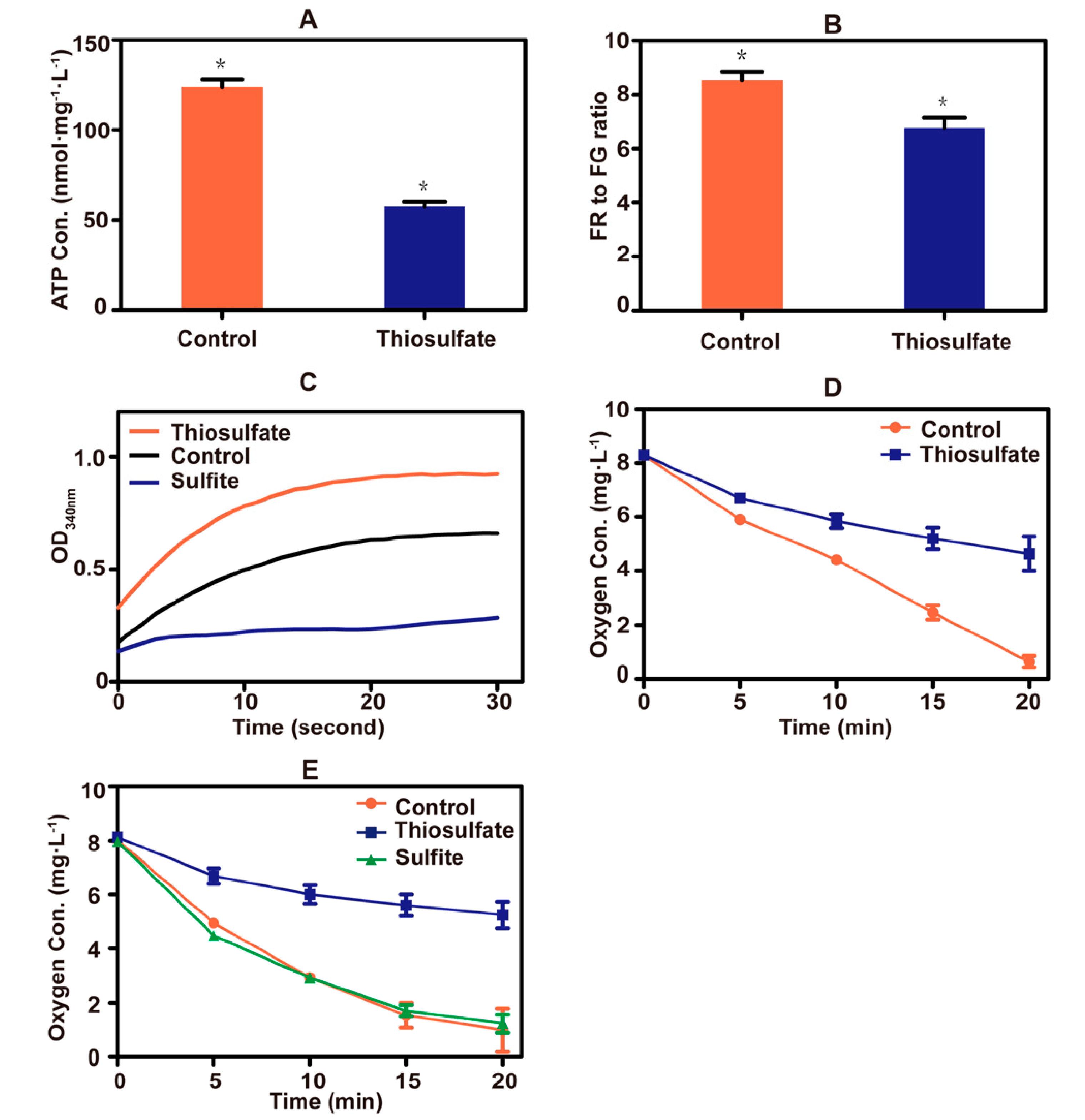
A sharp drop in cellular ATP concentration occurred after the addition of thiosulfate to yeast cells (Figure 5A). Thiosulfate also caused a decrease in the mitochondrial membrane potential (Figure 5B). These results indicate that thiosulfate perturbs the cellular bioenergetics. Since sulfite inhibits the activity of GAPDH [23], the effect of thiosulfate on the enzyme was tested. Thiosulfate did not inhibit the enzyme activity, but the sulfite control did (Figure 5C).
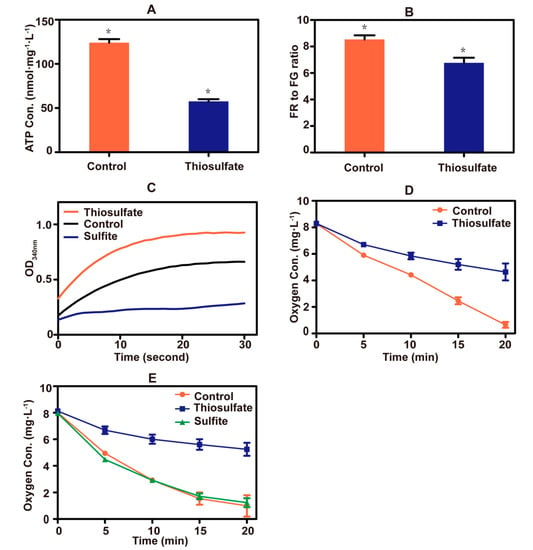
Figure 5.
Thiosulfate perturbs the bioenergetics. (A) The ATP concentrations of yeast cells. ATP in cell extracts was measured with firefly luciferase. (B) The mitochondrial membrane potential of yeast cells. The fluorescent probe JC-1 was used, and a high ratio of red fluorescence (RF) to green fluorescence (GF) represented a high mitochondrial membrane potential. (C) The GAPDH activity of yeast cells. The activity was monitored via NADH production. (D) Thiosulfate inhibition on oxygen consumption in PBS buffer with glucose addition. The sulfur-starved yeast cells were resuspended in sterile PBS buffer containing 2% glucose. Control, no thiosulfate addition; thiosulfate, + 10 mM thiosulfate. (E) Thiosulfate inhibition on oxygen consumption in PBS buffer with TMPD/ascorbate addition. The sulfur-starved yeast cells were resuspended in sterile PBS buffer containing 12.5 mM ascorbate and 1.4 mM TMPD. Control, no thiosulfate addition; thiosulfate, + 10 mM thiosulfate; sulfite, + 10 mM sulfite. The data are averages with standard deviations from three replicates for Figure 5A,B,D,E. * indicates p values < 0.01.
Copper (Cu2+) is an essential trace metal of the electron transport chain of yeast mitochondria [46]. When thiosulfate reacted with Cu2+, an intense color change occurred (Figure S4), likely due to the reduction of Cu2+ to Cu+ by thiosulfate as previously reported [47]. Thiosulfate also severely inhibited oxygen consumption by yeast cells (Figure 5D). TMPD and ascorbate are often used to donate electrons to cytochrome c, which is then oxidized by cytochrome c oxidase at the expense of O2 [48]. Replacing glucose with ascorbate/TMPD, thiosulfate still inhibited oxygen consumption, but sulfite did not (Figure 5E). These results suggested that excessive thiosulfate interferes with the function of the electron transport chain in mitochondria, likely by targeting Cu2+ in cytochrome c oxidase.
4. Discussion
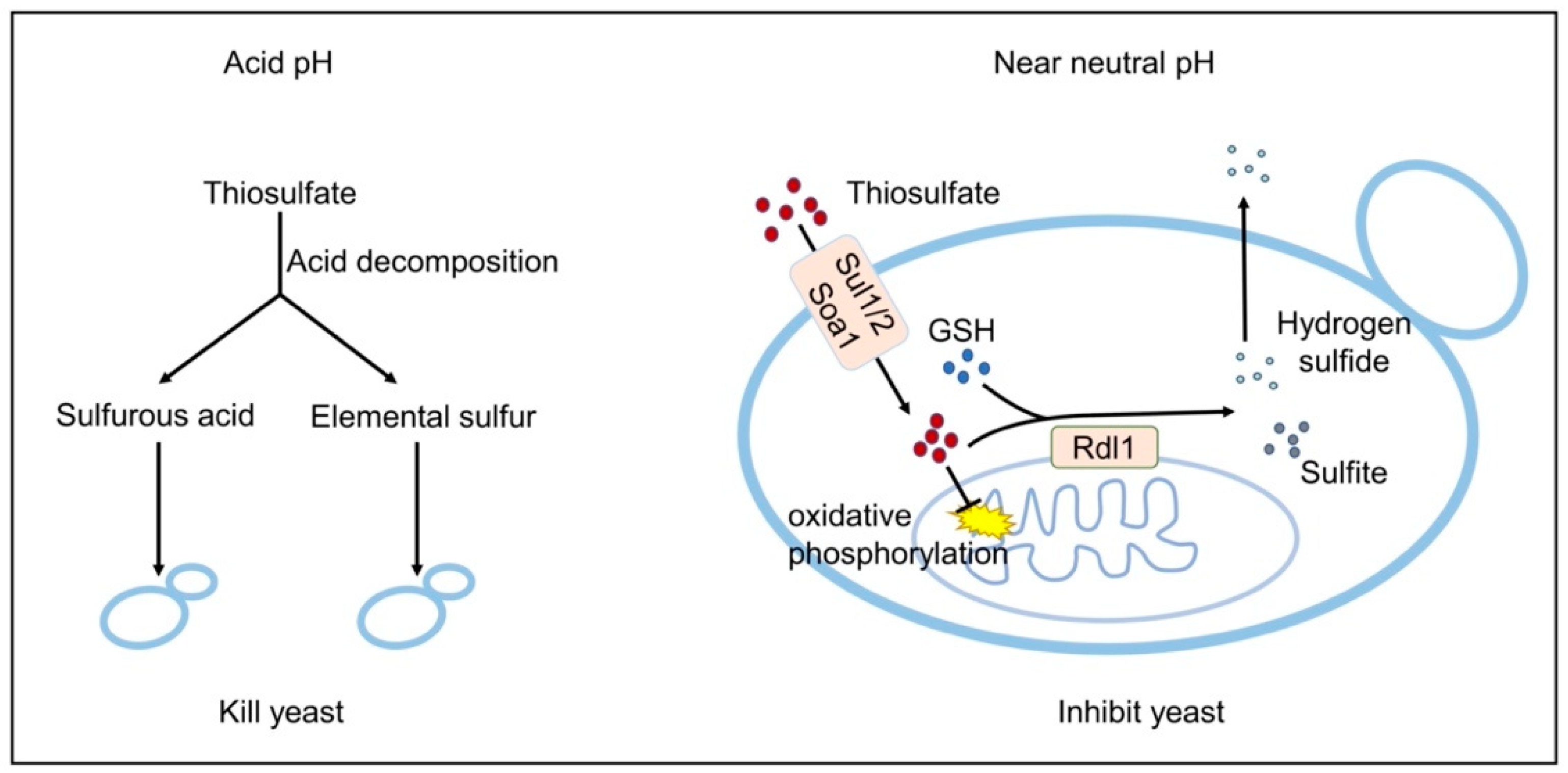
Thiosulfate is toxic to S. cerevisiae. The toxic mechanisms are different at neutral or acidic pHs (Figure 6). At an acidic pH, it slowly decomposes to S0 and sulfurous acid (Rolia et al., 1982), and in our test about 8% of 10 mM thiosulfate decomposed at pH 3.4 in 1 h. Both S0 and sulfurous acid are highly toxic to the yeast [20,29]. The decomposition likely occurs in the medium, as the pH inside yeast cells is relatively neutral even under acidic conditions [49]. Near neutral pH, thiosulfate is actively transported into the cells and accumulated inside the cell (Figure 3) [10]. The accumulated thiosulfate directly inhibits O2 consumption, which in turn lowers membrane potential and ATP levels (Figure 5). Yeast rhodaneses, primarily Rdl1, convert thiosulfate to H2S and sulfite, alleviating thiosulfate inhibition (Figure 4). The proposed mechanism of thiosulfate toxicity is summarized in Figure 6.
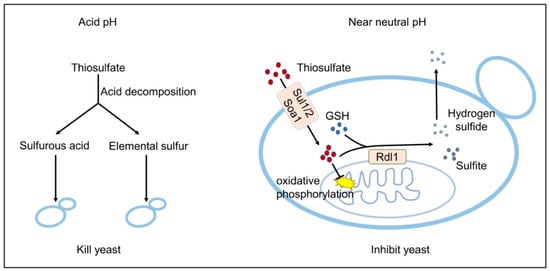
Figure 6.
The proposed mechanisms of thiosulfate toxicity to S. cerevisiae.
The direct target of thiosulfate is likely the electron transport chain (Figure 5), which is also the target of H2S. The cytochrome c oxidase of the mitochondrial electron transport chain contains two copper centers CuA and CuB that are involved in electron transfer [50]. Sulfide binds to CuB, making its re-oxidation difficult and blocking the electron flow [51]. Although thiosulfate is slowly converted into sulfite and H2S by Rdl1 in yeast [10], the produced H2S is likely not high enough to inhibit the yeast as the Δrdl1 mutant that produces much less H2S is more sensitive to thiosulfate (Figure 4A). Since both thiosulfate and sulfide are able to reduce Cu2+ to Cu+ (Figure S4) [47], thiosulfate also has the potential to bind to CuB to block the electron flow. The activity of cytochrome c oxidase can be directly assayed in whole cells with artificial electron donor TMPD, which is again reduced by ascorbate [48]. In the assay, thiosulfate inhibited cytochrome c oxidase (Figure 5E). This finding explains why thiosulfate decreased the oxygen consumption, mitochondrial membrane potential and ATP levels in S. cerevisiae (Figure 5).
The effect of thiosulfate is different from that of sulfite. Sulfite is widely used in fruit and vegetable preservation [16,17]. It is highly toxic at low pH, as undissociated H2SO3 is the effective agent, which easily kills yeasts [20]. At relatively neutral pH, sulfite is toxic only at high concentrations and is a known inhibitor of GAPDH, slowing down glycolysis and ATP production in S. cerevisiae [22,52,53]. However, sulfite does not inhibit oxidative phosphorylation in the mitochondria [23].
S0 is commonly used as a fungicide and employed widely in traditional agriculture as an eco-friendly fungicide to protect vineyards against Botrytis cinerea [54]. S0 exhibited antimicrobial activity (MIC = 5.47 µg/mL) against Staphylococcus aureus and had inhibitory effects on membrane lipids of Aspergillus niger [55,56]. However, its efficiency is impaired by its low solubility [28,29]. To circumvent the limitation, soluble organosulfur compounds that release S0 have been synthesized and used to treat antibiotic-resistant bacteria [57]. Thiosulfate may be used as a soluble inorganic source of S0 under acidic conditions to inhibit yeast and pathogenic microorganisms. A US patent has used a Lactobacillus strain with thiosulfate to treat urogenital yeast infections [58]. Although the mechanism is not reported, we can speculate that the Lactobacillus strain produces lactic acid and lowers pH that facilitates thiosulfate decomposition to release S0 and H2SO3. Thus, at an acidic pH thiosulfate is a good source of S0 and H2SO3, both of which are effective agents against S. cerevisiae.
5. Conclusions
Thiosulfate is an inhibitor of S. cerevisiae. It is more toxic than sulfite at near neutral pH, and it directly inhibits the electron transport chain. The inhibition is reversible, as the yeast recovered in fresh media without thiosulfate. The finding does not contradict the use of thiosulfate in treating cyanide poisoning in humans [34] since high doses of thiosulfate may only temporarily inhibit aerobic respiration. Thiosulfate should be rapidly released in the urine, and it has been shown to be safe in animal studies [59]. At an acidic pH, thiosulfate is a good source of S0 and H2SO3, both of which kill yeast (Figure 6). The finding may guide the appropriate use of thiosulfate as a preservative for foods or fruits or as an agent against pathogenic microorganisms.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antiox10050646/s1, Figure S1: The lethal effect of sulfur species on S. cerevisiae at different pH, Figure S2: The release of H2S from thiosulfate by yeast cells, Figure S3: Thiosulfate tolerance of different S. cerevisiae strains, Figure S4: The reaction of thiosulfate with Cu2+.
Author Contributions
Conceptualization, Z.C. and L.X.; methodology, Z.C.; validation, H.L. (Honglei Liu); investigation, Z.C.; resources, H.L. (Honglei Liu); data curation, Z.C. and H.L. (Honglei Liu); writing—original draft preparation, Z.C.; writing—review and editing, H.L. (Honglei Liu) and L.X.; visualization, Z.C.; supervision, Y.X. and H.L. (Huaiwei Liu); project administration, H.L. (Huaiwei Liu) and Y.X.; funding acquisition, H.L. (Honglei Liu), and Y.X. and L.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (91951202 & 31770126).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or supplementary material.
Acknowledgments
We thank Wenjun Guan of Zhejiang University for providing the S. cerevisiae strain BY4742.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Meyer, B. Elemental sulfur. Chem. Rev. 1976, 76, 367–388. [Google Scholar] [CrossRef]
- Jorgensen, B.B. A Thiosulfate Shunt in the Sulfur Cycle of Marine Sediments. Science 1990, 249, 152–154. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, H.; Cui, F.; Xun, L. Recombinant Escherichia coli with sulfide: Quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ. Microbiol. 2016, 18, 5123–5136. [Google Scholar] [CrossRef]
- Xin, Y.; Gao, R.; Cui, F.; Lu, C.; Liu, H.; Liu, H.; Xia, Y.; Xun, L. The heterotrophic bacterium Cupriavidus pinatubonensis JMP134 oxidizes sulfide to sulfate with thiosulfate as a key intermediate. Appl. Environ. Microbiol. 2020, 86, e01835-20. [Google Scholar] [CrossRef]
- Xia, Y.; Lu, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017, 11, 2754–2766. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, R.; Xi, S.; Cai, R.; Sun, C. A novel bacterial thiosulfate oxidation pathway provides a new clue about the formation of zero-valent sulfur in deep sea. ISME J. 2020, 14, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Rameez, M.J.; Pyne, P.; Mandal, S.; Chatterjee, S.; Ghosh, W.J.M.R. Two pathways for thiosulfate oxidation in the alphaproteobacterial chemolithotroph Paracoccus thiocyanatus SST. Microbiol. Res. 2019, 230, 126345. [Google Scholar] [CrossRef]
- Houghton, J.L.; Foustoukos, D.I.; Flynn, T.M.; Vetriani, C.; Bradley, A.S.; Fike, D.A. Thiosulfate oxidation by Thiomicrospira thermophila: Metabolic flexibility in response to ambient geochemistry. Environ. Microbiol. 2016, 18, 3057–3072. [Google Scholar] [CrossRef]
- Funahashi, E.; Saiki, K.; Honda, K.; Sugiura, Y.; Kawano, Y.; Ohtsu, I.; Watanabe, D.; Wakabayashi, Y.; Abe, T.; Nakanishi, T.; et al. Finding of thiosulfate pathway for synthesis of organic sulfur compounds in Saccharomyces cerevisiae and improvement of ethanol production. J. Biosci. Bioeng. 2015, 120, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, X.; Li, H.; Liu, H.; Xia, Y.; Xun, L. The complete pathway for thiosulfate utilization in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2018, 84, e01241-18. [Google Scholar] [CrossRef]
- Kawano, Y.; Onishi, F.; Shiroyama, M.; Miura, M.; Tanaka, N.; Oshiro, S.; Nonaka, G.; Nakanishi, T.; Ohtsu, I. Improved fermentative L-cysteine overproduction by enhancing a newly identified thiosulfate assimilation pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 6879–6889. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Yamagata, S.; Masui, R.; Inoue, Y.; Shibata, T.; Yokoyama, S.; Kuramitsu, S.; Iwama, T. Cloning and overexpression of the oah1 gene encoding O-acetyl-L-homoserine sulfhydrylase of Thermus thermophilus HB8 and characterization of the gene product. Biochim. Biophys. Acta 2001, 1549, 61–72. [Google Scholar] [CrossRef]
- Melideo, S.L.; Jackson, M.R.; Jorns, M.S. Biosynthesis of a central intermediate in hydrogen sulfide metabolism by a novel human sulfurtransferase and its yeast ortholog. Biochemistry 2014, 53, 4739–4753. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.W.; Walker, M.E.; Fedrizzi, B.; Gardner, R.C.; Jiranek, V. Hydrogen sulfide and its roles in Saccharomyces cerevisiae in a winemaking context. FEMS Yeast Res. 2017, 17, fox058. [Google Scholar] [CrossRef]
- Dinegar, R.H.; Smellie, R.H.; Mer, V.K.L. Kinetics of the acid decomposition of sodium thiosulfate in dilute solutions. J. Am. Chem. Soc. 1951, 73, 2050–2054. [Google Scholar] [CrossRef]
- Taylor, S.L.; Higley, N.A.; Bush, R.K. Sulfites in foods: Uses, analytical methods, residues, fate, exposure assessment, metabolism, toxicity, and hypersensitivity. Adv. Food Res. 1986, 30, 1–76. [Google Scholar]
- Liu, X.; Sang, M.; Zhang, X.; Zhang, T.; Zhang, H.; He, X.; Li, S.; Sun, X.; Zhang, Z. Enhancing expression of SSU1 genes in Saccharomyces uvarum leads to an increase in sulfite tolerance and transcriptome profiles change. FEMS Yeast Res. 2017, 17, fox023. [Google Scholar] [CrossRef]
- Chang, I.S.; Kim, B.H.; Shin, P.K.; Microbiology, A.E. Use of sulfite and hydrogen peroxide to control bacterial contamination in ethanol fermentation. Appl. Environ. Microbiol. 1997, 63, 1–6. [Google Scholar] [CrossRef]
- Nardi, T.; Corich, V.; Giacomini, A.; Blondin, B. A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology 2010, 156, 1686–1696. [Google Scholar] [CrossRef]
- King, A.D., Jr.; Ponting, J.D.; Sanshuck, D.W.; Jackson, R.; Mihara, K. Factors Affecting Death of Yeast by Sulfur Dioxide. J. Food Prot. 1981, 44, 92–97. [Google Scholar] [CrossRef]
- Schimz, K.L.; Holzer, H. Rapid decrease of ATP content in intact cells of Saccharomyces cerevisiae after incubation with low concentrations of sulfite. Arch. Microbiol. 1979, 121, 225–229. [Google Scholar] [CrossRef]
- Hinze, H.; Holzer, H. Analysis of the energy metabolism after incubation of Saccharomyces cerevisiae with sulfite or nitrite. Arch. Microbiol. 1986, 145, 27–31. [Google Scholar] [CrossRef]
- Steinacker, A. A suggested mechanism of action for sulfite sensitivity. J. Allergy Clin. Immunol. 1986, 77, 116–117. [Google Scholar] [CrossRef]
- Neta, P.; Huie, R.E. Free-radical chemistry of sulfite. Environ. Health Perspect. 1985, 64, 209–217. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.M.; Fridovich, I. The utility of superoxide dismutase in studying free radical reactions. I. Radicals generated by the interaction of sulfite, dimethyl sulfoxide, and oxygen. J. Biol. Chem. 1969, 244, 6056–6063. [Google Scholar] [CrossRef]
- Peiser, G.D.; Yang, S.F. Sulfite-mediated destruction of β-carotene. J. Agric. Food Chem. 1979, 27, 446–449. [Google Scholar] [CrossRef]
- Yang, S.F. Destruction of tryptophan during the aerobic oxidation of sulfite ions. Environ. Res. 1973, 6, 395–402. [Google Scholar] [CrossRef]
- Williams, J.S.; Cooper, R.M. The oldest fungicide and newest phytoalexin—A reappraisal of the fungitoxicity of elemental sulphur. Plant Pathol. 2010, 53, 263–279. [Google Scholar] [CrossRef]
- Rai, M.; Ingle, A.P.; Paralikar, P. Sulfur and sulfur nanoparticles as potential antimicrobials: From traditional medicine to nanomedicine. Expert Rev. Anti Infect. Ther. 2016, 14, 969–978. [Google Scholar] [CrossRef]
- Sato, I.; Shimatani, K.; Fujita, K.; Abe, T.; Shimizu, M.; Fujii, T.; Hoshino, T. Glutathione reductase/glutathione is responsible for cytotoxic elemental sulfur tolerance via polysulfide shuttle in fungi. J. Biol. Chem. 2011, 286, 20283–20291. [Google Scholar] [CrossRef]
- Islamov, R.A.; Bishimova, I.; Sabitov, A.N.; Ilin, A.I.; Burkitbaev, M.M. Lack of Mutagenic Activity of Sulfur Nanoparticles in Micronucleus Test on L5178Y Cell Culture. Cell Tissue Biol. 2018, 12, 27–32. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Mandal, A.; Ghosh, M.; Basu, S.; Chakravorty, D.; Goswami, A. Investigation of antimicrobial physiology of orthorhombic and monoclinic nanoallotropes of sulfur at the interface of transcriptome and metabolome. Appl. Microbiol. Biotechnol. 2013, 97, 5965–5978. [Google Scholar] [CrossRef]
- Cetkauskaite, A.; Pessala, P.; Sodergren, A. Elemental sulfur: Toxicity in vivo and in vitro to bacterial luciferase, in vitro yeast alcohol dehydrogenase, and bovine liver catalase. Environ. Toxicol. 2004, 19, 372–386. [Google Scholar] [CrossRef]
- De La Calzada-Jeanlouie, M.; Coombs, J.; Shaukat, N.; Olsen, D. Utility of sodium thiosulfate in acute cyanide toxicity. Ann. Emerg. Med. 2013, 61, 124–125. [Google Scholar] [CrossRef]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23–e30. [Google Scholar] [CrossRef]
- Xia, Y.; Chu, W.; Qi, Q.; Xun, L. New insights into the QuikChange™ process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res. 2015, 43, e12. [Google Scholar] [CrossRef]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]
- Togawa, T.; Ogawa, M.; Nawata, M.; Ogasawara, Y.; Kawanabe, K.; Tanabe, S. High performance liquid chromatographic determination of bound sulfide and sulfite and thiosulfate at their low levels in human serum by pre-column fluorescence derivatization with monobromobimane. Chem. Pharm. Bull. 1992, 40, 3000–3004. [Google Scholar] [CrossRef] [PubMed]
- Yaglom, J.; Linskens, M.; Sadis, S.; Rubin, D.M.; Futcher, B.; Finley, D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 1995, 15, 731–741. [Google Scholar] [CrossRef]
- Nash, R.; Tokiwa, G.; Anand, S.; Erickson, K.; Futcher, A. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988, 7, 4335–4346. [Google Scholar] [CrossRef]
- Reers, M.; Smith, T.W.; Chen, L.B. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry 1991, 30, 4480–4486. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, Y.; Zhang, Y.; Xie, Z.; Zhao, X.; Sun, Y.; Kong, W. The dual role of poly (ADP-ribose) polymerase-1 in modulating parthanatos and autophagy under oxidative stress in rat cochlear marginal cells of the stria vascularis. Redox Biol. 2018, 14, 361–370. [Google Scholar] [CrossRef]
- Su, B.; Ji, Y.; Sun, X.; Liu, X.; Chen, Z. Brain-derived neurotrophic factor (BDNF)-induced mitochondrial motility arrest and presynaptic docking contribute to BDNF-enhanced synaptic transmission. J. Biol. Chem. 2014, 289, 1213–1226. [Google Scholar] [CrossRef]
- Ferdinand, W. The isolation and specific activity of rabbit-muscle glyceraldehyde phosphate dehydrogenase. Biochem. J. 1964, 92, 578–585. [Google Scholar] [CrossRef]
- Rolia, E.; Chakrabarti, C.L. Kinetics of decomposition of tetrathionate, trithionate, and thiosulfate in alkaline media. Environ. Sci. Technol. 1982, 16, 852–857. [Google Scholar] [CrossRef]
- Ghezzi, D.; Zeviani, M. Assembly factors of human mitochondrial respiratory chain complexes: Physiology and pathophysiology. Adv. Exp. Med. Biol. 2012, 748, 65–106. [Google Scholar]
- Breuer, P.L.; Jeffrey, M.I. The reduction of copper (II) and the oxidation of thiosulfate and oxysulfur anions in gold leaching solutions. Hydrometallurgy 2003, 70, 163–173. [Google Scholar] [CrossRef]
- Packer, L.; Mustafa, M.G. Pathways of electron flow established by tetramethyl-phenylenediamine in mitochondria and ascites tumor cells. BBA-Enzymol. Biol. Oxid. 1966, 113, 1–12. [Google Scholar] [CrossRef]
- Valli, M.; Sauer, M.; Branduardi, P. Intracellular pH Distribution in Saccharomyces cerevisiae Cell Populations, Analyzed by Flow Cytometry. Appl. Environ. Microbiol. 2005, 71, 1515–1521. [Google Scholar] [CrossRef]
- Mansilla, N.; Racca, S.; Gras, D.E.; Gonzalez, D.H.; Welchen, E. The Complexity of Mitochondrial Complex IV: An Update of Cytochrome c Oxidase Biogenesis in Plants. Int. J. Mol. ENCES 2018, 19, 662–695. [Google Scholar] [CrossRef]
- Paul, B.D.; Snyder, S.H.; Kashfi, K. Effects of hydrogen sulfide on mitochondrial function and cellular bioenergetics. Redox. Biol. 2021, 38, 101772. [Google Scholar] [CrossRef] [PubMed]
- Schimz, K.L. The effect of sulfite on the yeast Saccharomyces cerevisiae. Arch. Microbiol. 1980, 125, 89–95. [Google Scholar] [CrossRef]
- Nadai, C.; Treu, L.; Campanaro, S.; Giacomini, A.; Corich, V. Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. Appl. Microbiol. Biotechnol. 2016, 100, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.M.; Williams, J.S. Elemental sulphur as an induced antifungal substance in plant defence. J. Exp. Bot. 2004, 55, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Ghosh, M.; Goswami, A.J.C.M. Inhibitory effects of sulfur nanoparticles on membrane lipids of Aspergillus niger: A novel route of fungistasis. Curr. Microbiol. 2012, 65, 91–97. [Google Scholar] [CrossRef]
- Suleiman, M.; Al-Masri, M.; Ali, A.A.; Aref, D.; Saadeddin, A.H.I.; Warad, I. Synthesis of Nano-sized Sulfur Nanoparticles and their Antibacterial Activities. J. Mater. Environ. Sci. 2015, 6, 513–518. [Google Scholar]
- Xu, Z.; Qiu, Z.; Liu, Q.; Huang, Y.; Li, D.; Shen, X.; Fan, K.; Xi, J.; Gu, Y.; Tang, Y.; et al. Converting organosulfur compounds to inorganic polysulfides against resistant bacterial infections. Nat. Commun. 2018, 9, 3713–3725. [Google Scholar] [CrossRef]
- Nivoliez, A. Use of Thiosulfate to Potentiate the Anti-Pathogenic Effect of Lactobacilli. U.S. Patent 15/899,273, 21 May 2015. [Google Scholar]
- Hall, A.H.; Rumack, B.H. Hydroxycobalamin/sodium thiosulfate as a cyanide antidote. J. Emerg. Med. 1987, 5, 115–121. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).