Functional Characterization of Arylalkylamine N-Acetyltransferase, a Pivotal Gene in Antioxidant Melatonin Biosynthesis from Chlamydomonas reinhardtii
Abstract
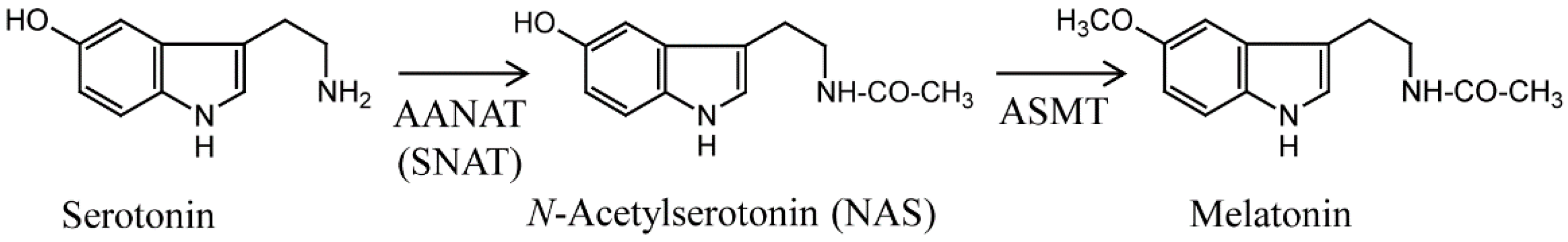
:1. Introduction
2. Materials and Methods
2.1. Synthesis of C. reinhardtii AANAT
2.2. Affinity Purification of Various Recombinant C. reinhardtii AANAT Proteins from Escherichia coli Expression
2.3. Purification of Recombinant CrAANAT Proteins
2.4. Measurement of Serotonin N-Acetyltransferase (SNAT) Enzyme Activity
2.5. Subcellular Localization of CrAANAT
2.6. Vector Construction and Production of CrAANAT-Overexpressing Transgenic Rice Plants
2.7. Plant Growth Conditions
2.8. Chemical Treatment
2.9. Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR) Analysis
2.10. Quantification of Melatonin
2.11. Statistical Analyses
3. Results
3.1. Codon-Optimized Synthesis of CrAANAT and Its Expression in Escherichia coli
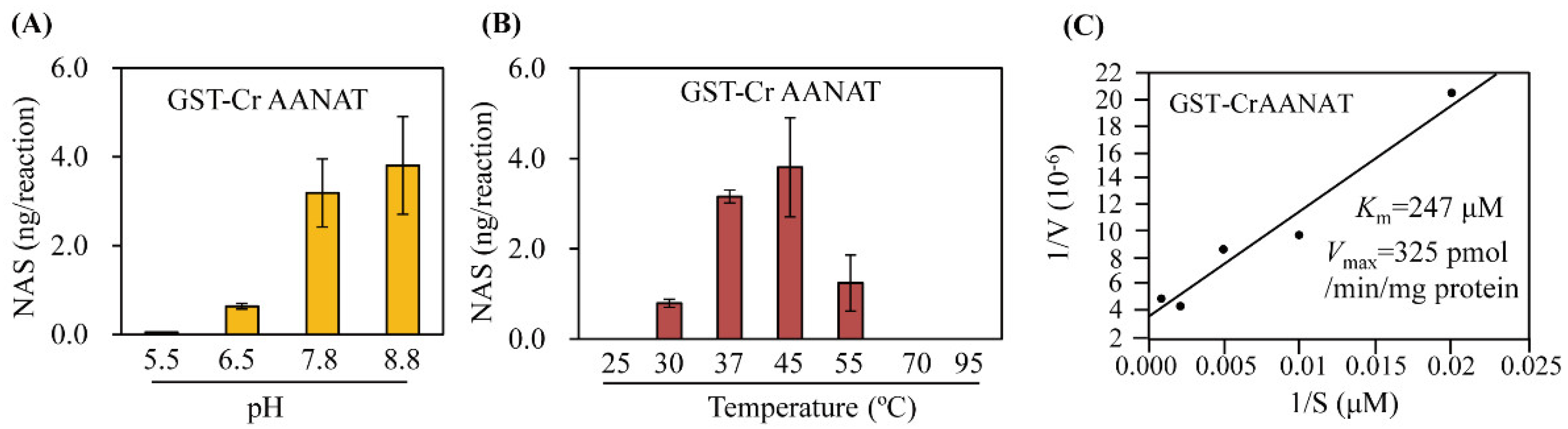
3.2. Kinetics of Recombinant CrAANAT
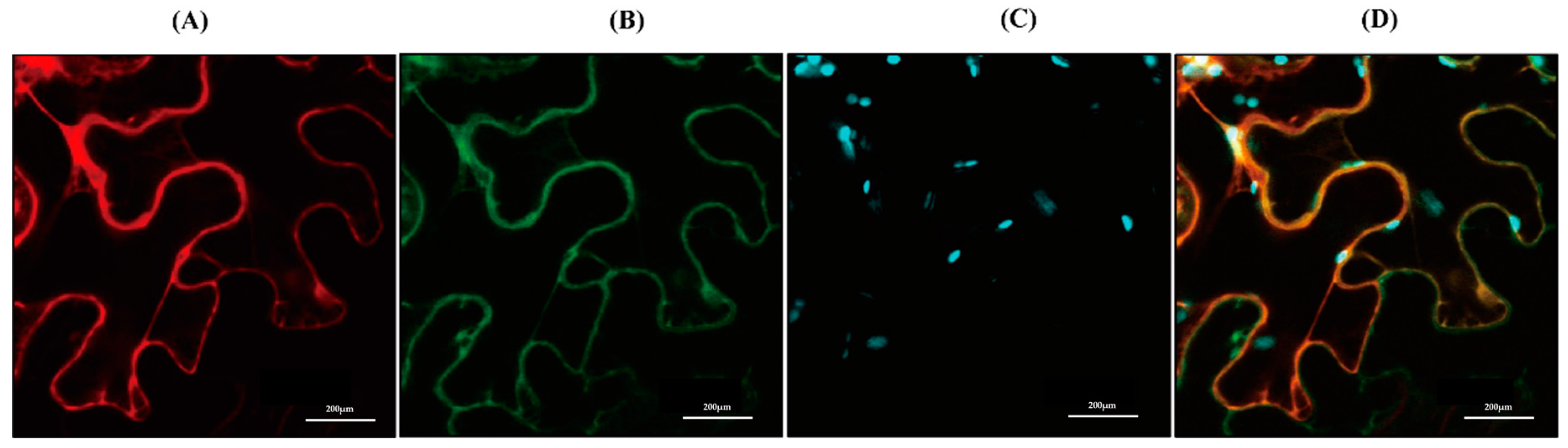
3.3. Subcellular Localization of CrAANAT
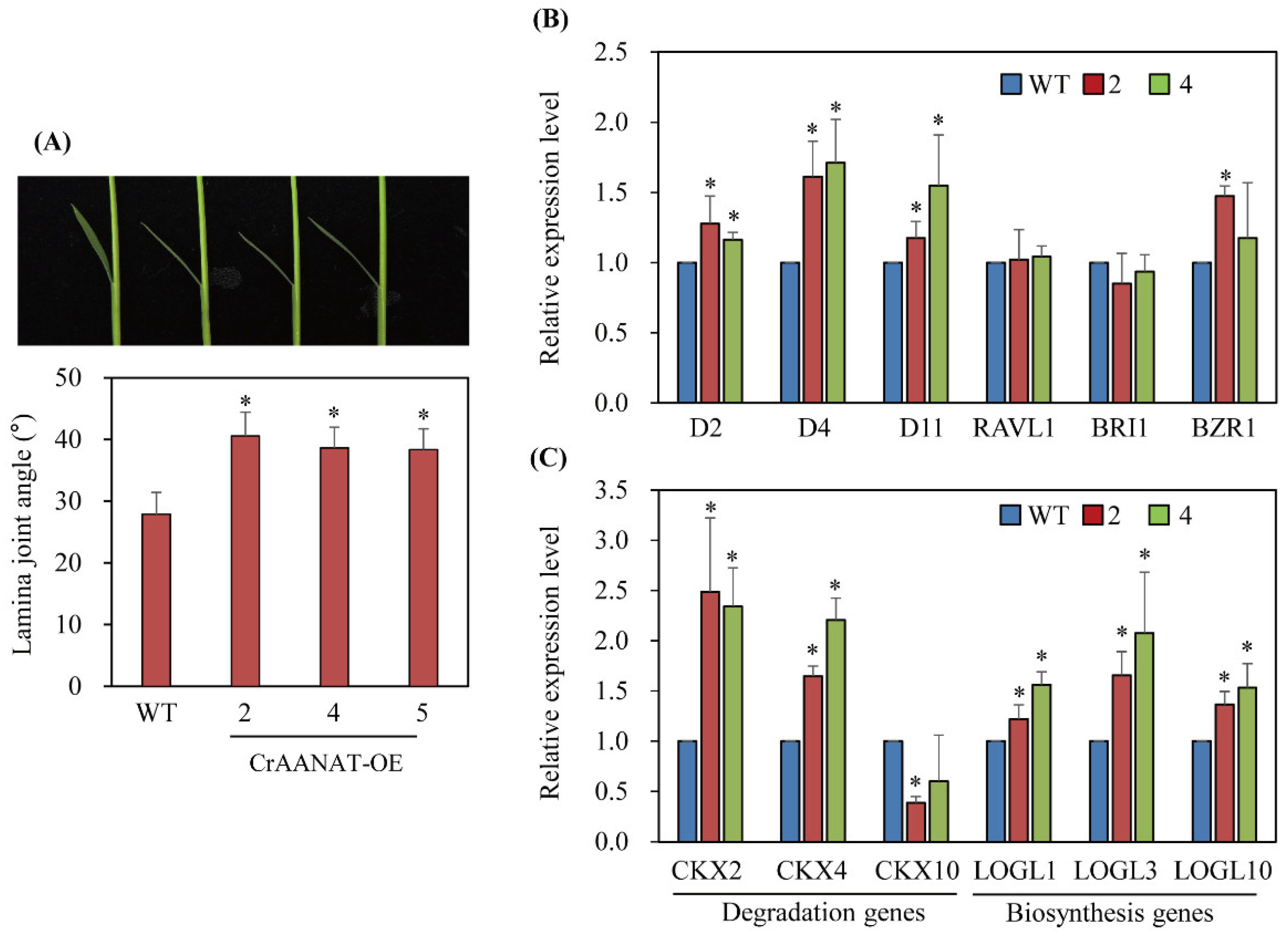
3.4. Transgenic Rice Plants Overexpressing CrAANAT (CrAANAT-OE)
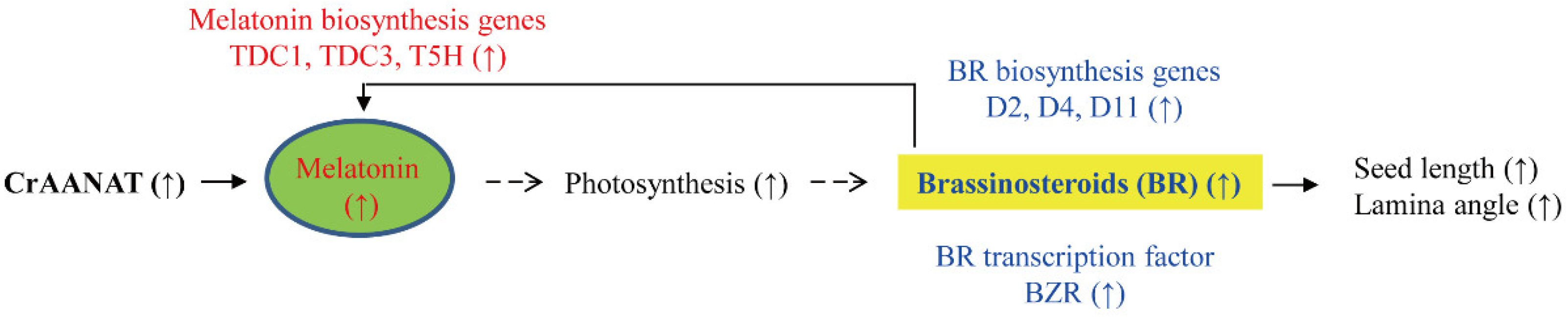
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin synthesis and function: Evolutionary history in animals and plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol. Cell. Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, L.; Zhou, Y.; Xu, Y.; Zhang, Y.; Liu, X.; Liu, B.; Chen, X.; Guo, Y.; Zeng, Z.; Zhao, Y. Structural and molecular dynamics analysis of plant serotonin N-acetyltransferase reveal an acid/base-assisted catalysis in melatonin biosynthesis. Angew. Chem Int. Ed. 2021, 60, 12020–12026. [Google Scholar] [CrossRef]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. From molecular manipulation of domestigated Chlamydomonas reinhardtii to survival in nature. eLife 2018, 7, e39233. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Voisin, P.; Namboodiri, M.A.; Klein, D.C. Arylamine N-acetyltransferase and arylalkylamine N-acetyltransferase in the mammalian pineal gland. J. Biol. Chem. 1984, 259, 10913–10918. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hwang, O.J.; Back, K. Functional characterization of tobacco (Nicotiana benthamiana) serotonin N-acetyltransferases (NbSNAT1 and NbSNAT2). Melatonin Res. 2021, 4, 507–521. [Google Scholar]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef]
- Arnao, M.B.; Cano, A.; Hernández-Ruiz, J. Phytomelatonin: An unexpected molecule with amazing performance in plants. J. Exp. Bot. 2022. [Google Scholar] [CrossRef]
- Yao, J.W.; Ma, Z.; Ma, Y.Q.; Zhu, Y.; Lei, M.Q.; Hao, C.Y.; Chen, L.Y.; Xu, Z.Q.; Huang, X. Role of melatonin in UV-B signaling pathway and UV-B stress resistance in Arabidopsis thaliana. Plant Cell Environ. 2021, 44, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Xie, Q.; Liu, Y.; Lv, H.; Bai, R.; Ma, R.; Li, X.; Zhang, X.; Guo, Y.D.; et al. SISNAT interacts with HSP40, a molecular chaperone, to regulate melatonin biosynthesis and promote thermotolerance in tomato. Plant Cell Physiol. 2020, 61, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Bian, L.; Jiao, Z.; Keke, Y.; Wan, Y.; Zhang, G.; Guo, D. Molecular cloning and characterization of a grapevine (Vitis vinifera L.) serotonin N-acetyltransferase (VvSNAT2) gene involved in plant defense. BMC Genom. 2019, 20, 880. [Google Scholar] [CrossRef]
- Zhao, G.; Yu, X.; Lou, W.; Wei, S.; Wang, R.; Wan, Q.; Shen, W. Transgenic Arabidopsis overexpressing MsSNAT enhances salt tolerance via the increase in autophagy, and the reestablishment of redox and ion homeostasis. Environ. Exp. Bot. 2019, 164, 20–28. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin induction and its role in high light stress tolerance in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12504. [Google Scholar] [CrossRef]
- Gu, Q.; Chen, Z.; Yu, X.; Cui, W.; Pan, J.; Zhao, G.; Xu, S.; Wang, R.; Shen, W. Melatonin confers plant tolerance against cadmium stress via the decrease of cadmium accumulation and reestablishment of microRNA-mediated redox homeostasis. Plant Sci. 2017, 261, 28–37. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Reiter, R.J.; He, C.; Liu, G.; Lei, Q.; Zuo, B.; Zheng, X.D.; Li, Q.; Kong, J. Change in melatonin levels in transgenic ‘Micro-Tom’ tomato over-expressing ovine AANAT and ovine HIOMT genes. J. Pineal Res. 2014, 56, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, D.E.; Jang, H.; Byeon, Y.; Kim, Y.S.; Back, K. Melatonin-rich transgenic rice plants exhibit resistance to herbicide-induced oxidative stress. J. Pineal Res. 2013, 54, 258–263. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Kim, Y.S.; Back, K. Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. J. Pineal Res. 2010, 49, 176–182. [Google Scholar] [CrossRef]
- Tan, D.X.; Manchester, L.C.; Terron, M.P.; Flores, L.J.; Reiter, R.J. One molecule, many derivatives: A never-ending interaction of melatonin with reactive oxygen and nitrogen species? J. Pineal Res. 2007, 42, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, M.; Higuchi, K.; Hahawa, Y.; Shiraiwa, Y.; Ezura, H. Cloning and characterization of a Chlamydomonas reinhardtii cDNA arylalkylamine N-acetyltransferase and its use in the genetic engineering of melatonin content in the Micro-Tom tomato. J. Pineal Res. 2009, 46, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef]
- Himmelbach, A.; Zierold, U.; Hensel, G.; Riechen, J.; Douchkov, D.; Schweizer, P.; Kumlehn, J. A set of modular binary vectors for transformation of cereals. Plant Physiol. 2007, 145, 1192–1200. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Lee, S.B.; Chung, J.S.; Han, S.U.; Han, O.; Guh, J.O.; Jeon, J.S.; An, G.; Back, K. Transgenic rice plants expressing a Bacillus subtilis protoporphyrinogen oxidase gene are resistant to diphenyl ether herbicide oxyfluorfen. Plant Cell Physiol. 2000, 41, 743–749. [Google Scholar] [CrossRef]
- Lee, K.; Choi, G.H.; Back, K. Cadmium-induced melatonin synthesis in rice requires light, hydrogen peroxide, and nitric oxide: Key regulatory roles for tryptophan decarboxylase and caffeic acid O-methyltransferase. J. Pineal Res. 2017, 63, e12441. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, K.; Back, K. Knockout of Arabidopsis serotonin N-acetyltransferase-2 reduces melatonin levels and delays flowering. Biomolecules 2019, 9, 712. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, J.; Gastel, J.; Klein, D.C.; Cole, P.A. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 1998, 273, 3045–3050. [Google Scholar] [CrossRef] [Green Version]
- Ferry, G.; Loynel, A.; Kucharczyk, N.; Bertin, S.; Rodriguez, M.; Delagrange, P.; Galizzi, J.-P.; Jacoby, E.; Volland, J.-P.; Lesieur, D.; et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem. 2000, 275, 8794–8805. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.; Lee, H.Y.; Choi, D.W.; Back, K. Chloroplast encoded serotonin N-acetyltransferase in the red alga Pyropia yezoensis: Gene transition to the nucleus from chloroplasts. J. Exp. Bot. 2015, 66, 709–717. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.; Lee, K.; Park, Y.I.; Park, S.; Back, K. Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 2013, 55, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Back, K. A rice chloroplast transit peptide sequence does not alter the cytoplasmic localization of sheep serotonin N-acetyltransferase expressed in transgenic rice plants. J. Pineal Res. 2014, 57, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Hardeland, R.; Back, K.; Manchester, L.C.; Alatorre-Jimenez, M.A.; Reiter, R.J. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine: Comparisons across species. J. Pineal Res. 2016, 61, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zhang, A.; Liu, X.; Chen, J. Grain size associated genes and the molecular regulatory mechanism in rice. Int. J. Mol. Sci. 2022, 23, 3169. [Google Scholar] [CrossRef]
- Zhang, C.; Bai, M.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef]
- Hwang, H.; Ryu, H.; Cho, H. Brassinosteroid signaling pathways interplaying with diverse signaling cues for crop enhancement. Agronomy 2022, 11, 556. [Google Scholar] [CrossRef]
- Coon, S.L.; Roseboom, P.H.; Baler, R.; Weller, J.L.; Nambroodiri, M.A.A.; Koonin, E.V.; Klein, D.C. Pineal serotonin N-acetyltransferase: Expression cloning and molecular analysis. Science 1995, 270, 1681–1683. [Google Scholar] [CrossRef]
- Borjigin, J.; Wang, M.M.; Snyder, S.H. Diurnal variation in mRNA encoding serotonin N-acetyltransferase in pineal gland. Nature 1995, 378, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Coon, S.L.; Bégay, V.; Deurloo, D.; Falcón, J.; Klein, D.C. Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. J. Biol. Chem. 1999, 274, 9076–9082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganguly, S.; Mummaneni, P.; Steinbach, P.J.; Klein, D.C.; Coon, S.L. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 2001, 276, 47239–47247. [Google Scholar] [CrossRef] [Green Version]
- Mehere, P.; Han, Q.; Christensen, B.M.; Li, J. Identification and characterization of two arylalkylamine N-acetyltransferases in the yellow fever mosquito, Ades aegypti. Insect Biochem. Mol. Biol. 2011, 41, 707–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan-Poe, X.; Craft, C.M. Biochemical characterization of recombinant serotonin N-acetyltransferase. J. Pineal Res. 1999, 27, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Wang, G.Y.; Muhammad, I.; Chi, Y.X.; Zeeshan, M.; Nasar, J.; Zhou, X.B. Interactive effects of melatonin and nitrogen improve drought tolerance of maize seedlings by regulating growth and physiological attributes. Antioxidants 2022, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Lamin-Samu, A.T.; Muhammad, I.; Farghal, M.; Khattak, A.M.; Jan, I.; Haq, S.U.; Khan, A.; Gong, Z.-H.; Lu, G. Melatonin mitigates the infection of Colletotrichum gloeosporioides via modulation of the chitinase gene and antioxidant activity in Capsicum annuum L. Antioxidants 2021, 10, 7. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Pareek, S.; Domínguez-Avila, J.A.; Gonzalez-Aguilar, G.A.; Valero, D.; Serrano, M. An exogenous pre-storage melatonin alleviates chilling injury in some mango fruit cultivars, by acting on the enzymatic and non-enzymatic antioxidant system. Antioxidants 2022, 11, 384. [Google Scholar] [CrossRef]
- Jiang, Y.; Huang, S.; Ma, L.; Kong, L.; Pan, S.; Tang, X.; Tian, H.; Duan, M.; Mo, Z. Effects of exogenous melatonin application on the grain yield and antioxidant capacity in aromatic rice under combined lead-cadmium stress. Antioxidants 2022, 11, 776. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, H.; Chen, S.; Yu, D.; Reiter, R.J. Phytomelatonin: An emerging regulator of plant biotic stress resistance. Trends Plant Sci. 2021, 26, 70–82. [Google Scholar] [CrossRef]
- Pardo-Hernández, M.; López-Delacalle, M.; Rivero, R.M. ROS and NO regulation by melatonin under abiotic stress in plants. Antioxidants 2020, 9, 1078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Y.; Ma, X.; Ouyang, Z.; Deng, L.; Shen, S.; Dong, X.; Du, N.; Dong, H.; Guo, Z.; et al. Melatonin alleviates copper toxicity via improving ROS metabolism and antioxidant defense response in tomato seedlings. Antioxidants 2022, 11, 758. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Hwang, O.J.; Back, K. Phytomelatonin as a signaling molecule for protein quality control via chaperone, autophagy, and ubiquitin–proteasome systems in plants. J. Exp. Bot. 2022. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to plant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Chen, J.; Ma, Y.; Huang, M.; Qiu, T.; Bian, H.; Han, N.; Wang, J. Function, mechanism, and application of plant melatonin: An update with a focus on the cereal crop, barley (Hordeum vulgare L.). Antioxidants 2022, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; You, J.; Li, J.; Wang, Y.; Chan, Z. Melatonin promotes Arabidopsis primary root growth in an IAA-dependent manner. J. Exp. Bot. 2021, 72, 5599–5611. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin regulates chloroplast protein quality control via a mitogen-activated protein kinase signaling pathway. Antioxidants 2021, 10, 511. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Melatonin is involved in skotomorphogenesis by regulating brassinosteroid biosynthesis in plants. J. Pineal Res. 2018, 65, e12495. [Google Scholar] [CrossRef]
- Altaf, M.A.; Shahid, R.; Ren, M.X.; Naz, S.; Altaf, M.M.; Khan, L.U.; Tiwari, R.K.; Lal, M.K.; Shahid, M.A.; Kumar, R.; et al. Melatonin improves drought tolerance of tomato by modulating plant growth, root architecture, photosynthesis, and antioxidant defense system. Antioxidants 2022, 11, 309. [Google Scholar] [CrossRef]
- Muhammad, I.; Yang, L.; Ahmad, S.; Mosaad, I.S.M.; Al-Ghamdi, A.A.; Abbasi, A.M.; Zhou, X.-B. Melatonin application alleviates stress-induced photosynthetic inhibition and oxidative damage by regulating antioxidant defense system of maize: A meta-analysis. Antioxidants 2022, 11, 512. [Google Scholar] [CrossRef]
- Huangfu, L.; Chen, R.; Lu, Y.; Zhang, E.; Miao, J.; Zuo, Z.; Zhao, Y.; Zhu, M.; Zhang, Z.; Li, P.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef] [PubMed]
- Hwang, O.J.; Back, K. Melatonin deficiency confers tolerance to multiple abiotic stresses in rice via decreased brassinosteroid levels. Int. J. Mol. Sci. 2019, 20, 5173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Choi, G.H.; Back, K. Functional charcacterization of serotonin N-acetyltransferase in archaeon Thermoplasma volcanium. Antioxidants 2022, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Hwang, O.J.; Back, K. Molecular regulation of antioxidant melatonin biosynthesis by brassinosteroid acting as an endogenous elicitor of melatonin induction in rice seedling. Antioxidant 2022, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Zheng, B. Melatonin mediated regulation of drought stress: Physiological and molecular aspects. Plants 2019, 8, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Yang, R.; Zheng, W.; Wu, L.; Zhang, C.; Zhang, H. Melatonin promotes SGT1oinvolved signals to ameliorate drought stress adaption in rice. Int. J. Mol. Sci. 2022, 23, 599. [Google Scholar] [CrossRef]
- Kuwabara, W.MT.; Gomes, P.RL.; Andrade-Silva, J.; Soares, J.M., Jr.; Amaral, F.G.; Cipolla-Neto, J. Melatonin and its ubiquitous effects on cell function and survival: A review. Melatonin Res. 2022, 5, 192–208. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, O.-J.; Back, K. Functional Characterization of Arylalkylamine N-Acetyltransferase, a Pivotal Gene in Antioxidant Melatonin Biosynthesis from Chlamydomonas reinhardtii. Antioxidants 2022, 11, 1531. https://doi.org/10.3390/antiox11081531
Hwang O-J, Back K. Functional Characterization of Arylalkylamine N-Acetyltransferase, a Pivotal Gene in Antioxidant Melatonin Biosynthesis from Chlamydomonas reinhardtii. Antioxidants. 2022; 11(8):1531. https://doi.org/10.3390/antiox11081531
Chicago/Turabian StyleHwang, Ok-Jin, and Kyoungwhan Back. 2022. "Functional Characterization of Arylalkylamine N-Acetyltransferase, a Pivotal Gene in Antioxidant Melatonin Biosynthesis from Chlamydomonas reinhardtii" Antioxidants 11, no. 8: 1531. https://doi.org/10.3390/antiox11081531






