Anti-Inflammatory Effect of Caffeine on Muscle under Lipopolysaccharide-Induced Inflammation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. LPS-Induced Inflammation
2.3. RNA Extraction and cDNA Synthesis
2.4. Real-Time Reverse Transcription and Quantitative PCR (RT-qPCR)
2.5. DNA Extraction
2.6. Enzymatic gDNA Treatment
2.7. Methylation-Specific qPCR (MS-qPCR)
2.8. Statistical Analysis
3. Results
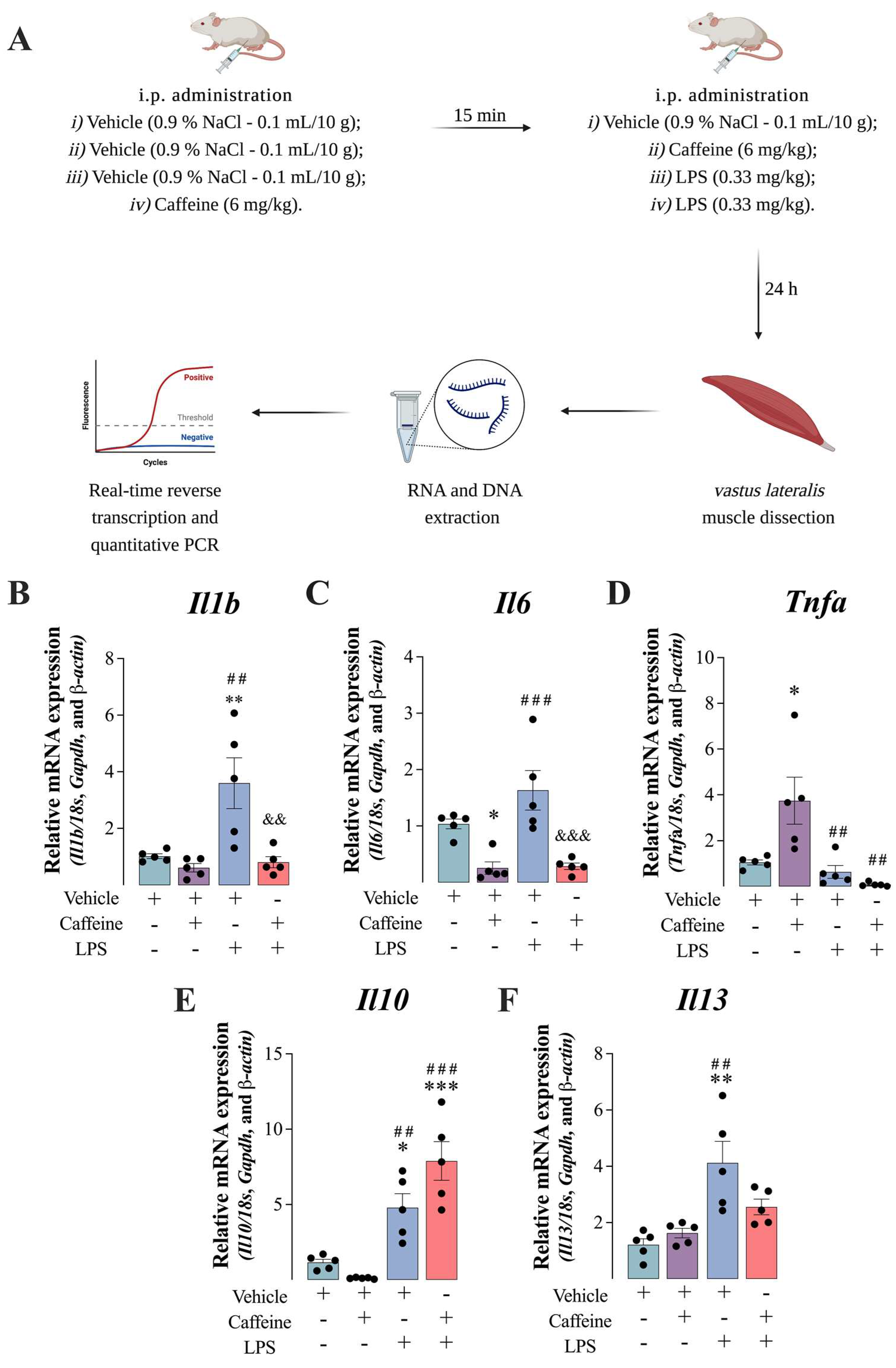
3.1. Caffeine Administration Reduced LPS-Mediated Inflammation in the Mouse Muscle
3.2. The Anti-Inflammatory Effect of Caffeine Was Mediated by Downregulating Nrlp3 Inflammasome Components
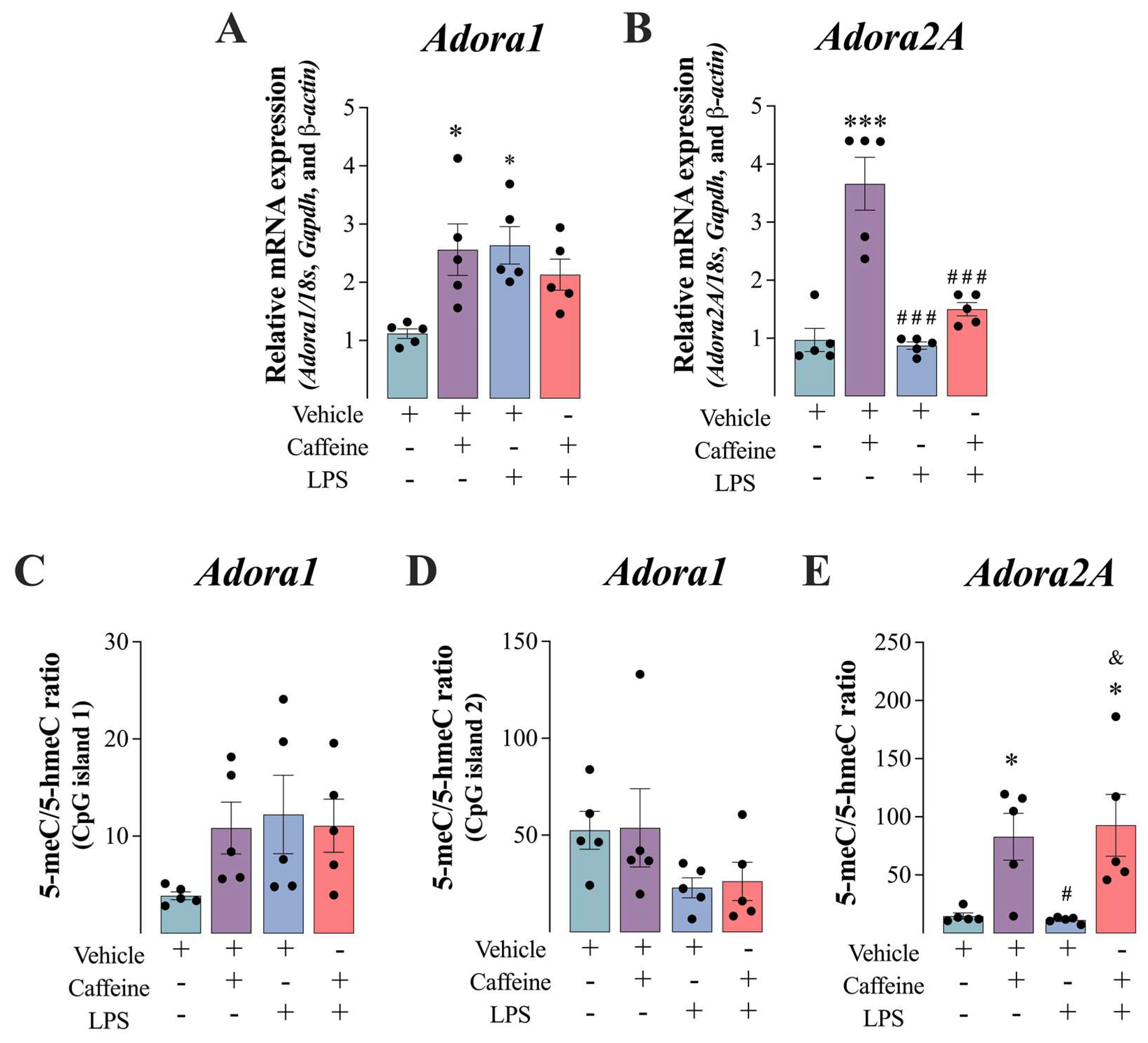
3.3. Caffeine Administration Enhanced the Expression of Adenosinergic Receptors in the Vastus Lateralis Muscle Mice
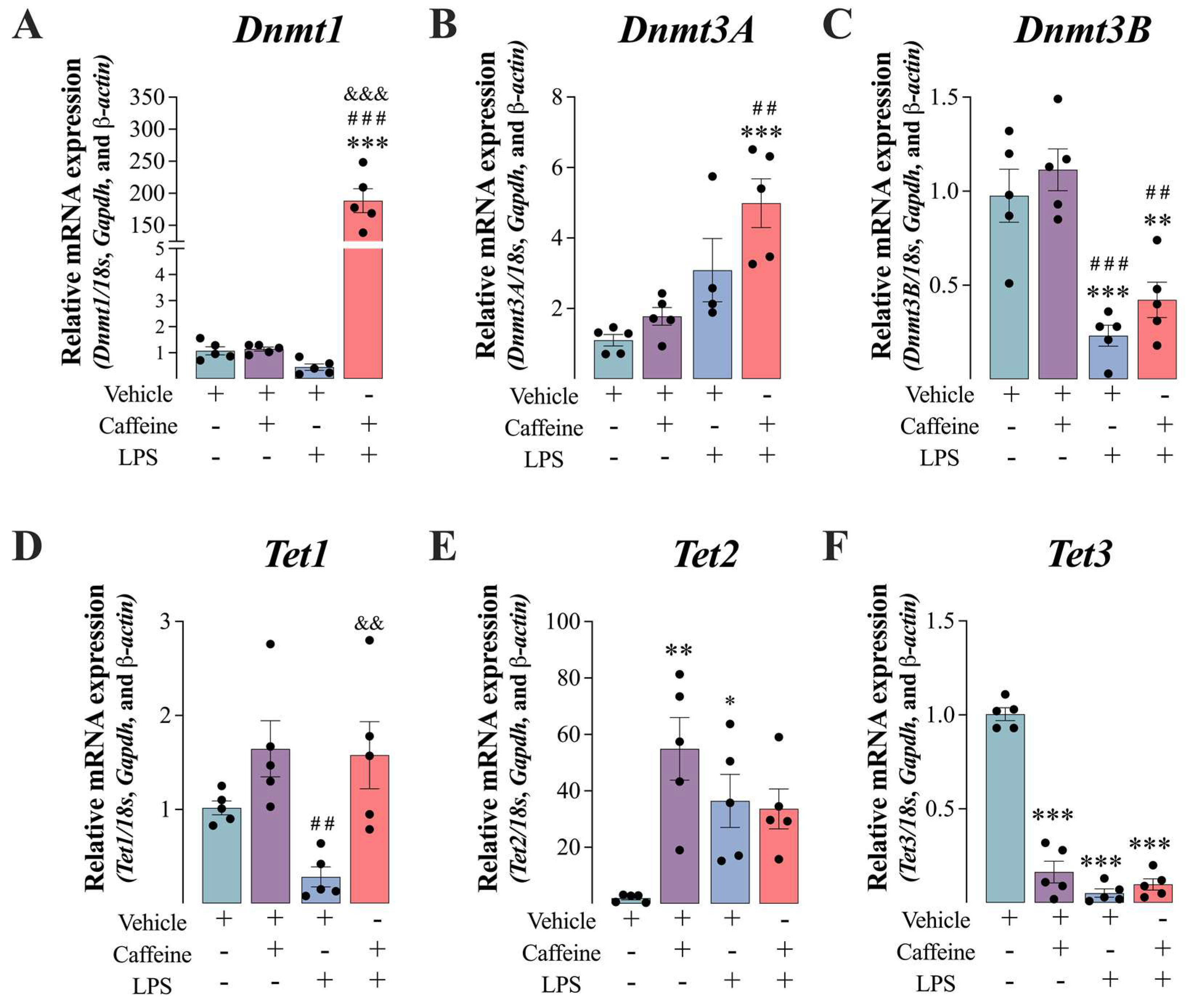
3.4. Caffeine+LPS Exposure Enhanced the De Novo DNA Methylation in the Vastus Lateralis Muscle Mice
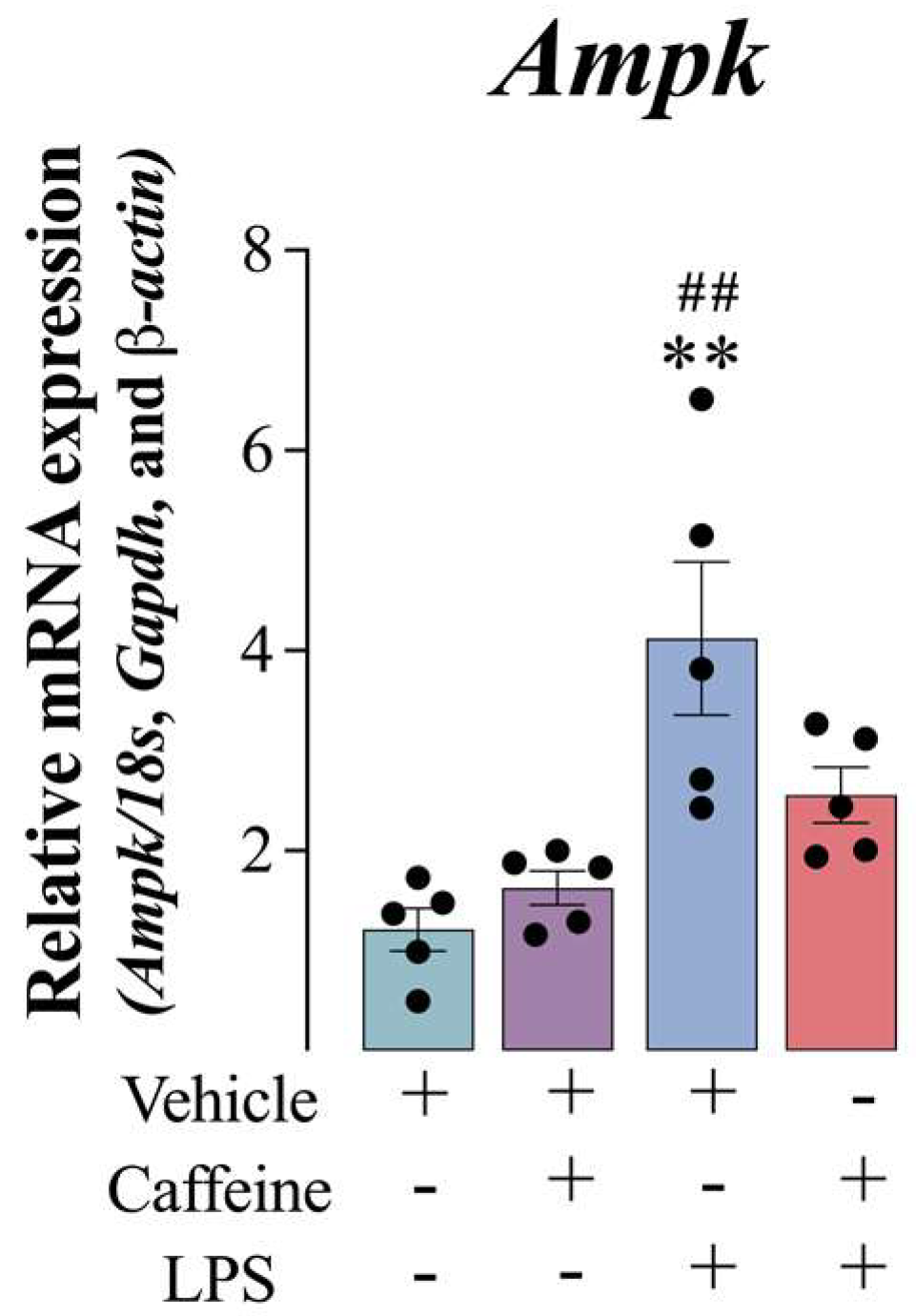
3.5. Caffeine Administration Attenuated the Catabolic State Induced by LPS Administration in the Mouse VASTUS lateralis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Luz Scheffer, D.; Latini, A. Exercise-Induced Immune System Response: Anti-Inflammatory Status on Peripheral and Central Organs. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165823. [Google Scholar] [CrossRef]
- Hargreaves, M.; Spriet, L.L. Skeletal Muscle Energy Metabolism during Exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef]
- Vital-Lopez, F.G.; Ramakrishnan, S.; Doty, T.J.; Balkin, T.J.; Reifman, J. Caffeine Dosing Strategies to Optimize Alertness during Sleep Loss. J. Sleep Res. 2018, 27, e12711. [Google Scholar] [CrossRef]
- Harpaz, E.; Tamir, S.; Weinstein, A.; Weinstein, Y. The Effect of Caffeine on Energy Balance. J. Basic Clin. Physiol. Pharmacol. 2017, 28, 1–10. [Google Scholar] [CrossRef]
- Astorino, T.A.; Rohmann, R.L.; Firth, K. Effect of Caffeine Ingestion on One-Repetition Maximum Muscular Strength. Eur. J. Appl. Physiol. 2007, 102, 127–132. [Google Scholar] [CrossRef]
- Graham, T.E.; Spriet, L.L. Performance and Metabolic Responses to a High Caffeine Dose during Prolonged Exercise. J. Appl. Physiol. 1991, 71, 2292–2298. [Google Scholar] [CrossRef]
- Spriet, L.L.; MacLean, D.A.; Dyck, D.J.; Hultman, E.; Cederblad, G.; Graham, T.E. Caffeine Ingestion and Muscle Metabolism during Prolonged Exercise in Humans. Am. J. Physiol. Metab. 1992, 262, E891–E898. [Google Scholar] [CrossRef]
- Schneiker, K.T.; Bishop, D.; Dawson, B.; Hackett, L.P. Effects of Caffeine on Prolonged Intermittent-Sprint Ability in Team-Sport Athletes. Med. Sci. Sport. Exerc. 2006, 38, 578–585. [Google Scholar] [CrossRef]
- Rodas, L.; Martinez, S.; Aguilo, A.; Tauler, P. Caffeine Supplementation Induces Higher IL-6 and IL-10 Plasma Levels in Response to a Treadmill Exercise Test. J. Int. Soc. Sports Nutr. 2020, 17, 47. [Google Scholar] [CrossRef]
- Tauler, P.; Martinez, S.; Martinez, P.; Lozano, L.; Moreno, C.; Aguiló, A. Effects of Caffeine Supplementation on Plasma and Blood Mononuclear Cell Interleukin-10 Levels After Exercise. Int. J. Sport Nutr. Exerc. Metab. 2016, 26, 8–16. [Google Scholar] [CrossRef]
- Tauler, P.; Martínez, S.; Moreno, C.; Monjo, M.; Martínez, P.; Aguiló, A. Effects of Caffeine on the Inflammatory Response Induced by a 15-Km Run Competition. Med. Sci. Sport. Exerc. 2013, 45, 1269–1276. [Google Scholar] [CrossRef]
- Farokhi-Sisakht, F.; Farhoudi, M.; Mahmoudi, J.; Farajdokht, F.; Kahfi-Ghaneh, R.; Sadigh-Eteghad, S. Effect of Intranasal Administration of Caffeine on MPFC Ischemia-induced Cognitive Impairment in BALB/c Mice. Acta Neurobiol. Exp. 2022, 82, 295–303. [Google Scholar] [CrossRef]
- Yang, L.; Yu, X.; Zhang, Y.; Liu, N.; Xue, X.; Fu, J. Caffeine Treatment Started before Injury Reduces Hypoxic-Ischemic White-Matter Damage in Neonatal Rats by Regulating Phenotypic Microglia Polarization. Pediatr. Res. 2022, 92, 1543–1554. [Google Scholar] [CrossRef]
- Jia, H.; Aw, W.; Egashira, K.; Takahashi, S.; Aoyama, S.; Saito, K.; Kishimoto, Y.; Kato, H. Coffee Intake Mitigated Inflammation and Obesity-Induced Insulin Resistance in Skeletal Muscle of High-Fat Diet-Induced Obese Mice. Genes Nutr. 2014, 9, 389. [Google Scholar] [CrossRef]
- Endesfelder, S.; Strauß, E.; Bendix, I.; Schmitz, T.; Bührer, C. Prevention of Oxygen-Induced Inflammatory Lung Injury by Caffeine in Neonatal Rats. Oxidative Med. Cell. Longev. 2020, 2020, 3840124. [Google Scholar] [CrossRef]
- Barcelos, R.P.; Lima, F.D.; Carvalho, N.R.; Bresciani, G.; Royes, L.F. Caffeine Effects on Systemic Metabolism, Oxidative-Inflammatory Pathways, and Exercise Performance. Nutr. Res. 2020, 80, 1–17. [Google Scholar] [CrossRef]
- Furman, D.; Chang, J.; Lartigue, L.; Bolen, C.R.; Haddad, F.; Gaudilliere, B.; Ganio, E.A.; Fragiadakis, G.K.; Spitzer, M.H.; Douchet, I.; et al. Expression of Specific Inflammasome Gene Modules Stratifies Older Individuals into Two Extreme Clinical and Immunological States. Nat. Med. 2017, 23, 174–184. [Google Scholar] [CrossRef]
- Egan, B.; O’Connor, P.L.; Zierath, J.R.; O’Gorman, D.J. Time Course Analysis Reveals Gene-Specific Transcript and Protein Kinetics of Adaptation to Short-Term Aerobic Exercise Training in Human Skeletal Muscle. PLoS ONE 2013, 8, e74098. [Google Scholar] [CrossRef]
- Egan, B.; Zierath, J.R. Exercise Metabolism and the Molecular Regulation of Skeletal Muscle Adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- Waddington, C.H. The Epigenotype. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef]
- Chuang, Y.-H.; Quach, A.; Absher, D.; Assimes, T.; Horvath, S.; Ritz, B. Coffee Consumption Is Associated with DNA Methylation Levels of Human Blood. Eur. J. Hum. Genet. 2017, 25, 608–616. [Google Scholar] [CrossRef]
- Remor, A.P.; da Silva, R.A.; de Matos, F.J.; Glaser, V.; de Paula Martins, R.; Ghisoni, K.; da Luz Scheffer, D.; Andia, D.C.; Portinho, D.; de Souza, A.P.; et al. Chronic Metabolic Derangement-Induced Cognitive Deficits and Neurotoxicity Are Associated with REST Inactivation. Mol. Neurobiol. 2018, 56, 1539–1557. [Google Scholar] [CrossRef]
- Ghisoni, K.; Martins, R.D.P.R.; de, P.; Barbeito, L.; Latini, A. Neopterin as a Potential Cytoprotective Brain Molecule. J. Psychiatr. Res. 2015, 71, 134–139. [Google Scholar] [CrossRef]
- De Paula Martins, R.; Glaser, V.; Aguiar, A.S.; de Paula Ferreira, P.M.; Ghisoni, K.; da Luz Scheffer, D.; Lanfumey, L.; Raisman-Vozari, R.; Corti, O.; De Paul, A.L.; et al. De Novo Tetrahydrobiopterin Biosynthesis Is Impaired in the Inflammed Striatum of Parkin(-/-) Mice. Cell Biol. Int. 2018, 42, 725–733. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Spriet, L.L. Exercise and Sport Performance with Low Doses of Caffeine. Sport. Med. 2014, 44, 175–184. [Google Scholar] [CrossRef]
- Graham, T.E.; Spriet, L.L. Metabolic, Catecholamine, and Exercise Performance Responses to Various Doses of Caffeine. J. Appl. Physiol. 1995, 78, 867–874. [Google Scholar] [CrossRef]
- Davis, J.K.; Green, J.M. Caffeine and Anaerobic Performance: Ergogenic Value and Mechanisms of Action. Sport. Med. 2009, 39, 813–832. [Google Scholar] [CrossRef]
- Van Soeren, M.H.; Sathasivam, P.; Spriet, L.L.; Graham, T.E. Caffeine Metabolism and Epinephrine Responses during Exercise in Users and Nonusers. J. Appl. Physiol. 1993, 75, 805–812. [Google Scholar] [CrossRef]
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Liu, H.; Xia, Y. Beneficial and Detrimental Role of Adenosine Signaling in Diseases and Therapy. J. Appl. Physiol. 2015, 119, 1173–1182. [Google Scholar] [CrossRef]
- Fredholm, B.B.; Bättig, K.; Holmén, J.; Nehlig, A.; Zvartau, E.E. Actions of Caffeine in the Brain with Special Reference to Factors That Contribute to Its Widespread Use. Pharmacol. Rev. 1999, 51, 83–133. [Google Scholar]
- Pasman, W.J.; Boessen, R.; Donner, Y.; Clabbers, N.; Boorsma, A. Effect of Caffeine on Attention and Alertness Measured in a Home-Setting, Using Web-Based Cognition Tests. JMIR Res. Protoc. 2017, 6, e6727. [Google Scholar] [CrossRef]
- Smith, A.P. Caffeine, Extraversion and Working Memory. J. Psychopharmacol. 2013, 27, 71–76. [Google Scholar] [CrossRef]
- Mielgo-Ayuso, J.; Marques-Jiménez, D.; Refoyo, I.; Del Coso, J.; León-Guereño, P.; Calleja-González, J. Effect of Caffeine Supplementation on Sports Performance Based on Differences Between Sexes: A Systematic Review. Nutrients 2019, 11, 2313. [Google Scholar] [CrossRef]
- Fisone, G.; Borgkvist, A.; Usiello, A. Caffeine as a Psychomotor Stimulant: Mechanism of Action. Cell. Mol. Life Sci. 2004, 61, 857–872. [Google Scholar] [CrossRef]
- Lorist, M.M.; Snel, J.; Kok, A.; Mulder, G. Influence of Caffeine on Selective Attention in Well-Rested and Fatigued Subjects. Psychophysiology 1994, 31, 525–534. [Google Scholar] [CrossRef]
- Childs, E.; de Wit, H. Enhanced Mood and Psychomotor Performance by a Caffeine-Containing Energy Capsule in Fatigued Individuals. Exp. Clin. Psychopharmacol. 2008, 16, 13–21. [Google Scholar] [CrossRef]
- Ralevic, V.; Burnstock, G. Receptors for Purines and Pyrimidines. Pharmacol. Rev. 1998, 50, 413–492. [Google Scholar]
- Fredholm, B.B.; IJzerman, A.P.; Jacobson, K.A.; Klotz, K.N.; Linden, J. International Union of Pharmacology. XXV. Nomenclature and Classification of Adenosine Receptors. Pharmacol. Rev. 2001, 53, 527–552. [Google Scholar]
- Ongini, E.; Fredholm, B.B. Pharmacology of Adenosine A2A Receptors. Trends Pharmacol. Sci. 1996, 17, 364–372. [Google Scholar] [CrossRef]
- Pasman, W.; van Baak, M.; Jeukendrup, A.; de Haan, A. The Effect of Different Dosages of Caffeine on Endurance Performance Time. Int. J. Sport. Med. 1995, 16, 225–230. [Google Scholar] [CrossRef]
- Costill, D.L.; Dalsky, G.P.; Fink, W.J. Effects of Caffeine Ingestion on Metabolism and Exercise Performance. Med. Sci. Sports 1978, 10, 155–158. [Google Scholar]
- Lim, B.-V.; Jang, M.-H.; Shin, M.-C.; Kim, H.-B.; Kim, Y.-J.; Kim, Y.-P.; Chung, J.-H.; Kim, H.; Shin, M.-S.; Kim, S.-S.; et al. Caffeine Inhibits Exercise-Induced Increase in Tryptophan Hydroxylase Expression in Dorsal and Median Raphe of Sprague–Dawley Rats. Neurosci. Lett. 2001, 308, 25–28. [Google Scholar] [CrossRef]
- Solano, A.F.; da Luz Scheffer, D.; de Bem Alves, A.C.; Silva Aguiar Jr, A.; Latini, A. Potential Pitfalls When Investigating the Ergogenic Effects of Caffeine in Mice. J. Syst. Integr. Neurosci. 2017, 3, 1–4. [Google Scholar] [CrossRef]
- Mohr, M.; Nielsen, J.J.; Bangsbo, J. Caffeine Intake Improves Intense Intermittent Exercise Performance and Reduces Muscle Interstitial Potassium Accumulation. J. Appl. Physiol. 2011, 111, 1372–1379. [Google Scholar] [CrossRef]
- Duncan, M.J.; Stanley, M.; Parkhouse, N.; Cook, K.; Smith, M. Acute Caffeine Ingestion Enhances Strength Performance and Reduces Perceived Exertion and Muscle Pain Perception during Resistance Exercise. Eur. J. Sport Sci. 2013, 13, 392–399. [Google Scholar] [CrossRef]
- Skulachev, M.V.; Antonenko, Y.N.; Anisimov, V.N.; Chernyak, B.V.; Cherepanov, D.A.; Chistyakov, V.A.; Egorov, M.V.; Kolosova, N.G.; Korshunova, G.A.; Lyamzaev, K.G.; et al. Mitochondrial-Targeted Plastoquinone Derivatives. Effect on Senescence and Acute Age-Related Pathologies. Curr. Drug Targets 2011, 12, 800–826. [Google Scholar] [CrossRef]
- Del Coso, J.; Muñoz, G.; Muñoz-Guerra, J. Prevalence of Caffeine Use in Elite Athletes Following Its Removal from the World Anti-Doping Agency List of Banned Substances. Appl. Physiol. Nutr. Metab. 2011, 36, 555–561. [Google Scholar] [CrossRef]
- Rodak, K.; Kokot, I.; Kratz, E.M. Caffeine as a Factor Influencing the Functioning of the Human Body—Friend or Foe? Nutrients 2021, 13, 3088. [Google Scholar] [CrossRef]
- Capuron, L.; Gumnick, J.F.; Musselman, D.L.; Lawson, D.H.; Reemsnyder, A.; Nemeroff, C.B.; Miller, A.H. Neurobehavioral Effects of Interferon-Alpha in Cancer Patients: Phenomenology and Paroxetine Responsiveness of Symptom Dimensions. Neuropsychopharmacology 2002, 26, 643–652. [Google Scholar] [CrossRef]
- Heesen, C.; Nawrath, L.; Reich, C.; Bauer, N.; Schulz, K.-H.; Gold, S.M. Fatigue in Multiple Sclerosis: An Example of Cytokine Mediated Sickness Behaviour? J. Neurol. Neurosurg. Psychiatr. 2006, 77, 34–39. [Google Scholar] [CrossRef]
- Cho, H.J.; Seeman, T.E.; Bower, J.E.; Kiefe, C.I.; Irwin, M.R. Prospective Association Between C-Reactive Protein and Fatigue in the Coronary Artery Risk Development in Young Adults Study. Biol. Psychiatr. 2009, 66, 871–878. [Google Scholar] [CrossRef]
- Goulart, F.O.; Godke, B.A.; Borges, V.; Azevedo-Silva, S.M.C.; Mendes, M.F.; Cendoroglo, M.S.; Ferraz, H.B. Fatigue in a Cohort of Geriatric Patients with and without Parkinson’s Disease. Brazilian J. Med. Biol. Res. 2009, 42, 771–775. [Google Scholar] [CrossRef]
- Kempf, K.; Herder, C.; Erlund, I.; Kolb, H.; Martin, S.; Carstensen, M.; Koenig, W.; Sundvall, J.; Bidel, S.; Kuha, S.; et al. Effects of Coffee Consumption on Subclinical Inflammation and Other Risk Factors for Type 2 Diabetes: A Clinical Trial. Am. J. Clin. Nutr. 2010, 91, 950–957. [Google Scholar] [CrossRef]
- Xiao, C.; Beitler, J.J.; Higgins, K.A.; Conneely, K.; Dwivedi, B.; Felger, J.; Wommack, E.C.; Shin, D.M.; Saba, N.F.; Ong, L.Y.; et al. Fatigue Is Associated with Inflammation in Patients with Head and Neck Cancer before and after Intensity-Modulated Radiation Therapy. Brain. Behav. Immun. 2016, 52, 145–152. [Google Scholar] [CrossRef]
- Lee, A.; Lim, W.; Kim, S.; Khil, H.; Cheon, E.; An, S.; Hong, S.; Lee, D.H.; Kang, S.-S.; Oh, H.; et al. Coffee Intake and Obesity: A Meta-Analysis. Nutrients 2019, 11, 1274. [Google Scholar] [CrossRef]
- Cao, C.; Loewenstein, D.A.; Lin, X.; Zhang, C.; Wang, L.; Duara, R.; Wu, Y.; Giannini, A.; Bai, G.; Cai, J.; et al. High Blood Caffeine Levels in MCI Linked to Lack of Progression to Dementia. J. Alzheimer’s Dis. 2012, 30, 559–572. [Google Scholar] [CrossRef]
- Jokela, M.; Virtanen, M.; Batty, G.D.; Kivimäki, M. Inflammation and Specific Symptoms of Depression. JAMA Psychiatr. 2016, 73, 87–88. [Google Scholar] [CrossRef]
- DellaGioia, N.; Devine, L.; Pittman, B.; Hannestad, J. Bupropion Pre-Treatment of Endotoxin-Induced Depressive Symptoms. Brain. Behav. Immun. 2013, 31, 197–204. [Google Scholar] [CrossRef]
- Ding, M.; Bhupathiraju, S.N.; Chen, M.; van Dam, R.M.; Hu, F.B. Caffeinated and Decaffeinated Coffee Consumption and Risk of Type 2 Diabetes: A Systematic Review and a Dose-Response Meta-Analysis. Diabetes Care 2014, 37, 569–586. [Google Scholar] [CrossRef]
- Shang, F.; Li, X.; Jiang, X. Coffee Consumption and Risk of the Metabolic Syndrome: A Meta-Analysis. Diabetes Metab. 2016, 42, 80–87. [Google Scholar] [CrossRef]
- Taylor, J.L.; Todd, G.; Gandevia, S.C. Evidence for a Supraspinal Contribution to Human Muscle Fatigue. Clin. Exp. Pharmacol. Physiol. 2006, 33, 400–405. [Google Scholar] [CrossRef]
- Lawrence, T. The Nuclear Factor NF-KappaB Pathway in Inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Abkenar, I.K.; Rahmani-nia, F.; Lombardi, G. The Effects of Acute and Chronic Aerobic Activity on the Signaling Pathway of the Inflammasome NLRP3 Complex in Young Men. Medicina 2019, 55, 105. [Google Scholar] [CrossRef]
- Li, H.; Miao, W.; Ma, J.; Xv, Z.; Bo, H.; Li, J.; Zhang, Y.; Ji, L.L. Acute Exercise-Induced Mitochondrial Stress Triggers an Inflammatory Response in the Myocardium via NLRP3 Inflammasome Activation with Mitophagy. Oxidative Med. Cell. Longev. 2016, 2016, 1987149. [Google Scholar] [CrossRef]
- Ostrowski, K.; Rohde, T.; Asp, S.; Schjerling, P.; Pedersen, B.K. Pro- and Anti-inflammatory Cytokine Balance in Strenuous Exercise in Humans. J. Physiol. 1999, 515, 287–291. [Google Scholar] [CrossRef]
- Kovács, E.G.; Alatshan, A.; Budai, M.M.; Czimmerer, Z.; Bíró, E.; Benkő, S. Caffeine Has Different Immunomodulatory Effect on the Cytokine Expression and NLRP3 Inflammasome Function in Various Human Macrophage Subpopulations. Nutrients 2021, 13, 2409. [Google Scholar] [CrossRef]
- Pandit, M.; Timilshina, M.; Gu, Y.; Acharya, S.; Chung, Y.; Seo, S.-U.; Chang, J.-H. AMPK Suppresses Th2 Cell Responses by Repressing MTORC2. Exp. Mol. Med. 2022, 54, 1214–1224. [Google Scholar] [CrossRef]
- Egawa, T.; Hamada, T.; Kameda, N.; Karaike, K.; Ma, X.; Masuda, S.; Iwanaka, N.; Hayashi, T. Caffeine Acutely Activates 5′adenosine Monophosphate–Activated Protein Kinase and Increases Insulin-Independent Glucose Transport in Rat Skeletal Muscles. Metabolism 2009, 58, 1609–1617. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Horrigan, L.A.; Kelly, J.P.; Connor, T.J. Caffeine Suppresses TNF-α Production via Activation of the Cyclic AMP/Protein Kinase A Pathway. Int. Immunopharmacol. 2004, 4, 1409–1417. [Google Scholar] [CrossRef]
- Ritter, M.; Hohenberger, K.; Alter, P.; Herzum, M.; Tebbe, J.; Maisch, M. Caffeine Inhibits Cytokine Expression in Lymphocytes. Cytokine 2005, 30, 177–181. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and Characterization of a Family of Novel Mammalian DNA (Cytosine-5) Methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(Cytosine-C5)-Methyltransferase Methylates DNA Processively with High Preference for Hemimethylated Target Sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet Proteins in 5mC to 5hmC Conversion, ES-Cell Self-Renewal and Inner Cell Mass Specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Monda, K.L.; Yu, K.; Paynter, N.; Azzato, E.M.; Bennett, S.N.; Berndt, S.I.; Boerwinkle, E.; Chanock, S.; Chatterjee, N.; et al. Genome-Wide Meta-Analysis Identifies Regions on 7p21 (AHR) and 15q24 (CYP1A2) As Determinants of Habitual Caffeine Consumption. PLoS Genet. 2011, 7, e1002033. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Byrne, E.M.; Esko, T.; Nalls, M.A.; Ganna, A.; Paynter, N.; Monda, K.L.; Amin, N.; Fischer, K.; Renstrom, F.; et al. Genome-Wide Meta-Analysis Identifies Six Novel Loci Associated with Habitual Coffee Consumption. Mol. Psychiatr. 2015, 20, 647–656. [Google Scholar] [CrossRef]

| Gene (ID) | Primer | 5′-3′Sequence | Reaction Condition | Product Size (bp) |
|---|---|---|---|---|
| Il1b (16176) | Forward | GAC CTT GGA TGA GGA CA | 95 °C-15 s; 60 °C-30 s; 72 °C-30 s | 183 |
| Reverse | AGC TCA TAT GGG TCC GAC AG | |||
| Il6 (16193) | Forward | AGT TCG CTT CTT GGG ACT GA | 95 °C-15 s; 60 °C-30 s; 72 °C-30 s | 191 |
| Reverse | CAG AAT TGC CAT TGC ACA AC | |||
| Tnfa (11647) | Forward | CCA CAT CTC CCT CCA GAA AA | 95 °C-15 s; 60 °C-30 s; 72 °C-30 s | 259 |
| Reverse | AGG GTC TGG GCC ATA GAA CT | |||
| Il10 (21926) | Forward | CCA AGC CCT TAT CGG AAA TGA | 95 °C-15 s; 60 °C-30 s; 72 °C-30 s | 163 |
| Reverse | TTT TCA CAG GGG AGA AAT CG | |||
| Il13 (16163) | Forward | CAG TCC TGG CTC TTG CTT G | 95 °C-15 s; 60 °C-30 s; 72 °C-30 s | 165 |
| Reverse | CCA GGT CCA CAC TCC ATA CC | |||
| Adora1 (11539) | Forward | AGA ACC ACC TCC ACC CTT CT | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 227 |
| Reverse | TAC TCT GGG TGG TGG TCA CA | |||
| Adora2A (11540) | Forward | ATC CCT CAGAGA AGG GAA GC | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 300 |
| Reverse | AGC TTC CCA AAG GCT TTC TC | |||
| Dnmt1 (13433) | Forward | CCT TTG TGG GAA CCT GGA A | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 240 |
| Reverse | CTG TCG TCT GCG GTG ATT | |||
| Dnmt3A (13435) | Forward | GAG GGA ACT GAG ACC CCA C | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 216 |
| Reverse | CTG GAA GGT GAG TCT TGG CA | |||
| Dnmt3B (113436) | Forward | AGC GGG TAT GAG GAG TGC AT | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 91 |
| Reverse | GGG AGC ATC CTT CGT GTC TG | |||
| Tet1 (52463) | Forward | GAG CCT GTT CCT CGA TGT GG | 95 °C-15 s; 65 °C-30 s; 72 °C-30 s | 367 |
| Reverse | CAA ACC CAC CTG AGG CTG TT | |||
| Tet2 (214133) | Forward | AAC CTG GCT ACT GTC ATT GCT CCA | 95 °C-15 s; 65 °C-30 s; 72 °C-30 s | 211 |
| Reverse | ATG TTC TGC TGG TCT CTG TGG GAA | |||
| Tet3 (194388) | Forward | GTC TCC CCA AGT CCT ACC TCC G | 95 °C-15 s; 63 °C-30 s; 72 °C-30 s | 137 |
| Reverse | GTC AGT GCC CCA CGC TTC A | |||
| b-actin (11461) | Forward | TCT TGG GTA TGG AAT CCT GTG | 95 °C-15 s; 58 °C-30 s; 72 °C-30 s | 82 |
| Reverse | AGG TCT TTA CGG ATG TCA ACG | |||
| Gapdh (14433) | Forward | AGG CCG GTG CTG AGT ATG TC | 95 °C-15 s; 58 °C-30 s; 72 °C-30 s | 530 |
| Reverse | TGC CTG CTT CAC CAC CTT CT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eichwald, T.; Solano, A.F.; Souza, J.; de Miranda, T.B.; Carvalho, L.B.; dos Santos Sanna, P.L.; da Silva, R.A.F.; Latini, A. Anti-Inflammatory Effect of Caffeine on Muscle under Lipopolysaccharide-Induced Inflammation. Antioxidants 2023, 12, 554. https://doi.org/10.3390/antiox12030554
Eichwald T, Solano AF, Souza J, de Miranda TB, Carvalho LB, dos Santos Sanna PL, da Silva RAF, Latini A. Anti-Inflammatory Effect of Caffeine on Muscle under Lipopolysaccharide-Induced Inflammation. Antioxidants. 2023; 12(3):554. https://doi.org/10.3390/antiox12030554
Chicago/Turabian StyleEichwald, Tuany, Alexandre Francisco Solano, Jennyffer Souza, Taís Browne de Miranda, Liebert Bernardes Carvalho, Paula Lemes dos Santos Sanna, Rodrigo A. Foganholi da Silva, and Alexandra Latini. 2023. "Anti-Inflammatory Effect of Caffeine on Muscle under Lipopolysaccharide-Induced Inflammation" Antioxidants 12, no. 3: 554. https://doi.org/10.3390/antiox12030554
APA StyleEichwald, T., Solano, A. F., Souza, J., de Miranda, T. B., Carvalho, L. B., dos Santos Sanna, P. L., da Silva, R. A. F., & Latini, A. (2023). Anti-Inflammatory Effect of Caffeine on Muscle under Lipopolysaccharide-Induced Inflammation. Antioxidants, 12(3), 554. https://doi.org/10.3390/antiox12030554







