Effect-Directed, Chemical and Taxonomic Profiling of Peppermint Proprietary Varieties and Corresponding Leaf Extracts †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
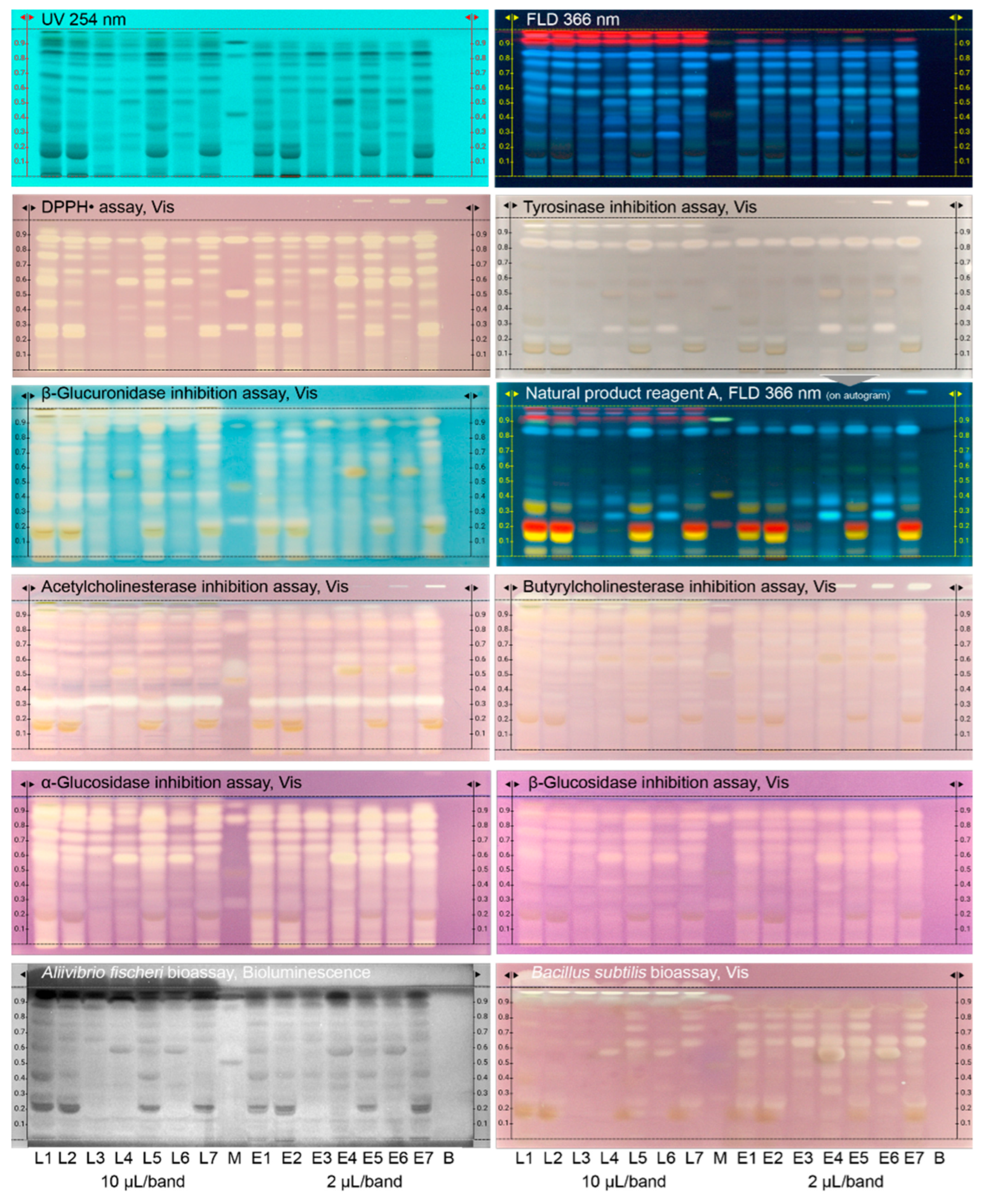
2.1.1. HPTLC–UV/Vis/FLD–EDA
2.1.2. HPLC–PDA/MS
2.1.3. Caenorhabditis elegans Strain and Maintenance
2.1.4. Genetic Assessment
2.2. Solutions Prepared
2.2.1. HPTLC–UV/Vis/FLD–EDA
2.2.2. HPLC–PDA/MS
2.3. Sample Preparation
2.3.1. HPTLC–UV/Vis/FLD–EDA
2.3.2. HPLC–PDA/MS
2.3.3. Headspace SPME–GC–FID/MS
2.3.4. Caenorhabditis elegans Method
2.3.5. Genetic Assessment
2.4. Methods
2.4.1. HPTLC–UV/Vis/FLD–EDA
- (1)
- (2)
- For the tyrosinase inhibition assay [21], 2.0 mL levodopa substrate solution (4.5 mg/mL in phosphate buffer, 20 mM, pH 6.8, plus 2.5 mg CHAPS and 7.5 mg PEG 8000), and after drying (1 min), 2.0 mL tyrosinase solution (400 U/mL in phosphate buffer, 20 mM, pH 6.8) were sprayed (blue nozzle), followed by incubation at room temperature for 15–20 min. The PC was kojic acid (1, 3 and 6 µL/band, 0.1 mg/mL in ethanol).
- (3)
- For the β-glucuronidase inhibition assay [28], 2.0 mL β-glucuronidase solution (50 U/mL in potassium phosphate buffer, pH 7.0), and after incubation at 37 °C for 15 min, 1.5 mL X-glucuronide solution (2 mg/mL in water) were sprayed (yellow, then red nozzle), and incubated at 37 °C for 1 h. The PC was D-saccharolactone (0.8, 1.5 and 3 µL/band, 0.1 mg/mL in water).
- (4)
- For the acetylcholinesterase inhibition assay [21], 3.0 mL acetylcholinesterase solution (6.66 U/mL in Tris–HCl buffer plus 1 mg bovine serum albumin), followed by incubation at 37 °C for 25 min, and then 0.5 mL substrate solution (1:2 each of 3 mg/mL 1-naphthyl acetate solution in ethanol and Fast Blue B salt solution in water) were sprayed (green nozzle). The PC was rivastigmine (2, 4 and 8 µL/band, 0.1 mg/mL in methanol).
- (5)
- For the butyrylcholinesterase inhibition assay, the same workflow was applied as in (4) but the enzyme solution was 3.34 U/mL.
- (6)
- For the α-glucosidase inhibition assay [21], 2 mL 2-naphthyl-α-D-glucopyranoside substrate solution (12 mg in 10 mL ethanol with 10% 10 mM sodium chloride solution) and after drying (2 min), 2.5 mL α-glucosidase solution (10 U/mL in sodium acetate buffer, pH 7.5) were sprayed (yellow nozzle), followed by incubation at 37 °C for 15 min. Then, 0.75 mL chromogenic reagent Fast Blue B salt solution (2 mg/mL in water) were sprayed. The PC was acarbose (1, 3 and 6 µL/band, 3 mg/mL in methanol).
- (7)
- For the β-glucosidase inhibition assay, the same workflow was applied as in (6) except using the 2-naphthyl-β-D-glucopyranoside substrate, β-glucosidase solution (1000 U/mL) and an incubation for 30 min. The PC was imidazole (2, 5 and 8 µL/band, 1 mg/mL in ethanol).
- (8)
- For the A. fischeri bioassay [20,21], 3.5 mL bacterial suspension (150 µL bacterial cryostock incubated in 20 mL medium as specified [11] at 75 rpm and room temperature for 18–24 h) were sprayed (red nozzle). Before the green–blue bacterial luminescence was visually proven to be ready for use by shaking the culture flask in a dark room. Ten images of the still humid plate were recorded over 30 min (exposure time 60 s, trigger interval 3.0 min, BioLuminizer). The PC was caffeine (0.5, 1.5 and 3 µL/band, 1 mg/mL in methanol).
- (9)
- For the B. subtilis bioassay [21], 3.5 mL bacterial suspension (100 µL bacterial cryostock in 20 mL 2.3% Müller–Hinton broth, 0.8 optical density at 600 nm) were sprayed (red nozzle), followed by incubation at 37 °C for 2 h. Then 400 µL 0.2% MTT solution in Dulbecco´s phosphate-buffered saline were sprayed (blue nozzle), followed by incubation at 37 °C for 45 min and drying at 50 °C for 5 min (TLC Plate Heater). The PC was tetracycline (0.4, 0.8 and 1.2 µL/band, 0.01 mg/mL in ethanol).
- (10)
- For the duplex planar yeast antagonist estrogen screen (HPTLCfix–pYAES–FLD) bioassay [15], the agonist 17β-estradiol solution (5 µL, 0.4 ng/µL in ethanol) was applied as stripe (0.1 mm × 80 mm, width × high, FreeMode software option) along each separated sample track, followed by drying (2 min). Afterward, the plate was horizontally immersed (manually) into a Degalan solution (0.25% in n-hexane), dried at room temperature for 8 min and sprayed (blue nozzle) with 2.5 mL Tween 20 solution (0.05% in ethanol with 10% 10 mM sodium chloride solution). Then 2.8 mL Saccharomyces cerevisiae BJ3505 suspension (1 mL cryostock incubated in 29 mL medium at 100 rpm (rotatory horizontal shaker SM-30, Edmund Bühler, Bodelshausen, Germany) and 30 °C for 16 h and adjusted to 0.8 × 108 cells/mL) were sprayed (red nozzle), followed by 3 h incubation at 30 °C. Then, 2 mL fluorescein di(β-D-galactopyranoside) substrate solution (5 mg in 1 mL dimethyl sulfoxide; thereof 25 µL in 2.5 mL phosphate buffer pH 7.0) was sprayed (yellow nozzle), followed by incubation at 37 °C for 0.5 h.
- (11)
- For the duplex planar yeast antagonist androgen screen (HPTLCfix–pYAAS–FLD) bioassay [15], the same workflow was applied as in (10) except for Saccharomyces cerevisiae BJ1991 cells, testosterone solution (2 µL, 15 ng/µL in ethanol) for stripe application, 39 mL medium and 4 h incubation.
- (12)
- For the SOS-Umu-C bioassay [16,17], the Salmonella TA1535/pSK1002 suspension, i.e., 25 µL cryostock in 35 mL medium (20 g/L lysogeny broth; thereof 3 mL added with 1 g D-glucose and 106 mg ampicillin; 37 °C, 16 h, 75 rpm) with 0.2 optical density at 660 nm, was sprayed (red nozzle), followed by incubation at 37 °C for 3 h and drying (4 min). Then 2.5 mL fluorescein di(β-D-galactopyranoside) substrate solution (5 mg in 1 mL dimethyl sulfoxide; thereof 25 µL in 2.5 mL phosphate buffer pH 7.0) was sprayed (yellow nozzle), followed by incubation at 37 °C for 0.5 h.
2.4.2. HPLC–PDA/MS
2.4.3. Headspace SPME–GC–FID/MS
2.4.4. Caenorhabditis elegans Method
2.4.5. Genetic Assessment
3. Results
3.1. Development of the Effect-Directed Profiling (HPTLC–UV/Vis/FLD–EDA)
3.2. Effect-Directed Profiles via HPTLC–UV/Vis/FLD–EDA
3.3. Results of HPLC–PDA/MS Analysis
3.4. Results of Headspace SPME–GC–FID/MS Analysis
3.5. Antioxidant Activity in Caenorhabditis elegans Assay
3.6. Enhanced Resistance of Caenorhabditis elegans to Staphylococcus aureus Pathogen Infection
3.7. Genetic Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dines, P.C.; Falé, P.L.; Madeira, P.J.A.; Florêncio, M.H.; Serralheiro, M.L. Acetylcholinesterase Inhibitory Activity After In Vitro Gastrointestinal Digestion of Infusions of Mentha Species. Eur. J. Med. Plants 2012, 3, 381–393. [Google Scholar] [CrossRef]
- Silva, H.A. Descriptive Overview of the Medical Uses Given to Mentha Aromatic Herbs throughout History. Biology 2020, 9, 484. [Google Scholar] [CrossRef]
- Tyler, V.E.; Brady, L.R.; Robbers, J.E. Pharmacognosy, 9th ed.; Lea and Fabiger: Philadelphia, PA, USA, 1988. [Google Scholar]
- Almatroodi, S.A.; Alsahli, M.A.; Almatroudi, A.; Khan, A.A.; Rahmani, A.H. Peppermint, (Mentha × piperita): Role in Management of Diseases through Modulating Various Biological Activities. Pharmacogn. J. 2021, 13, 822–827. [Google Scholar] [CrossRef]
- McKay, D.L.; Blumberg, J.B. A review of the bioactivity and potential health benefits of peppermint tea (Mentha piperita L.). Phytother. Res. 2006, 20, 619–633. [Google Scholar] [CrossRef]
- Mahendran, G.; Rahman, L.-U. Ethnomedicinal, phytochemical and pharmacological updates on Peppermint (Mentha × piperita L.)—A review. Phytother. Res. 2020, 34, 2088–2139. [Google Scholar] [CrossRef]
- Bunse, M.; Daniels, R.; Gründemann, C.; Heilmann, J.; Kammerer, D.R.; Keusgen, M.; Lindequist, U.; Melzig, M.F.; Morlock, G.; Schulz, H.; et al. Essential oils as multicomponent mixtures and their potential for human health and well-being. Front. Pharmacol. 2022, 13, 956541. [Google Scholar] [CrossRef]
- Nair, B. Final report on the safety assessment of Mentha piperita (peppermint) oil, Mentha piperita (peppermint) leaf extract, Mentha piperita (peppermint) leaf, and Mentha piperita (peppermint) leaf water. Int. J. Toxicol. 2001, 20, 61–73. [Google Scholar]
- Dhanarasu, S.; Selvam, M.; Al-Shammari, N.K.A. Evaluating the pharmacological dose (Oral LD50) and antibacterial activity of leaf extracts of Mentha piperita Linn. grown in Kingdom of Saudi Arabia: A pilot study for nephrotoxicity. Int. J. Pharmacol. 2016, 12, 195–200. [Google Scholar] [CrossRef]
- Peppermint Oil Market Snapshot (2022–2032). Available online: https://www.futuremarketinsights.com/reports/peppermint-oil-market (accessed on 25 December 2022).
- Etter, P.D.; Johnson, E. RAD paired-end sequencing for local de novo assembly and SNP discovery in non-model organisms. Methods. Mol. Biol. 2012, 888, 135–151. [Google Scholar]
- Maslo, R.; Morlock, G.; Sievers, H.; Preußker, A.; Bauer, K.; Gabhardt, M.; Pahne, N.; Schneider, S.; Schlagintweit, B.; Terlinden, S. Fundamental Paper on Plant Extracts in Foods from LChG. Eur. Food Feed Law Rev. 2022, 17, 284–299. [Google Scholar]
- McDonnell, D.P.; Nawaz, Z.; Densmore, C.; Weigel, N.L.; Pham, T.A.; Clark, J.H.; O’Malley, B.W. High level expression of biologically active estrogen receptor in Saccharomyces cerevisiae. J. Steroid Biochem. Mol. Biol. 1991, 39, 291–297. [Google Scholar] [CrossRef]
- McDonnell, D.P.; Nawaz, Z.; O’Malley, B.W. In situ distinction between steroid receptor binding and transactivation at a target gene. Mol. Cell. Biol. 1991, 11, 4350–4355. [Google Scholar]
- Ronzheimer, A.; Schreiner, T.; Morlock, G.E. Multiplex planar bioassay detecting phytoestrogens and verified antiestrogens as sharp zones on normal phase. Phytomedicine 2022, 103, 154230. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Hockamp, N.; Morlock, G.E. Non-targeted detection and differentiation of agonists versus antagonists, directly in bioprofiles of everyday products. Anal. Chim. Acta 2020, 1125, 288–298. [Google Scholar] [CrossRef]
- Schreiner, T.; Ronzheimer, A.; Friz, M.; Morlock, G.E. Multiplex planar bioassay with reduced diffusion on normal phase, identifying androgens, verified antiandrogens and synergists in botanicals via 12D hyphenation. Food Chem. 2022, 395, 133610. [Google Scholar] [CrossRef]
- Meyer, D.; Marin-Kuan, M.; Debon, E.; Serrant, P.; Cottet-Fontannaz, C.; Schilter, B.; Morlock, G.E. Detection of low levels of genotoxic compounds in food contact materials using an alternative HPTLC-SOS-Umu-C Assay. ALTEX—Altern. Anim. Exp. 2021, 38, 387–397. [Google Scholar] [CrossRef]
- Debon, E.; Rogeboz, P.; Latado, H.; Morlock, G.E.; Meyer, D.; Cottet-Fontannaz, C.; Scholz, G.; Schilter, B.; Marin-Kuan, M. Incorporation of metabolic activation in the HPTLC-SOS-Umu-C bioassay to detect low levels of genotoxic chemicals in food contact materials. Toxics 2022, 10, 501. [Google Scholar] [CrossRef]
- DIN EN ISO 11348–1; Water Quality—Determination of the Inhibitory Effect of Water Samples on the Light Emission of Vibrio Fischeri (Luminescent Bacteria Test)—Part 1: Method Using Freshly Prepared Bacteria. Beuth Verlag: Berlin, Germany, 2009.
- Schreiner, T.; Sauter, D.; Friz, M.; Heil, J.; Morlock, G.E. Is our Natural Food our Homeostasis? Array of A Thousand Ef-fect-Directed Profiles of 68 Herbs and Spices. Front. Pharmacol. 2021, 12, 755941. [Google Scholar] [CrossRef]
- Tajima, F.; Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984, 1, 269–285. [Google Scholar]
- Andrew, H.P.; Curt, L.B.; Jonathan, F.W. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 1993, 11, 122–127. [Google Scholar]
- Morlock, G. Background mass signals in TLC/HPTLC-ESI-MS and practical advices for use of the TLC-MS Interface. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 2892–2914. [Google Scholar] [CrossRef]
- Krüger, S.; Hüsken, L.; Fornasari, R.; Scainelli, I.; Morlock, G.E. Effect-directed fingerprints of 77 botanicals via a generic high-performance thin-layer chromatography method combined with assays and mass spectrometry. J. Chromatogr. A 2017, 1529, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Morlock, G.E.; Heil, J.; Inarejos-García, A.M.; Maeder, J. Effect-directed profiling of powdered tea extracts for catechins, theaflavins, flavonols and caffeine. Antioxidants 2021, 10, 117. [Google Scholar] [CrossRef]
- Gawande, V.; Morlock, G. Effect-directed profiling of Ficus religiosa leaf extracts for multipotent compounds via 12 ef-fect-directed assays. J. Chromatogr. A 2021, 1637, 461836. [Google Scholar] [CrossRef] [PubMed]
- Mahran, E.; Keusgen, M.; Morlock, G.E. New planar assay for a streamlined detection and quantification of β-glucuronidase inhibitors and application to botanical extracts. Anal. Chim. Acta X 2020, 4, 100039. [Google Scholar]
- Fecka, I.; Turek, S. Determination of Water-Soluble Polyphenolic Compounds in Commercial Herbal Teas from Lamiaceae: Peppermint, Melissa, and Sage. Agric. Food Chem. 2007, 55, 10908–10917. [Google Scholar] [CrossRef]
- Inarejos-García, A.M.; Helbig, I.; Klette, P.; Weber, S.; Maeder, J.; Morlock, G.E. Authentication of Commercial Powdered Tea Extracts (Camellia sinensis L.) by Gas Chromatography. ACS Food Sci. Technol. 2021, 1, 596–604. [Google Scholar] [CrossRef]
- Martorell, P.; Forment, J.V.; de Llanos, R.; Montón, F.; Llopis, S.; González, N.; Genovés, S.; Cienfuegos, E.; Monzó, H.; Ramón, D. Use of Saccharomyces cerevisiae and Caenorhabditis elegans as model organisms to study the effect of cocoa polyphenols in the resistance to oxidative stress. J. Agric. Food Chem. 2011, 59, 2077–2085. [Google Scholar] [CrossRef]
- Yang, H.; Wie, C.-L.; Liu, H.-W.; Wu, J.-L.; Li, Z.-G.; Zhang, L.; Jian, J.-B.; Li, Y.-Y.; Tai, Y.-L.; Zhang, J.; et al. Genetic Divergence between Camellia sinensis and Its Wild Relatives Revealed via Genome-Wide SNPs from RAD Sequencing. PLoS ONE 2016, 11, e0151424. [Google Scholar] [CrossRef]
- Tamura, K.M.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Morlock, G.E.; Busso, M.; Tomeba, S.; Sighicelli, A. Effect-directed analysis of 32 vanilla products, characterization of mul-ti-potent compounds and quantification of vanillin and ethyl vanillin. J. Chromatogr. A 2021, 1652, 462377. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.E.; Bharadwaj, S.; Sahoo, A.K.; Yadava, U.; Kang, S.G. Determination of tyrosinase-cyanidin-3-O-glucoside and (−/+)-catechin binding modes reveal mechanistic differences in tyrosinase inhibition. Sci. Rep. 2021, 11, 24494. [Google Scholar] [CrossRef]
- Wu, Z.; Tan, B.; Liu, Y.; Dunn, J.; Guerola, P.M.; Tortajada, M.; Cao, Z.; Ji, P. Chemical Composition and Antioxidant Properties of Essential Oils from Peppermint, Native Spearmint and Scotch Spearmint. Molecules 2019, 24, 2825. [Google Scholar] [CrossRef]
- Kapp, K.; Hakala, E.; Orav, A.; Pohjala, L.; Vuorela, P.; Püssa, T.; Vuorela, H.; Raal, A. Commercial peppermint (Mentha × piperita L.) teas: Antichlamydial effect and polyphenolic composition. Food Res. Int. 2013, 53, 758–766. [Google Scholar] [CrossRef]
- Luță, E.A.; Ghica, M.; Costea, T.; Gîrd, C.E. Phytosociological study and its influence on the biosynthesis of active compounds of two medicinal plants Mentha piperita L. and Melissa officinalis L. Farmacia 2020, 68, 919–924. [Google Scholar] [CrossRef]
- Marchetti, L.; Rossi, M.C.; Pellati, F.; Benvenuti, S.; Bertelli, D. HR-1H NMR spectroscopy and multivariate statistical analysis to determine the composition of herbal mixtures for infusions. Phytochem. Anal. 2020, 32, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Stojanović-Radić, Z.; Matejić, J.; Sharopov, F.; Antolak, H.; Kregiel, D.; Sen, S.; Sharifi-Rad, M.; Acharya, K.; Shari-fi-Rad, R.; et al. Plants of Genus Mentha: From Farm to Food Factory. Plants 2018, 7, 70. [Google Scholar] [CrossRef] [Green Version]
- Scariolo, F.; Palumbo, F.; Vannozzi, A.; Sacilotto, G.B.; Gazzola, M.; Barcaccia, G. Genotyping Analysis by RAD-Seq Reads Is Useful to Assess the Genetic Identity and Relationships of Breeding Lines in Lavender Species Aimed at Managing Plant Variety Protection. Genes 2021, 12, 1656. [Google Scholar] [CrossRef]
- Petrovic, M.; Suznjevic, D.; Pastor, F.; Veljovic, M.; Pezo, L.; Antic, M.; Gorjanovic, S. Antioxidant Capacity Determination of Complex Samples and Individual Phenolics-Multilateral Approach. Comb. Chem. High Throughput Screen. 2016, 19, 58–65. [Google Scholar] [CrossRef]
- Wong, S.P.; Leong, L.P.; Koh, J.H.W. Antioxidant activities of aqueous extracts of selected plants. Food Chem. 2006, 99, 775–783. [Google Scholar] [CrossRef]
- Gorjanović, S.; Komes, D.; Pastor, F.T.; Belščak-Cvitanović, A.; Pezo, L.; Hečimović, I.; Sužnjević, D. Antioxidant Capacity of Teas and Herbal Infusions: Polarographic Assessment. Antioxidant Capacity of Teas and Herbal Infusions: Polarographic Assessment. J. Agric. Food Chem. 2012, 60, 9573–9580. [Google Scholar] [CrossRef]
- Dinh, J.; Angeloni, J.T.; Pederson, D.B.; Wang, X.; Cao, M.; Dong, Y. Cranberry extract standardized for proanthocyanidins promotes the immune response of Caenorhabditis elegans to Vibrio cholerae through the p38 MAPK pathway and HSF-1. PLoS ONE 2014, 9, e103290. [Google Scholar] [CrossRef] [PubMed]
- Roxo, M.; Peixoto, H.; Wetterauer, P.; Lima, E.; Wink, M. Piquiá Shells (Caryocar villosum): A Fruit by-Product with Antioxidant and Antiaging Properties in Caenorhabditis elegans. Oxid. Med. Cell. Longev. 2020, 2020, 7590707. [Google Scholar] [CrossRef]
- Parham, S.; Kharazi, A.Z.; Bakhsheshi-Rad, H.R.; Nur, H.; Ismail, A.F.; Sharif, S.; RamaKrishna, S.; Berto, F. Antioxidant, Antimicrobial and Antiviral Properties of Herbal Materials. Antioxidants 2020, 9, 1309. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Kim, D.; Kim, S.; Lee, J.; Chon, J.; Song, K.; Bae, D.; Kim, J.; Kim, H.; Seo, K. Antimicrobial Effect of Mentha piperita (Peppermint) Oil against Bacillus cereus, Staphylococcus aureus, Cronobacter sakazakii, and Salmonella enteritidis in Various Dairy Foods: Preliminary Study. J. Milk Sci. Biotechnol. 2018, 36, 146–154. [Google Scholar] [CrossRef]
- Işcan, G.; Kirimer, N.; Kürkcüoğlu, M.; Başer, K.H.; Demirci, F. Antimicrobial screening of Mentha piperita essential oils. J. Agric. Food Chem. 2002, 50, 3943–3946. [Google Scholar] [CrossRef]
- Mimica-Dukic, N.; Bozin, B.; Mentha, L. Species (Lamiaceae) as promising sources of bioactive secondary metabolites. Curr. Pharm. Des. 2008, 14, 3141–3150. [Google Scholar] [CrossRef]





| ID | Declaration of Peppermint Products |
|---|---|
| L1 | EU Mint leaves 130004707 |
| L2 | EU Mint leaves 1502100065 |
| L3 | USA Mint leaves CS28 |
| L4 | USA Mint leaves MA6 |
| L5 | USA Mint leaves MP2 |
| L6 | USA Mint leaves MP11 |
| L7 | USA Mint leaves MP13 |
| L8 | EU Mint leaves ME-22/0054 |
| E1 | Extract from EU Mint leaves 130004707 |
| E2 | Extract from EU Mint leaves 1502100065 |
| E3 | Extract from USA Mint leaves CS28 |
| E4 | Extract from USA Mint leaves MA6 |
| E5 | Extract from USA Mint leaves MP2 |
| E6 | Extract from USA Mint leaves MP11 |
| E7 | Extract from USA Mint leaves MP13 |
| E8 | Extract ME-22/0054 Ref35219000560000 |
| Phenolic Compound | HPLC–ESI-QTOF-MS/MS | HPLC–PDA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Score | Formula | Intensity | Expected m/z | Found at m/z | Fragment Ion m/z | RT (min) | RT (min) | UV Max (nm) | |
| Caffeic acid | 90% | C9H8O4 | 2844 | 179.0350 | 1.79.0355 | - | 17.51 | 9.01 | 218. 323 |
| Eriocitrin | 90% | C27H32O15 | 577,510 | 595.1668 | 595.1665 | 150.9669, 287.0000 | 19.84 | 12.336 | 216. 283 |
| Luteolin-7-O-rutinoside | 91% | C27H30O16 | 9361 | 609.1461 | 609.1459 | 270.9649, 299.9681 | 20.02 | 12.747 | 216. 348 |
| Eriodictyol-7-O-glucoside | 93% | C21H22O11 | 8672 | 449.1089 | 449.1085 | 135.0124, 150.9654 | 21.67 | 13.42 | 217. 283 |
| Luteolin-7-O-glucuronide | 93% | C21H18O12 | 99,000 | 461.0725 | 461.0721 | 284.9838 | 21.92 | 13.64 | 255. 348 |
| Isorhoifolin | 90% | C27H30O14 | 220,895 | 577.1563 | 577.1567 | 268.9917 | 22.33 | 15.014 | 266. 336 |
| Rosmarinic acid | 90% | C18H16O8 | 21,842 | 359.0772 | 359.0775 | 123.0144, 132.9933, 160.9828 | 27.81 | 17.845 | 218. 328 |
| Components (%, Dry Basis) | USA | Europe | ||||||
|---|---|---|---|---|---|---|---|---|
| in Leaves | L3 | L4 | L5 | L6 | L7 | L8 | L2 | L1 |
| Eriocitrin | 0.06 ± 0.00 | 0.00 ± 0.00 | 0.62 ± 0.12 | 0.00 ± 0.00 | 1.16 ± 0.03 | 1.43 ± 0.05 | 1.40 ± 0.00 | 1.30 ± 0.42 |
| Luteolin-7-O-rutinoside | 0.16 ± 0.00 | 0.04 ± 0.00 | 0.65 ± 0.14 | 0.05 ± 0.01 | 0.78 ± 0.03 | 0.47 ± 0.01 | 0.82 ± 0.00 | 0.70 ± 0.32 |
| Eriodictyol-7-O-glucoside | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.13 ± 0.03 | 0.01 ± 0.01 | 0.04 ± 0.01 | 0.21 ± 0.00 | 0.07 ± 0.00 | 0.10 ± 0.03 |
| Luteolin-7-O-glucuronide | 0.11 ± 0.00 | 0.02 ± 0.00 | 0.05 ± 0.01 | 0.02 ± 0.00 | 0.22 ± 0.00 | 0.05 ± 0.00 | 0.26 ± 0.00 | 0.18 ± 0.16 |
| Luteolin-7-O-glucoside | 1.04 ± 0.01 | 2.88 ± 0.12 | 0.12 ± 0.01 | 2.06 ± 0.14 | 0.38 ± 0.01 | 0.05 ± 0.01 | 0.09 ± 0.00 | 0.11 ± 0.00 |
| Isohoifolin | 1.81 ± 0.01 | 1.27 ± 0.07 | 0.18 ± 0.03 | 0.52 ± 0.00 | 1.19 ± 0.06 | 0.13 ± 0.01 | 0.24 ± 0.00 | 0.30 ± 0.02 |
| Rosmarinic acid | 0.75 ± 0.01 | 0.22 ± 0.00 | 0.60 ± 0.13 | 0.30 ± 0.05 | 0.75 ± 0.05 | 0.49 ± 0.02 | 0.27 ± 0.01 | 0.35 ± 0.05 |
| in Extracts | E3 | E4 | E5 | E6 | E7 | E8 | E2 | E1 |
| Eriocitrin | 0.14 ± 0.01 | 0.02 ± 0.01 | 2.05 ± 0.00 | 0.01 ± 0.00 | 3.78 ± 0.01 | 3.81 ± 0.04 | 6.38 ± 1.80 | 4.04 ± 0.63 |
| Luteolin-7-O-rutinoside | 0.30 ± 0.02 | 0.05 ± 0.01 | 2.29 ± 0.04 | 0.02 ± 0.00 | 2.39 ± 0.01 | 1.51 ± 0.01 | 3.85 ± 1.11 | 2.09 ± 0.34 |
| Eriodictyol-7-O-glucoside | 0.04 ± 0.00 | 0.02 ± 0.01 | 0.57 ± 0.00 | 0.00 ± 0.00 | 0.21 ± 0.03 | 0.78 ± 0.01 | 0.41 ± 0.13 | 0.75 ± 0.12 |
| Luteolin-7-O-glucuronide | 0.10 ± 0.00 | 0.01 ± 0.00 | 0.26 ± 0.00 | 0.01 ± 0.00 | 0.61 ± 0.03 | 0.24 ± 0.00 | 1.26 ± 0.39 | 0.36 ± 0.07 |
| Luteolin-7-O-glucoside | 0.05 ± 0.00 | 0.03 ± 0.01 | 0.34 ± 0.00 | 0.01 ± 0.00 | 0.24 ± 0.02 | 0.11 ± 0.03 | 0.27 ± 0.08 | 0.25 ± 0.04 |
| Isohoifolin | 1.46 ± 0.02 | 0.19 ± 0.05 | 0.54 ± 0.00 | 0.04 ± 0.00 | 0.90 ± 0.00 | 0.31 ± 0.00 | 1.05 ± 0.29 | 0.74 ± 0.12 |
| Rosmarinic acid | 2.32 ± 0.00 | 0.45 ± 0.12 | 2.46 ± 0.03 | 0.37 ± 0.00 | 2.57 ± 0.01 | 1.71 ± 0.01 | 1.40 ± 0.43 | 1.68 ± 0.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inarejos-Garcia, A.M.; Heil, J.; Martorell, P.; Álvarez, B.; Llopis, S.; Helbig, I.; Liu, J.; Quebbeman, B.; Nemeth, T.; Holmgren, D.; et al. Effect-Directed, Chemical and Taxonomic Profiling of Peppermint Proprietary Varieties and Corresponding Leaf Extracts. Antioxidants 2023, 12, 476. https://doi.org/10.3390/antiox12020476
Inarejos-Garcia AM, Heil J, Martorell P, Álvarez B, Llopis S, Helbig I, Liu J, Quebbeman B, Nemeth T, Holmgren D, et al. Effect-Directed, Chemical and Taxonomic Profiling of Peppermint Proprietary Varieties and Corresponding Leaf Extracts. Antioxidants. 2023; 12(2):476. https://doi.org/10.3390/antiox12020476
Chicago/Turabian StyleInarejos-Garcia, Antonio M., Julia Heil, Patricia Martorell, Beatriz Álvarez, Silvia Llopis, Ines Helbig, Jie Liu, Bryon Quebbeman, Tim Nemeth, Deven Holmgren, and et al. 2023. "Effect-Directed, Chemical and Taxonomic Profiling of Peppermint Proprietary Varieties and Corresponding Leaf Extracts" Antioxidants 12, no. 2: 476. https://doi.org/10.3390/antiox12020476
APA StyleInarejos-Garcia, A. M., Heil, J., Martorell, P., Álvarez, B., Llopis, S., Helbig, I., Liu, J., Quebbeman, B., Nemeth, T., Holmgren, D., & Morlock, G. E. (2023). Effect-Directed, Chemical and Taxonomic Profiling of Peppermint Proprietary Varieties and Corresponding Leaf Extracts. Antioxidants, 12(2), 476. https://doi.org/10.3390/antiox12020476







