Antiaging Effect of 4-N-Furfurylcytosine in Yeast Model Manifests through Enhancement of Mitochondrial Activity and ROS Reduction
Abstract
:1. Introduction
2. Materials and Methods
2.1. 4-N-Furfurylcytosine
2.2. Yeast Strain and Media
2.3. CLS Analysis
2.4. Growth Kinetics
2.5. Flow Cytometry Analyses
2.5.1. Effect of FC on MMP
2.5.2. Protective Effect against H2O2
2.5.3. Regenerative Effect after H2O2 Treatment
2.5.4. ROS Measurement
2.6. Confocal Microscopy Analyses
2.6.1. FC Effect on the Yeast Cell Viability
2.6.2. Mitochondrial Membrane Potential
2.6.3. Comparative Analysis of Maturity and Cell Size after FC Treatment
2.7. Respiration Rate Analysis
2.8. Real-Time PCR
2.8.1. Total RNA Isolation
2.8.2. cDNA Synthesis and qPCR
2.9. Statistical Analysis
3. Results
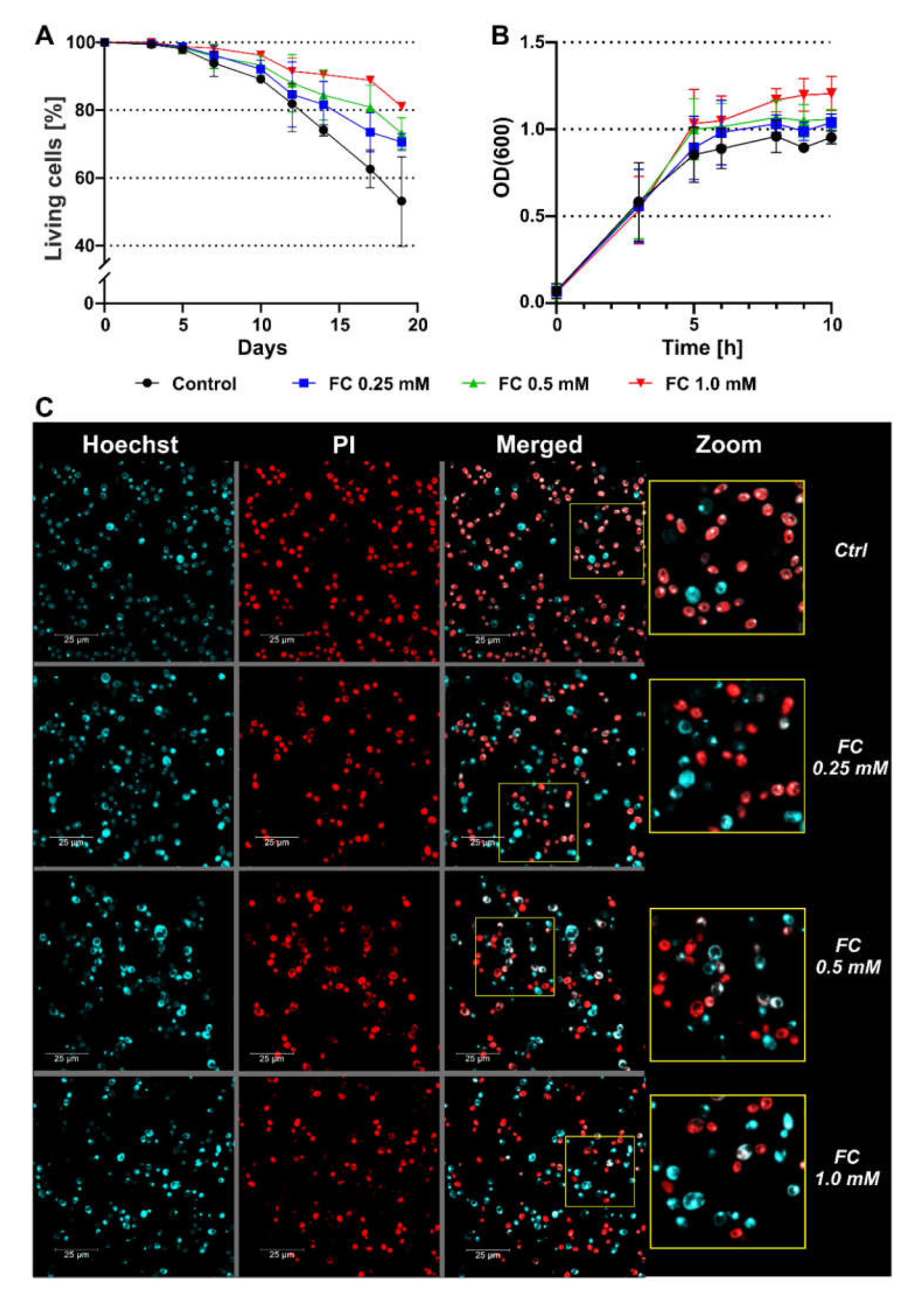
3.1. FC Supplementation Extends Yeast Survival
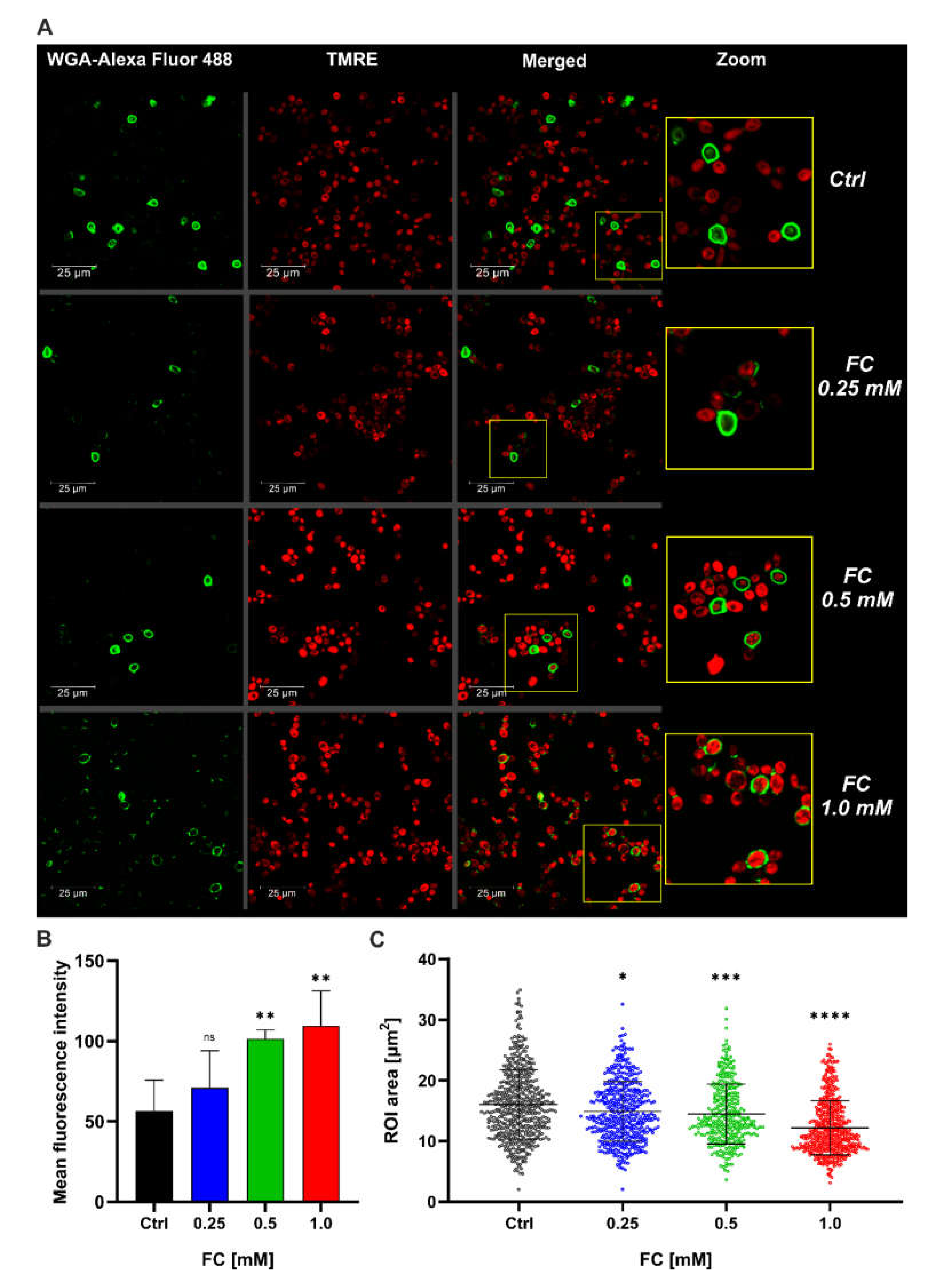
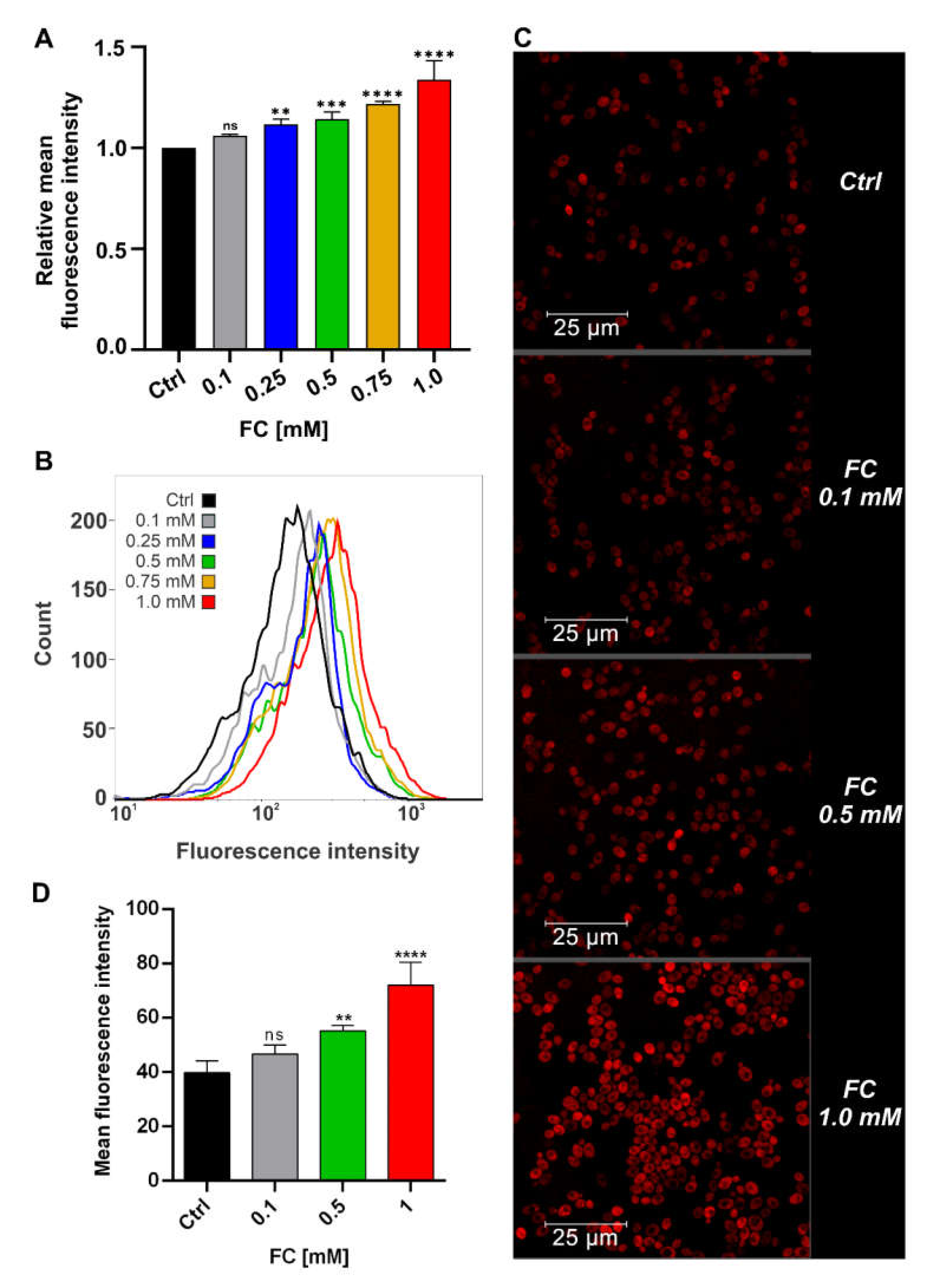
3.2. FC Strongly Enhances Mitochondrial Performance in Glucose Medium and Also Reduces ROS Level
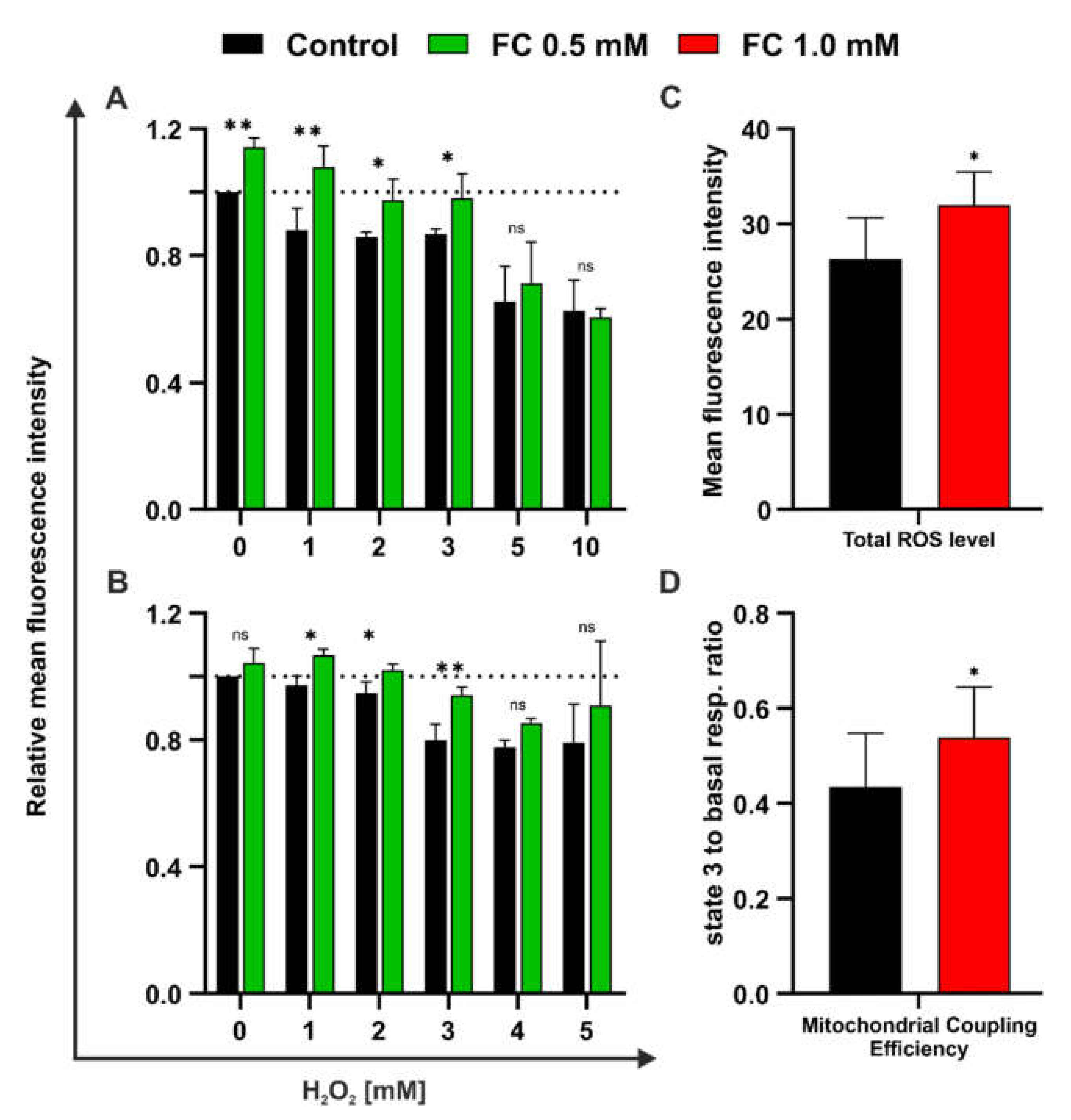
3.3. FC Exerts Both Protective and Regenerative Effects on Yeast Treated with H2O2
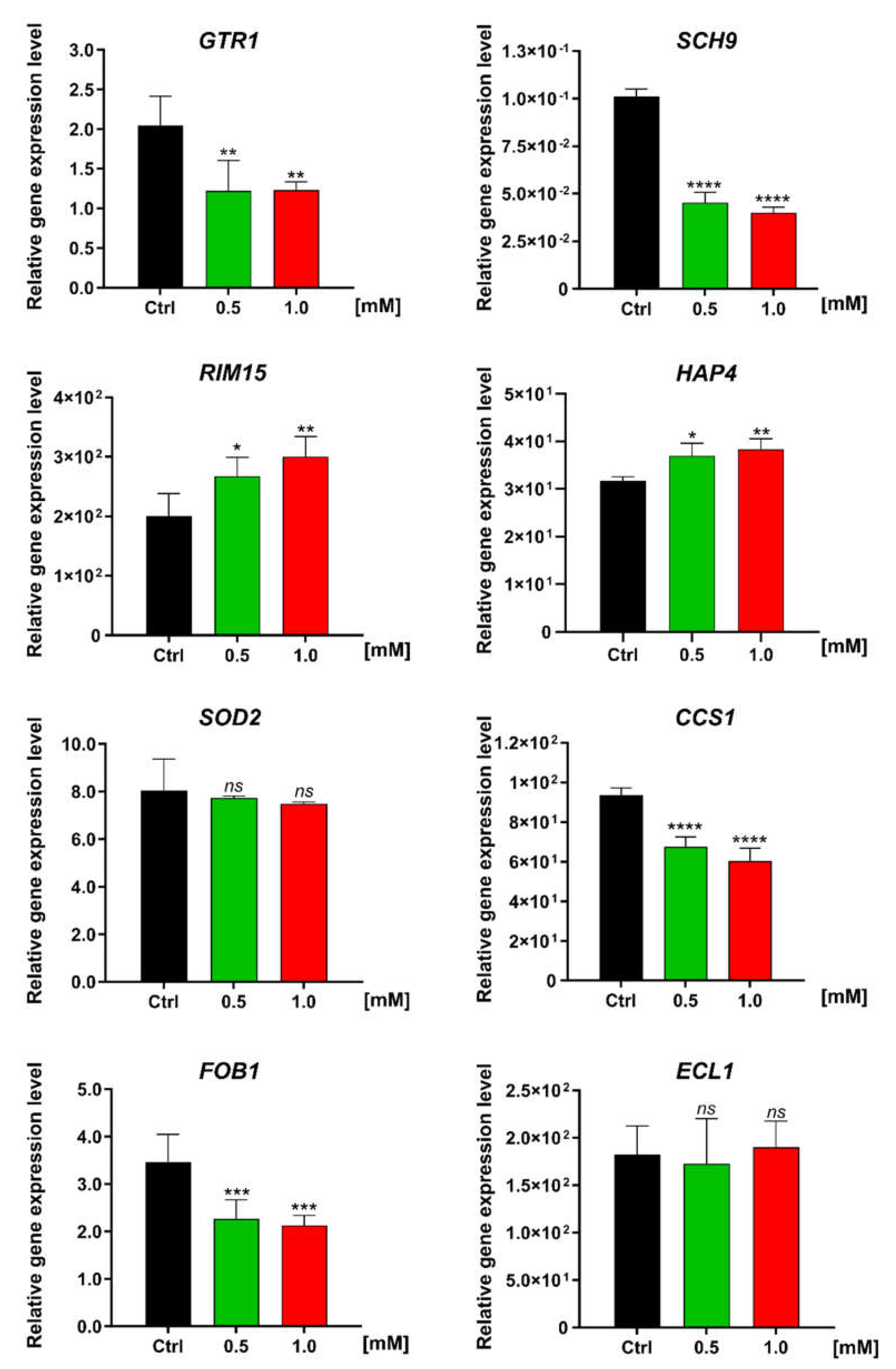
3.4. Effect of FC on the Expression Level of Genes Involved in Metabolism and Stress Resistance in Yeast Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zheng, Y.; Chen, X. Drugs for Autoimmune Inflammatory Diseases: From Small Molecule Compounds to Anti-TNF Biologics. Front. Pharmacol. 2017, 8, 460. [Google Scholar] [CrossRef] [PubMed]
- Teiten, M.H.; Gaascht, F.; Dicato, M.; Diederich, M. Anticancer bioactivity of compounds from medicinal plants used in European medieval traditions. Biochem. Pharmacol. 2013, 86, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xiao, J.; Liu, X.; Jiang, Z.; Zhan, Y.; Yin, T.; He, L.; Zhang, F.; Xing, S.; Chen, B.; et al. AICD: An integrated anti-inflammatory compounds database for drug discovery. Sci. Rep. 2019, 9, 7737. [Google Scholar] [CrossRef]
- Zawirska-Wojtasiak, R.; Fedoruk-Wyszomirska, A.; Piechowska, P.; Mildner-Szkudlarz, S.; Bajerska, J.; Wojtowicz, E.; Przygoński, K.; Gurda, D.; Kubicka, W.; Wyszko, E. β-Carbolines in Experiments on Laboratory Animals. Int. J. Mol. Sci. 2020, 21, 5245. [Google Scholar] [CrossRef]
- Sidorova, Y.A.; Volcho, K.P.; Salakhutdinov, N.F. Neuroregeneration in Parkinson’s Disease: From Proteins to Small Molecules. Curr. Neuropharmacol. 2019, 17, 268–287. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, S.; Li, J.; Jiang, T.; Feng, L.; Pei, J.; Wang, G.; Ouyang, L.; Liu, B. Targeting autophagy using small-molecule compounds to improve potential therapy of Parkinson’s disease. Acta Pharm. Sin. B 2021, 11, 3015–3034. [Google Scholar] [CrossRef]
- Iraji, A.; Khoshneviszadeh, M.; Firuzi, O.; Khoshneviszadeh, M.; Edraki, N. Novel small molecule therapeutic agents for Alzheimer disease: Focusing on BACE1 and multi-target directed ligands. Bioorg. Chem. 2020, 97, 103649. [Google Scholar] [CrossRef]
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [Green Version]
- Alexeyev, M.F. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009, 276, 5768–5787. [Google Scholar] [CrossRef] [Green Version]
- Bokov, A.; Chaudhuri, A.; Richardson, A. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 2004, 125, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Lees, H.; Walters, H.; Cox, L.S. Animal and human models to understand ageing. Maturitas 2016, 93, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Arlia-Ciommo, A.; Svistkova, V.; Mohtashami, S.; Titorenko, V.I. A novel approach to the discovery of anti-tumor pharmaceuticals: Searching for activators of liponecrosis. Oncotarget 2016, 7, 5204–5225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.D.; Fabrizio, P. Regulation of longevity and stress resistance: A molecular strategy conserved from yeast to humans? Cell Mol. Life Sci. 2002, 59, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Lasserre, J.P.; Dautant, A.; Aiyar, R.S.; Kucharczyk, R.; Glatigny, A.; Tribouillard-Tanvier, D.; Rytka, J.; Blondel, M.; Skoczen, N.; Reynier, P.; et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model. Mech. 2015, 8, 509–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, D.A. Studying the replicative life span of yeast cells. Methods Mol. Biol. 2013, 1048, 49–63. [Google Scholar] [PubMed] [Green Version]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef]
- Kadlecová, A.; Maková, B.; Artal-Sanz, M.; Strnad, M.; Voller, J. The plant hormone kinetin in disease therapy and healthy aging. Ageing Res. Rev. 2019, 55, 100958. [Google Scholar] [CrossRef]
- Orlicka-Płocka, M.; Fedoruk-Wyszomirska, A.; Gurda-Woźna, D.; Pawelczak, P.; Krawczyk, P.; Giel-Pietraszuk, M.; Framski, G.; Ostrowski, T.; Wyszko, E. Implications of Oxidative Stress in Glioblastoma Multiforme Following Treatment with Purine Derivatives. Antioxidants 2021, 10, 950. [Google Scholar] [CrossRef]
- Karachitos, A.; Jordan, J.; Kmita, H. Cytoprotective activity of minocycline includes improvement of mitochondrial coupling: The importance of minocycline concentration and the presence of VDAC. J. Bioenerg. Biomembr. 2012, 44, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Fabrizio, P.; Longo, V.D. The chronological life span of Saccharomyces cerevisiae. Methods Mol. Biol. 2007, 371, 89–95. [Google Scholar] [PubMed]
- He, C.; Zhou, C.; Kennedy, B.K. The yeast replicative aging model. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864 Pt A, 2690–2696. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrugia, G.; Balzan, R. Oxidative Stress and Programmed Cell Death in Yeast. Front. Oncol. 2012, 2, 64. [Google Scholar] [CrossRef] [Green Version]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, E.; Beauvoit, B.; Blondin, B.; Salmon, J.M. Oxygen consumption by anaerobic Saccharomyces cerevisiae under enological conditions: Effect on fermentation kinetics. Appl. Environ. Microbiol. 2003, 69, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Gasmi, N.; Jacques, P.E.; Klimova, N.; Guo, X.; Ricciardi, A.; Robert, F.; Turcotte, B. The switch from fermentation to respiration in Saccharomyces cerevisiae is regulated by the Ert1 transcriptional activator/repressor. Genetics 2014, 198, 547–560. [Google Scholar] [CrossRef]
- Turcotte, B.; Liang, X.B.; Robert, F.; Soontorngun, N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010, 10, 2–13. [Google Scholar] [CrossRef] [Green Version]
- Reddi, A.R.; Culotta, V.C. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 2013, 152, 224–235. [Google Scholar] [CrossRef] [Green Version]
- M, M.F.; Boyd, S.D.; Winkler, D.D.; Winge, D.R. Oxygen-dependent activation of Cu,Zn-superoxide dismutase-1. Metallomics 2017, 9, 1047–1059. [Google Scholar]
- Unlu, E.S.; Koc, A. Effects of deleting mitochondrial antioxidant genes on life span. Ann. N. Y. Acad. Sci. 2007, 1100, 505–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. emBo J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deprez, M.A.; Eskes, E.; Winderickx, J.; Wilms, T. The TORC1-Sch9 pathway as a crucial mediator of chronological lifespan in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy048. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, E.; Ghillebert, R.; Wilms, T.; Winderickx, J. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Fabrizio, P.; Pozza, F.; Pletcher, S.D.; Gendron, C.M.; Longo, V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 2001, 292, 288–290. [Google Scholar] [CrossRef] [Green Version]
- Defossez, P.A.; Prusty, R.; Kaeberlein, M.; Lin, S.J.; Ferrigno, P.; Silver, P.A.; Keil, R.L.; Guarente, L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 1999, 3, 447–455. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Zou, Y.; Chen, Y.; Ying, H. Overexpression of THI4 and HAP4 Improves Glucose Metabolism and Ethanol Production in Saccharomyces cerevisiae. Front. Microbiol. 2018, 9, 1444. [Google Scholar] [CrossRef] [Green Version]
- Azuma, K.; Ohtsuka, H.; Murakami, H.; Aiba, H. Extension of chronological lifespan by ScEcl1 depends on mitochondria in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2012, 76, 1938–1942. [Google Scholar] [CrossRef] [Green Version]
- Garmany, A.; Yamada, S.; Terzic, A. Longevity leap: Mind the healthspan gap. npj Regen MedI. 2021, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Ferri, E.; Marzetti, E.; Calvani, R.; Picca, A.; Cesari, M.; Arosio, B. Role of Age-Related Mitochondrial Dysfunction in Sarcopenia. Int. J. Mol. Sci. 2020, 21, 5236. [Google Scholar] [CrossRef] [PubMed]
- Miquel, J.; Economos, A.C.; Fleming, J.; Johnson, J.E., Jr. Mitochondrial role in cell aging. Exp. Gerontol. 1980, 15, 575–591. [Google Scholar] [CrossRef]
- Wickens, A.P. Ageing and the free radical theory. Respir. Physiol. 2001, 128, 379–391. [Google Scholar] [CrossRef]
- Warraich, U.E.; Hussain, F.; Kayani, H.U.R. Aging—Oxidative stress, antioxidants and computational modeling. Heliyon 2020, 6, e04107. [Google Scholar] [CrossRef] [PubMed]
- Polidori, M.C.; Mecocci, P. Modeling the dynamics of energy imbalance: The free radical theory of aging and frailty revisited. Free Radic. Biol. Med. 2022, 181, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Viña, J.; Borras, C.; Gomez-Cabrera, M.C. A free radical theory of frailty. Free Radic. Biol. Med. 2018, 124, 358–363. [Google Scholar] [CrossRef]
- Arauna, D.; Furrianca, M.; Espinosa-Parrilla, Y.; Fuentes, E.; Alarcón, M.; Palomo, I. Natural Bioactive Compounds As Protectors Of Mitochondrial Dysfunction In Cardiovascular Diseases And Aging. Molecules 2019, 24, 4259. [Google Scholar] [CrossRef] [Green Version]
- Picca, A.; Guerra, F.; Calvani, R.; Bucci, C.; Lo Monaco, M.R.; Bentivoglio, A.R.; Coelho-Júnior, H.J.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial Dysfunction and Aging: Insights from the Analysis of Extracellular Vesicles. Int. J. Mol. Sci. 2019, 20, 805. [Google Scholar] [CrossRef] [Green Version]
- Kaeberlein, M. The Biology of Aging: Citizen Scientists and Their Pets as a Bridge between Research on Model Organisms and Human Subjects. Vet. Pathol. 2016, 53, 291–298. [Google Scholar] [CrossRef] [Green Version]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef]
- Weichhart, T. mTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Di Tano, M.; Mattson, M.P.; Guidi, N. Intermittent and periodic fasting, longevity and disease. Nat. Aging 2021, 1, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, A.; Kurup, K.; Salmon, A.B.; Richardson, A. Is Rapamycin a Dietary Restriction Mimetic? J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, 4–13. [Google Scholar] [CrossRef]
- Yalcin, G.; Lee, C.-K. Recent studies on anti-aging compounds with Saccharomyces cerevisiae as a model organism. Transl. Med. Aging 2019, 3, 109–115. [Google Scholar] [CrossRef]
- Masoro, E.J. Caloric restriction and aging: Controversial issues. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 14–19. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Liu, S.Q.; Huang, D. Dietary restriction depends on nutrient composition to extend chronological lifespan in budding yeast Saccharomyces cerevisiae. PLoS ONE 2013, 8, e64448. [Google Scholar] [CrossRef] [Green Version]
- Minois, N.; Lagona, F.; Frajnt, M.; Vaupel, J.W. Plasticity of death rates in stationary phase in Saccharomyces cerevisiae. Aging Cell. 2009, 8, 36–44. [Google Scholar] [CrossRef]
- Goldberg, A.A.; Bourque, S.D.; Kyryakov, P.; Gregg, C.; Boukh-Viner, T.; Beach, A.; Burstein, M.T.; Machkalyan, G.; Richard, V.; Rampersad, S.; et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp. Gerontol. 2009, 44, 555–571. [Google Scholar] [CrossRef]
- Bayliak, M.M.; Lushchak, V.I. The golden root, Rhodiola rosea, prolongs lifespan but decreases oxidative stress resistance in yeast Saccharomyces cerevisiae. Phytomedicine 2011, 18, 1262–1268. [Google Scholar] [CrossRef]
- Choi, K.M.; Hong, S.J.; van Deursen, J.M.; Kim, S.; Kim, K.H.; Lee, C.K. Caloric Restriction and Rapamycin Differentially Alter Energy Metabolism in Yeast. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 73, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Aerts, A.M.; Zabrocki, P.; Govaert, G.; Mathys, J.; Carmona-Gutierrez, D.; Madeo, F.; Winderickx, J.; Cammue, B.P.; Thevissen, K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009, 583, 113–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, D.-G.; Hong, S.; Huh, W.-K. Mitochondrial dysfunction reduces yeast replicative lifespan by elevating RAS-dependent ROS production by the ER-localized NADPH oxidase Yno1. PLoS ONE 2018, 13, e0198619. [Google Scholar] [CrossRef] [Green Version]
- Barros, M.H.; Bandy, B.; Tahara, E.B.; Kowaltowski, A.J. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 49883–49888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, P.; Fovez, Q.; Germain, N.; Khamari, R.; Kluza, J. Mitochondrial spare respiratory capacity: Mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J. 2020, 34, 13106–13124. [Google Scholar] [CrossRef] [PubMed]
- Tungmunnithum, D.; Drouet, S.; Hano, C. Flavonoids from Sacred Lotus Stamen Extract Slows Chronological Aging in Yeast Model by Reducing Oxidative Stress and Maintaining Cellular Metabolism. Cells 2022, 11, 599. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, I.; Stamerra, G.; Vai, M. Altered Expression of Mitochondrial NAD+ Carriers Influences Yeast Chronological Lifespan by Modulating Cytosolic and Mitochondrial Metabolism. Front. Genet. 2018, 9, 676. [Google Scholar] [CrossRef] [Green Version]
- Bonawitz, N.D.; Chatenay-Lapointe, M.; Pan, Y.; Shadel, G.S. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007, 5, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.M.; Weindruch, R. Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol. Metab. 2010, 21, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Hanson, R.W.; Berger, N.A.; Trubitsyn, A. Reprogramming of energy metabolism as a driver of aging. Oncotarget 2016, 7, 15410–15420. [Google Scholar] [CrossRef] [Green Version]
- Akbari, M.; Kirkwood, T.B.L.; Bohr, V.A. Mitochondria in the signaling pathways that control longevity and health span. Ageing Res. Rev. 2019, 54, 100940. [Google Scholar] [CrossRef]
- Kwon, Y.Y.; Lee, S.K.; Lee, C.K. Caloric Restriction-Induced Extension of Chronological Lifespan Requires Intact Respiration in Budding Yeast. Mol. Cells. 2017, 40, 307–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofer, S.J.; Davinelli, S.; Bergmann, M.; Scapagnini, G.; Madeo, F. Caloric Restriction Mimetics in Nutrition and Clinical Trials. Front. Nutr. 2021, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Carmona-Gutierrez, D.; Hofer, S.J.; Kroemer, G. Caloric Restriction Mimetics against Age-Associated Disease: Targets, Mechanisms, and Therapeutic Potential. Cell Metab. 2019, 29, 592–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocampo, A.; Liu, J.; Schroeder, E.A.; Shadel, G.S.; Barrientos, A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012, 16, 55–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunthonkun, P.; Palajai, R.; Somboon, P.; Suan, C.L.; Ungsurangsri, M.; Soontorngun, N. Life-span extension by pigmented rice bran in the model yeast Saccharomyces cerevisiae. Sci. Rep. 2019, 9, 18061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavoie, H.; Whiteway, M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell 2008, 7, 1127–1135. [Google Scholar] [CrossRef] [Green Version]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, e13. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, S.; Rallis, C. The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front. Cell Dev. Biol. 2017, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Maslanka, R.; Kwolek-Mirek, M.; Zadrag-Tecza, R. Consequences of calorie restriction and calorie excess for the physiological parameters of the yeast Saccharomyces cerevisiae cells. FEMS Yeast Res. 2017, 17, fox087. [Google Scholar] [CrossRef]
- Maslanka, R.; Zadrag-Tecza, R.; Kwolek-Mirek, M. Linkage between Carbon Metabolism, Redox Status and Cellular Physiology in the Yeast Saccharomyces cerevisiae Devoid of SOD1 or SOD2 Gene. Genes 2020, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Okamoto, K. The TORC1 signaling pathway regulates respiration-induced mitophagy in yeast. Biochem. Biophys. Res. Commun. 2018, 502, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Molinet, J.; Salinas, F.; Guillamón, J.M.; Martínez, C. GTR1 Affects Nitrogen Consumption and TORC1 Activity in Saccharomyces cerevisiae Under Fermentation Conditions. Front. Genet. 2020, 11, 519. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.P.; Zhang, Z.; Paczia, N.; Jaeger, C.; Ignac, T.; May, P.; Linster, C.L. Natural variation of chronological aging in the Saccharomyces cerevisiae species reveals diet-dependent mechanisms of life span control. NPJ Aging Mech. Dis. 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.-J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.-A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wu, M.; Li, S.; Gao, Q.; Zeng, Q. Artemisinin mimics calorie restriction to extend yeast lifespan via a dual-phase mode: A conclusion drawn from global transcriptome profiling. Sci. China Life Sci. 2015, 58, 451–465. [Google Scholar] [CrossRef]
- Capó, X.; Martorell, M.; Ferrer, M.D.; Sureda, A.; Pons, V.; Domingo, J.C.; Drobnic, F.; Martínez-Rodríguez, A.; Leyva-Vela, B.; Sarabia, J.M.; et al. Calorie Restriction Improves Physical Performance and Modulates the Antioxidant and Inflammatory Responses to Acute Exercise. Nutrients 2020, 12, 930. [Google Scholar] [CrossRef] [Green Version]
- Perrone, G.G.; Tan, S.-X.; Dawes, I.W. Reactive oxygen species and yeast apoptosis. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008, 1783, 1354–1368. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.M.; De Marco, M.; Barnes, K.; Shaw, P.J.; Ferraiuolo, L.; Blackburn, D.J.; Mortiboys, H.; Venneri, A. Deficits in Mitochondrial Spare Respiratory Capacity Contribute to the Neuropsychological Changes of Alzheimer’s Disease. J. Pers. Med. 2020, 10, 32. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, Y.; Paxman, J.; O’Laughlin, R.; Klepin, S.; Zhu, Y.; Pillus, L.; Tsimring, L.S.; Hasty, J.; Hao, N. A programmable fate decision landscape underlies single-cell aging in yeast. Science 2020, 369, 325–329. [Google Scholar] [CrossRef]
- Xiberras, J.; Klein, M.; Nevoigt, E. Glycerol as a substrate for Saccharomyces cerevisiae based bioprocesses—Knowledge gaps regarding the central carbon catabolism of this ‘non-fermentable’ carbon source. Biotechnol. Adv. 2019, 37, 107378. [Google Scholar] [CrossRef] [PubMed]



| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | UPL |
|---|---|---|---|
| CCS1 | TGAACCACCCAGAAAACGA | GCAATGACTCCCAGAAATGAG | 69 |
| ECL1 | AATAAGTTATATTGCTCCGAAGATTGT | TTTTATGCAAGTGGGATAATAATTTCT | 161 |
| FOB1 | CGAAACGTAAACCTGTGCAA | CTCTCTTAGTCTCAAACTTGGCATT | 85 |
| GTR1 | AAGATGGATCTTGTTCAGTTGGA | CAGGTTTTTCATCATGATTTGG | 17 |
| HAP4 | TCAGGATGAAAGCGCTGATT | AGCCAAAGTGATTTCTGAACCT | 9 |
| RIM15 | TTTCCCTCCTTGATATTTCTCG | TCGAATTTGTGGGATTTGCT | 63 |
| SCH9 | TTGGGATCTCACTCGGAATTA | GGATACAGTCTAACTTGGCCTAAGAA | 44 |
| SOD2 | AAACCACTGTCTATTCTGGGAAA | CTCGTCGATTGCCTTTGC | 36 |
| ACT1 * | TTCCAGATGGTCAAGTCATCA | AAACAGAAGGATGGAACAAAGC | 45 |
| ALG9 * | CAAGAGCATGCTTAGGCTTTTT | CCCGGATTAAACAATTGGAA | 27 |
| TAF10 * | GAGGAGATTCTAGAGATGATGGACA | AGTCTATTACTGCATCGGGAATG | 63 |
| TFC1 * | CTCAAATGCCATAGAGGAGGA | TGGCGCCATTATCATCAAA | 9 |
| UBC6 * | AGGCTCACAAGAGATTGACGA | GGGCGAGCAAGAATATATGG | 31 |
| Day of CLS | FC (mM) | |||
|---|---|---|---|---|
| Ctrl | 0.25 | 0.5 | 1.0 | |
| 3 | 99.4 ± 0.001 | 99.6 ± 0.007 | 99.7 ± 0.002 | 100.0 ± 0.000 |
| 5 | 98.1 ± 1.285 | 98.8 ± 0.957 | 98.4 ± 1.603 | 98.8 ± 1.341 |
| 7 | 93.9 ± 3.893 | 96.3 ± 2.813 | 95.9 ± 3.553 | 98.3 ± 1.192 |
| 10 | 92.1 ± 2.616 | 92.1 ± 2.616 | 93.3 ± 2.951 | 96.2 ± 1.259 |
| 12 | 81.9 ± 6.879 | 84.6 ± 9.620 | 88.0 ± 8.443 | 91.5 ± 3.888 * |
| 14 | 74.1 ± 1.401 | 81.7 ± 6.798 | 84.5 ± 7.386 * | 90.5 ± 1.050 **** |
| 17 | 62.6 ± 6.130 | 73.5 ± 5.770 ** | 80.8 ± 6.503 **** | 88.8 ± 0.656 **** |
| 19 | 53.1 ± 14.623 | 70.6 ± 2.512 **** | 73.2 ± 4.595 **** | 81.1 ± 0.403 **** |
| Cellular Significance | Gene | Changes in Expression Level | Function | |
|---|---|---|---|---|
| Antioxidant defense | CCS1 | Copper chaperone for SOD1 | ↓↓ | Involved in oxidative stress protection; increase in protein abundance in response to DNA replication stress [32] |
| SOD2 | Superoxide dismutase | No change | Mitochondrial manganese superoxide dismutase; protects cells against oxygen toxicity and oxidative stress [32] | |
| Nutrient sensing and TORC1 signaling pathway | GTR1 | GTP-binding protein resemblance | ↓↓↓ | GTPase that activates TORC1 in response to amino acid stimulation [33] |
| RIM15 | Regulator of IME2 | ↑↑ | Protein kinase involved in cell proliferation in response to nutrients; plays a crucial role in the entry of cells into the stationary phase upon nutrient starvation conditions [34,35,36] | |
| SCH9 | Serine/threonine protein kinase | ↓↓↓ | Direct downstream protein kinase of TORC1 [34,35,36] | |
| Replication process | FOB1 | Fork blocking less | ↓↓ | Required for replication fork blocking; involved in ERCs generation [37] |
| Respiratory activity | HAP4 | Heme activator protein | ↑ | Transcriptional activator and global regulator of respiratory gene expression; plays a central role in shifting cells from fermentative to respiratory growth [38,39] |
| Unknown function | ECL1 | Extender of the chronological lifespan | No change | Mitochondrial-dependent role in the extension of CLS; unknown exact molecular function [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawelczak, P.; Fedoruk-Wyszomirska, A.; Wyszko, E. Antiaging Effect of 4-N-Furfurylcytosine in Yeast Model Manifests through Enhancement of Mitochondrial Activity and ROS Reduction. Antioxidants 2022, 11, 850. https://doi.org/10.3390/antiox11050850
Pawelczak P, Fedoruk-Wyszomirska A, Wyszko E. Antiaging Effect of 4-N-Furfurylcytosine in Yeast Model Manifests through Enhancement of Mitochondrial Activity and ROS Reduction. Antioxidants. 2022; 11(5):850. https://doi.org/10.3390/antiox11050850
Chicago/Turabian StylePawelczak, Paweł, Agnieszka Fedoruk-Wyszomirska, and Eliza Wyszko. 2022. "Antiaging Effect of 4-N-Furfurylcytosine in Yeast Model Manifests through Enhancement of Mitochondrial Activity and ROS Reduction" Antioxidants 11, no. 5: 850. https://doi.org/10.3390/antiox11050850







