Platinum Nanoparticles: The Potential Antioxidant in the Human Lung Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Platinum Nanoparticles and Hydrogen Peroxide
2.2. Treatment Procedure
2.3. Cell Culture
2.4. Ferric Reducing Antioxidant Power (FRAP) Assay
2.5. MTS Assay
2.6. Nanoparticles Uptake
2.7. Comet Assay
2.8. Reactive Oxygen Species (ROS) Production
2.9. Antioxidant Enzymes
2.10. Statistical Analysis
3. Results
3.1. Antioxidant Capacity of PtNPs was Lower Than Ascorbic Acid
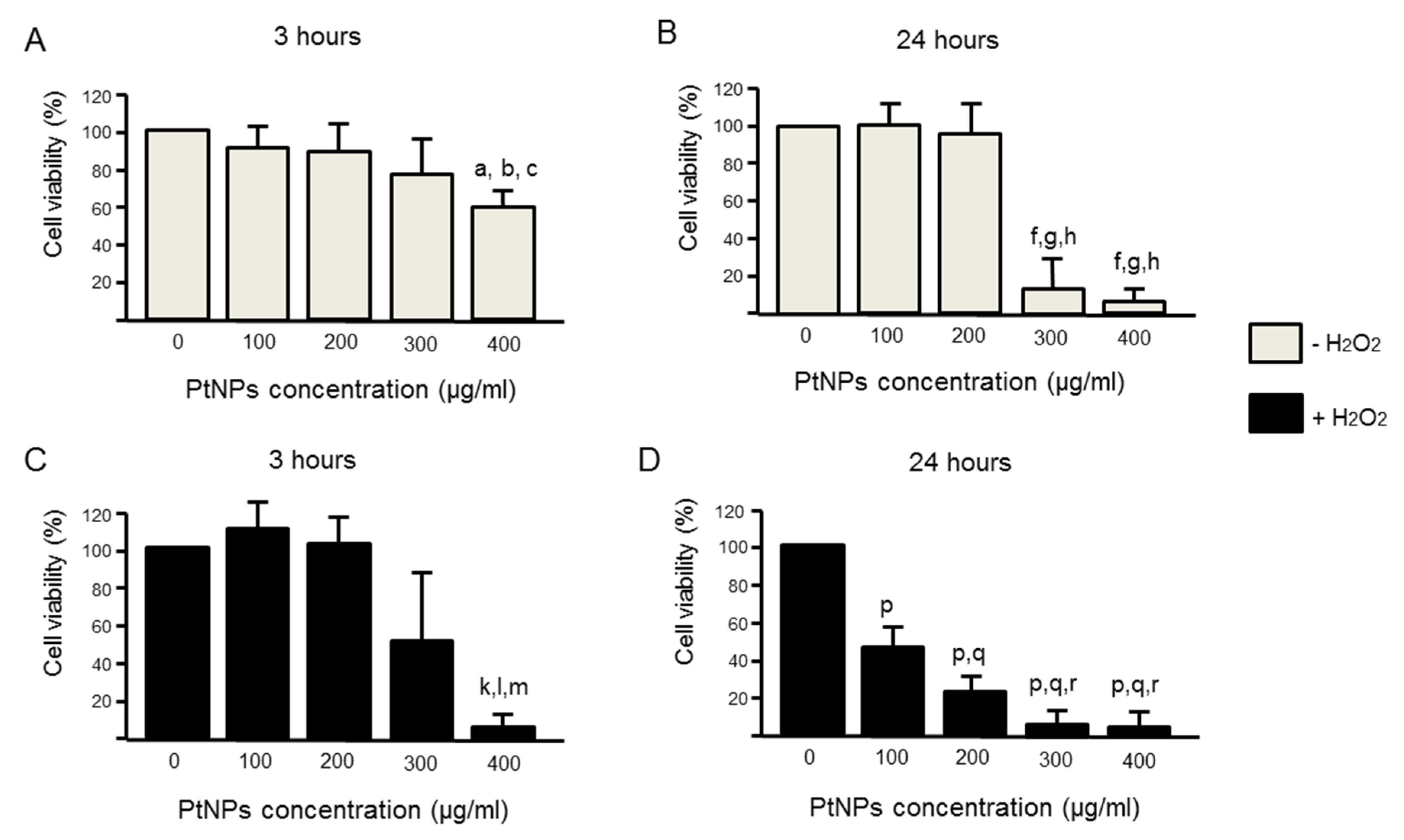
3.2. Cell Viability Was Better in Acute PtNPs
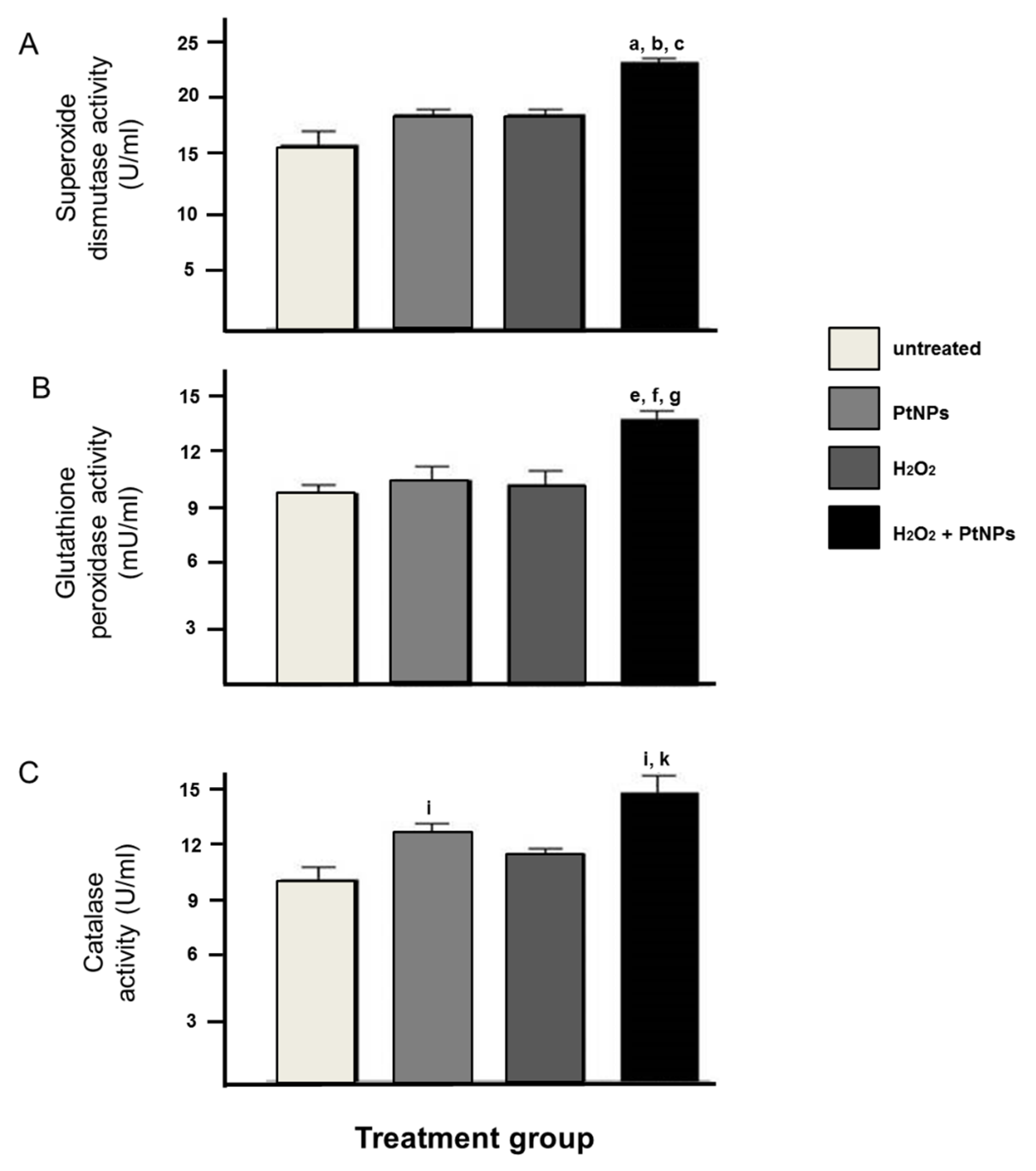
3.3. The Level of Antioxidant Enzymes was Elevated in the Presence of PtNPs
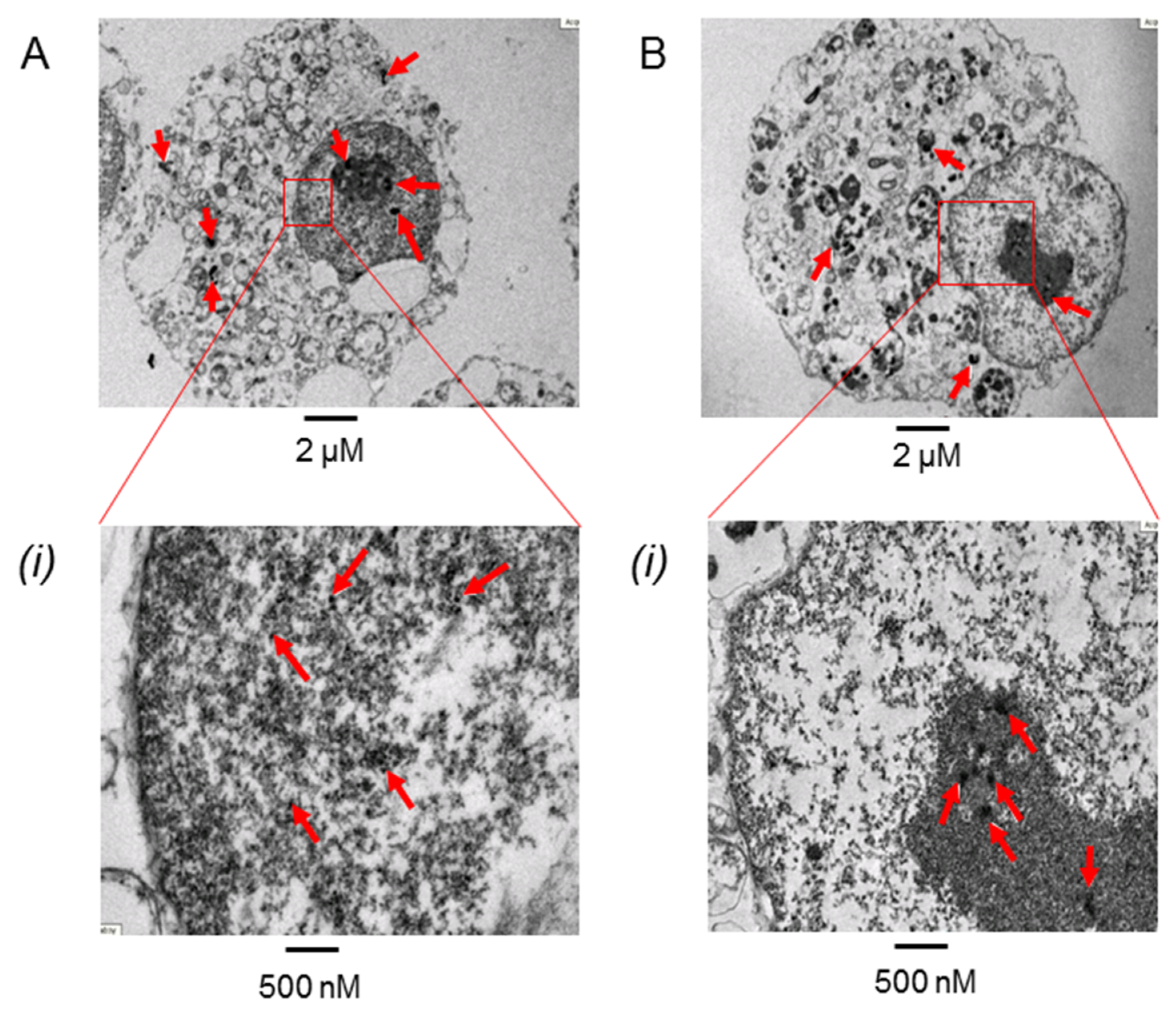
3.4. PtNPs Uptake was Seen in the Nucleus
3.5. PtNPs Can Reduce Oxidative Condition in H2O2 Treated Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Humphries, K.M.; Szweda, P.A.; Szweda, L.I. Aging: A shift from redox regulation to oxidative damage. Free Radic. Res. 2006, 40, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yotnda, P. Production and detection of reactive oxygen species (ROS) in cancers. J. Vis. Exp. 2011, 57, e3357. [Google Scholar] [CrossRef]
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- York-Duran, M.J.; Godoy-Gallardo, M.; Jansman, M.M.T.; Hosta-Rigau, L. A dual-component carrier with both non-enzymatic and enzymatic antioxidant activity towards ROS depletion. Biomater. Sci. 2019, 7, 4813–4826. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef]
- Xu, D.-P.; Li, Y.; Meng, X.; Zhou, T.; Zhou, Y.; Zheng, J.; Zhang, J.-J.; Li, H.-B. Natural Antioxidants in Foods and Medicinal Plants: Extraction, Assessment and Resources. Int. J. Mol. Sci. 2017, 18, 96. [Google Scholar] [CrossRef]
- Harris, I.S.; DeNicola, G.M. The Complex Interplay between Antioxidants and ROS in Cancer. Trends Cell Biol. 2020, 30, 440–451. [Google Scholar] [CrossRef]
- El-Sheikh, A.; Khired, Z. Interactions of Analgesics with Cisplatin: Modulation of Anticancer Efficacy and Potential Organ Toxicity. Medicina 2022, 58, 46. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Shibuya, S.; Ozawa, Y.; Watanabe, K.; Izuo, N.; Toda, T.; Yokote, K.; Shimizu, T. Palladium and platinum nanoparticles attenuate aging-like skin atrophy via antioxidant activity in mice. PLoS ONE 2014, 9, e109288. [Google Scholar] [CrossRef] [PubMed]
- Onizawa, S.; Aoshiba, K.; Kajita, M.; Miyamoto, Y.; Nagai, A. Platinum nanoparticle antioxidants inhibit pulmonary inflammation in mice exposed to cigarette smoke. Pulm. Pharmacol. Ther. 2009, 22, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Zimmermann, S.; Krug, H.F.; Sures, B. Influence of platinum, palladium and rhodium as compared with cadmium, nickel and chromium on cell viability and oxidative stress in human bronchial epithelial cells. Environ. Int. 2007, 33, 385–390. [Google Scholar] [CrossRef]
- Shakibaie, M.; Torabi-Shamsabad, R.; Forootanfar, H.; Amiri-Moghadam, P.; Amirheidari, B.; Adeli-Sardou, M.; Ameri, A. Rapid microwave-assisted biosynthesis of platinum nanoparticles and evaluation of their antioxidant properties and cytotoxic effects against MCF-7 and A549 cell lines. 3 Biotech 2021, 11, 511. [Google Scholar] [CrossRef]
- Kajita, M.; Hikosaka, K.; Iitsuka, M.; Kanayama, A.; Toshima, N.; Miyamoto, Y. Platinum nanoparticle is a useful scavenger of superoxide anion and hydrogen peroxide. Free Radic. Res. 2007, 41, 615–626. [Google Scholar] [CrossRef]
- Hikosaka, K.; Kim, J.; Kajita, M.; Kanayama, A.; Miyamoto, Y. Platinum nanoparticles have an activity similar to mitochondrial NADH:ubiquinone oxidoreductase. Colloids Surf. B Biointerfaces 2008, 66, 195–200. [Google Scholar] [CrossRef]
- Alshatwi, A.A.; Athinarayanan, J.; Vaiyapuri Subbarayan, P. Green synthesis of platinum nanoparticles that induce cell death and G2/M-phase cell cycle arrest in human cervical cancer cells. J. Mater. Sci. Mater. Med. 2015, 26, 5330. [Google Scholar] [CrossRef]
- Hullo, M.; Grall, R.; Perrot, Y.; Mathé, C.; Ménard, V.; Yang, X.; Lacombe, S.; Porcel, E.; Villagrasa, C.; Chevillard, S.; et al. Radiation Enhancer Effect of Platinum Nanoparticles in Breast Cancer Cell Lines: In Vitro and In Silico Analyses. Int. J. Mol. Sci. 2021, 22, 4436. [Google Scholar] [CrossRef]
- Pelka, J.; Gehrke, H.; Esselen, M.; Türk, M.; Crone, M.; Bräse, S.; Muller, T.; Blank, H.; Send, W.; Zibat, V.; et al. Cellular uptake of platinum nanoparticles in human colon carcinoma cells and their impact on cellular redox systems and DNA integrity. Chem. Res. Toxicol. 2009, 22, 649–659. [Google Scholar] [CrossRef]
- Gehrke, H.; Pelka, J.; Hartinger, C.G.; Blank, H.; Bleimund, F.; Schneider, R.; Gerthsen, D.; Bräse, S.; Crone, M.; Türk, M.; et al. Platinum nanoparticles and their cellular uptake and DNA platination at non-cytotoxic concentrations. Arch. Toxicol. 2011, 85, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, T.; Kashiwagi, T.; Imada, T.; Nakamichi, N.; Aramaki, S.; Toh, K.; Morisawa, S.; Shimakoshi, H.; Hisaeda, Y.; Shirahata, S. Kinetic analysis of superoxide anion radical-scavenging and hydroxyl radical-scavenging activities of platinum nanoparticles. Langmuir 2008, 24, 7354–7364. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.F.F.; Strain, J.J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell viability assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2016. [Google Scholar]
- Graham, L.; Orenstein, J.M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007, 2, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef]
- Wojtala, A.; Bonora, M.; Malinska, D.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Chapter Thirteen—Methods to Monitor ROS Production by Fluorescence Microscopy and Fluorometry. In Methods in Enzymology; Galluzzi, L., Kroemer, G., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 542, pp. 243–262. [Google Scholar]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Elder, A.; Yang, H.; Gwiazda, R.; Teng, X.; Thurston, S.; He, H.; Oberdörster, G. Testing nanomaterials of unknown toxicity: An example based on platinum nanoparticles of different shapes. Adv. Mater. 2007, 19, 3124–3129. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, H.; Lee, Y.S.; Jung, D.Y. Enhanced catalytic activity of platinum nanoparticles by exfoliated metal hydroxide nanosheets. ChemCatChem 2014, 6, 113–118. [Google Scholar] [CrossRef]
- Yoshihisa, Y.; Zhao, Q.L.; Hassan, M.A.; Wei, Z.L.; Furuichi, M.; Miyamoto, Y.; Kondo, T.; Shimizu, T. SOD/catalase mimetic platinum nanoparticles inhibit heat-induced apoptosis in human lymphoma U937 and HH cells. Free Radic. Res. 2011, 45, 326–335. [Google Scholar] [CrossRef]
- Nakanishi, H.; Hamasaki, T.; Kinjo, T.; Yan, H.; Nakamichi, N.; Kabayama, S.; Teruya, K.; Shirahata, S. Low Concentration Platinum Nanoparticles Effectively Scavenge Reactive Oxygen Species in Rat Skeletal L6 Cells. Nano Biomed. Eng. 2013, 5, 76–85. [Google Scholar] [CrossRef][Green Version]
- Wen, T.; Yang, A.; Piao, L.; Hao, S.; Du, L.; Meng, J.; Liu, J.; Xu, H. Comparative study of in vitro effects of different nanoparticles at non-cytotoxic concentration on the adherens junction of human vascular endothelial cells. Int. J. Nanomed. 2019, 14, 4475. [Google Scholar] [CrossRef]
- Gurunathan, S.; Jeyaraj, M.; La, H.; Yoo, H.; Choi, Y.; Do, J.T.; Park, C.; Kim, J.-H.; Hong, K. Anisotropic Platinum Nanoparticle-Induced Cytotoxicity, Apoptosis, Inflammatory Response, and Transcriptomic and Molecular Pathways in Human Acute Monocytic Leukemia Cells. Int. J. Mol. Sci. 2020, 21, 440. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Xu, J.; Luo, Q.; Hou, C.; Liu, J. Rational Design and Biological Application of Antioxidant Nanozymes. Front. Chem. 2020, 8, 831. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, P.; Dzięgielewski, K.; Drozd, M.; Skorupska, S.; Grabowska-Jadach, I.; Pietrzak, M. Nanoparticles of chosen noble metals as reactive oxygen species scavengers. Nanotechnology 2021, 32, 055704. [Google Scholar] [CrossRef]
- Zheng, W.; Jiang, B.; Hao, Y.; Zhao, Y.; Zhang, W.; Jiang, X. Screening reactive oxygen species scavenging properties of platinum nanoparticles on a microfluidic chip. Biofabrication 2014, 6, 045004. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; Lima, I.; Guidelli, É.J.; Baffa Filho, O. Antioxidative activity of gold and platinum nanoparticles assessed through electron spin resonance. Eclética Química 2021, 46, 68–74. [Google Scholar] [CrossRef]
- Bloch, K.; Pardesi, K.; Satriano, C.; Ghosh, S. Bacteriogenic Platinum Nanoparticles for Application in Nanomedicine. Front. Chem. 2021, 9, 624344. [Google Scholar] [CrossRef]
- Jan, H.; Gul, R.; Andleeb, A.; Ullah, S.; Shah, M.; Khanum, M.; Ullah, I.; Hano, C.; Abbasi, B.H. A detailed review on biosynthesis of platinum nanoparticles (PtNPs), their potential antimicrobial and biomedical applications. J. Saudi Chem. Soc. 2021, 25, 101297. [Google Scholar] [CrossRef]
- Jeyaraj, M.; Gurunathan, S.; Qasim, M.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on the Synthesis, Characterization, and Biomedical Application of Platinum Nanoparticles. Nanomaterials 2019, 9, 1719. [Google Scholar] [CrossRef]
- Abed, A.; Derakhshan, M.; Karimi, M.; Shirazinia, M.; Mahjoubin-Tehran, M.; Homayonfal, M.; Hamblin, M.R.; Mirzaei, S.A.; Soleimanpour, H.; Dehghani, S.; et al. Platinum Nanoparticles in Biomedicine: Preparation, Anti-Cancer Activity, and Drug Delivery Vehicles. Front. Pharmacol. 2022, 13, 797804. [Google Scholar] [CrossRef]
- Horie, M.; Kato, H.; Endoh, S.; Fujita, K.; Nishio, K.; Komaba, L.K.; Fukui, H.; Nakamura, A.; Miyauchi, A.; Nakazato, T.; et al. Evaluation of cellular influences of platinum nanoparticles by stable medium dispersion. Metallomics 2011, 3, 1244–1252. [Google Scholar] [CrossRef]
- Torrano, A.A.; Herrmann, R.; Strobel, C.; Rennhak, M.; Engelke, H.; Reller, A.; Hilger, I.; Wixforth, A.; Bräuchle, C. Cell membrane penetration and mitochondrial targeting by platinum-decorated ceria nanoparticles. Nanoscale 2016, 8, 13352–13367. [Google Scholar] [CrossRef] [PubMed]
- Allouni, Z.E.; Cimpan, M.R.; Høl, P.J.; Skodvin, T.; Gjerdet, N.R. Agglomeration and sedimentation of TiO2 nanoparticles in cell culture medium. Colloids Surf. B Biointerfaces 2009, 68, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Bendale, Y.; Bendale, V.; Natu, R.; Paul, S. Biosynthesized Platinum Nanoparticles Inhibit the Proliferation of Human Lung-Cancer Cells in vitro and Delay the Growth of a Human Lung-Tumor Xenograft in vivo: -In vitro and in vivo Anticancer Activity of bio-Pt NPs. J. Pharmacopunct. 2016, 19, 114–121. [Google Scholar] [CrossRef]
- Javed, R.; Zia, M.; Naz, S.; Aisida, S.O.; Ain, N.u.; Ao, Q. Role of capping agents in the application of nanoparticles in biomedicine and environmental remediation: Recent trends and future prospects. J. Nanobiotechnol. 2020, 18, 172. [Google Scholar] [CrossRef]
- Ullah, S.; Ahmad, A.; Wang, A.; Raza, M.; Jan, A.U.; Tahir, K.; Rahman, A.U.; Qipeng, Y. Bio-fabrication of catalytic platinum nanoparticles and their in vitro efficac.cy against lungs cancer cells line (A549). J. Photochem. Photobiol. B 2017, 173, 368–375. [Google Scholar] [CrossRef]
- Zhao, N.; Xin, H.; Zhang, L. Advanced Biomedical Applications of Reactive Oxygen Species-Based Nanomaterials in Lung Cancer. Front. Chem. 2021, 9, 649772. [Google Scholar] [CrossRef]
- Pawar, A.A.; Sahoo, J.; Verma, A.; Lodh, A.; Lakkakula, J. Usage of Platinum Nanoparticles for Anticancer Therapy over Last Decade: A Review. Part. Part. Syst. Charact. 2021, 38, 2100115. [Google Scholar] [CrossRef]
- Akbari, E.; Mousazadeh, H.; Hanifehpour, Y.; Mostafavi, E.; Gorabi, A.M.; Nejati, K.; Keyhanvar, P.; Pazoki-Toroudi, H.; Mohammadhosseini, M.; Akbarzadeh, A. Co-Loading of Cisplatin and Methotrexate in Nanoparticle-Based PCL-PEG System Enhances Lung Cancer Chemotherapy Effects. J. Clust. Sci. 2019, 4, 209–219. [Google Scholar] [CrossRef]
- Esim, O.; Bakirhan, N.K.; Yildirim, N.; Sarper, M.; Savaser, A.; Ozkan, S.A.; Ozkan, Y. Development, optimization and in vitro evaluation of oxaliplatin loaded nanoparticles in non-small cell lung cancer. Daru 2020, 28, 673–684. [Google Scholar] [CrossRef]
- Guo, X.; Zhuang, Q.; Ji, T.; Zhang, Y.; Li, C.; Wang, Y.; Li, H.; Jia, H.; Liu, Y.; Du, L. Multi-functionalized chitosan nanoparticles for enhanced chemotherapy in lung cancer. Carbohydr. Polym. 2018, 195, 311–320. [Google Scholar] [CrossRef]
- Fahmy, S.A.; Preis, E.; Bakowsky, U.; Azzazy, H.M.E.-S. Platinum Nanoparticles: Green Synthesis and Biomedical Applications. Molecules 2020, 25, 4981. [Google Scholar] [CrossRef] [PubMed]
- Hosny, M.; Fawzy, M.; El-Fakharany, E.M.; Omer, A.M.; El-Monaem, E.M.A.; Khalifa, R.E.; Eltaweil, A.S. Biogenic synthesis, characterization, antimicrobial, antioxidant, antidiabetic, and catalytic applications of platinum nanoparticles synthesized from Polygonum salicifolium leaves. J. Environ. Chem. Eng. 2022, 10, 106806. [Google Scholar] [CrossRef]
- Eltaweil, A.S.; Fawzy, M.; Hosny, M.; Abd El-Monaem, E.M.; Tamer, T.M.; Omer, A.M. Green synthesis of platinum nanoparticles using Atriplex halimus leaves for potential antimicrobial, antioxidant, and catalytic applications. Arab. J. Chem. 2022, 15, 103517. [Google Scholar] [CrossRef]
- Rajendran, K.; Sen, S. Effect of capping agent on antimicrobial activity of nanoparticles. Der Pharm. Lett. 2015, 7, 37–42. [Google Scholar]
- Javed, R.; Usman, M.; Tabassum, S.; Zia, M. Effect of capping agents: Structural, optical and biological properties of ZnO nanoparticles. Appl. Surf. Sci. 2016, 386, 319–326. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, N.A.S.; Lee, J.X.; Yusof, F. Platinum Nanoparticles: The Potential Antioxidant in the Human Lung Cancer Cells. Antioxidants 2022, 11, 986. https://doi.org/10.3390/antiox11050986
Ismail NAS, Lee JX, Yusof F. Platinum Nanoparticles: The Potential Antioxidant in the Human Lung Cancer Cells. Antioxidants. 2022; 11(5):986. https://doi.org/10.3390/antiox11050986
Chicago/Turabian StyleIsmail, Noor Akmal Shareela, Jun Xin Lee, and Fatimah Yusof. 2022. "Platinum Nanoparticles: The Potential Antioxidant in the Human Lung Cancer Cells" Antioxidants 11, no. 5: 986. https://doi.org/10.3390/antiox11050986
APA StyleIsmail, N. A. S., Lee, J. X., & Yusof, F. (2022). Platinum Nanoparticles: The Potential Antioxidant in the Human Lung Cancer Cells. Antioxidants, 11(5), 986. https://doi.org/10.3390/antiox11050986






