Ten “Cheat Codes” for Measuring Oxidative Stress in Humans
Abstract
:1. Introduction
1.1. Oxygen
1.2. ROS
- Superoxide is not necessarily super. The name “superoxide” originated from the odd stoichiometry of a chemical reaction in 1934 [19]. It had nothing to do with any special “super” biochemical reactivity as an oxidant [20]. Sawyer and Valentine commented that the probability of superoxide oxidising a molecule to yield the peroxide dianion is nil. Moreover, McCord and Fridovich discovered superoxide dismutase (SOD) by observing that superoxide reduced ferric cytochrome c [21,22].
- To quote Sies and Jones, “ROS is a term, not a molecule” [30].
1.3. Antioxidants
- There is no individual “best” antioxidant. Just like ROS, they are all different (see Figure 2).
- A few “frontline” enzymes like SOD perform most of the redox “heavy lifting”. Ordinarily, SOD isoforms consume most of the superoxide produced in a cell [38]. So, only picomoles remain for other molecules, such as vitamin C, to “scavenge”. For example, 3.01 × 1018 (5 µM) superoxide molecules can be produced per second in Escherichia coli, but SOD limits [superoxide] by 4-logs to 6.0 × 1014 molecules corresponding to 0.0009 µM or 900 picomoles [39].
- Some antioxidant enzymes react with many species. Like superoxide also reacts with (and inactivates) CAT and glutathione peroxidase (GPX) [45,46], emerging evidence demonstrates that SOD metabolizes hydrogen sulphide [47], which complements prior evidence of reactivity, albeit kinetically slow, with hydrogen peroxide [48].
1.4. Oxidative Stress
- The “ROS” in the sample will have inevitably disappeared before one can measure them. They ephemerally flit in and out of existence on nanosecond timescales (10−9 of a second). So, what is one really measuring? Potentially, the rate of artificial ROS production in a heavily oxygenated sample [37].
- Although it is possible to minimise the above (e.g., degassing the sample and rapidly adding a probe), it is arduous. Even if artificial generation were minimised, a superoxide probe, for example, must still compete with SOD [89], hampering the ability to detect all of the molecules in the sample. There is always the possibility of inadvertent artefacts, such as the release of redox-active iron ions from the haemolysis of erythrocytes in blood samples.
- Many lysis buffers contain ROS, such as hydrogen peroxide and lipid hydroperoxides (LOOH) [90].
- Cutting-edge genetically encoded probes cannot be used in humans [91].
- It is an artificial redox challenge imposed on ex vivo biological material and may have questionable relevance to the ability of said material to “defend” against other species, such as superoxide.
- It can be useful with aqueous antioxidants, like vitamin C, in so far as confirming nutrient loading, when combined with assays to measure the nutrient content, and potential for redox-activity. The potential is non-equivalent to the actual activity.
- The actual antioxidant activity of blood plasma is influenced by erythrocytes and surrounding tissues, such as the endothelium.
- There are many commercial “kits” for TAC. Please carefully consider their use and properly report their information. Statements like “TAC was measured with X-kit” without detailing the procedure are discouraged.
- Use other assays to better interpret TAC in plasma (see cheat codes 4–9) and refrain from measuring it in tissues; as a general rule one would be better advised to measure antioxidant enzymes.
- The word total, unless carefully qualified (as in non-enzymatic capacity against peroxyl radical), is a misleading misnomer [123].
- Do consider the assay biochemistry. For instance, some SOD assays are prone to artefacts arising from other molecules able to reduce ferric cytochrome c and the complex biochemistry of assay reporter molecules, such as nitro-blue tetrazolium [141].
- Do quantify the systemic release of antioxidant enzymes by ELISA and immunoblot [142], especially in exosomes [143]. Do not measure GSH or antioxidant enzyme activity (e.g., GPX) in plasma/serum [4]. The concentrations of GSH, glutathione reductase, and NADPH needed to sustain appreciable plasma GPX activity are minimal.
- Do consider the possibility that enzyme activities measured ex vivo may not reflect what is possible in vivo. For example, for thioredoxin reductase, much would depend on the continual supply of NAPDH [148].
- Do consider that there is no one true “best” antioxidant (i.e., there is no master antioxidant ring to rule them all).

| n | Name | Description |
|---|---|---|
| 1 | Lipidomics | The discovery of ferroptosis—lipid peroxidation and iron-dependent cell death [153,154,155]—spurred interest in sophisticated technologies for measuring the myriads of oxidised lipid products at scale by using mass spectrometry (MS) [156]. One can even tell apart enzymatic from non-enzymatic F2-isoprostanes [157,158]. Commercial lipidomic analytical services are available. |
| 2 | HPLC | HPLC can measure lipid peroxidation products [31], such as F2-isoprostanes and MDA. In particular, the HPLC analysis of MDA is a valid and highly sensitive biomarker of exercise-induced lipid peroxidation [159]. |
| 3 | ELISA | ELISA kits can measure some lipid peroxidation products [160], notably F2-isoprostanes. While they can be insufficiently specific in some cases [161], they have illuminated individual responses to exercise in humans [139,162,163,164]. |
| 4 | LOOH | The ferrous oxidation−xylenol orange (FOX) assay developed by Wolff [165] can measure lipid hydroperoxides (LOOH). It can be combined with assays for lipid soluble antioxidants, such as vitamin E [166]. Specificity concerns can plague the FOX assay when certain types of highly oxidised lipids are analysed [167]. |
| 5 | Immunoblot | Some lipid peroxidation products, such as 4-hydrononeneal (4-HNE), react with DNA and proteins [168]. Antibodies recognising 4-HNE-conjugated protein epitopes, formed secondary to Michael addition reactions, are available [169]. The global immunoblots can infer lipid peroxidation in human samples. |
| 6 | Targeted approach | In a variant of 5, a specific protein can be immunopurified for targeted analysis of protein-specific lipid peroxidation [170,171]. For example, by measuring a mitochondrial protein located near a membrane, this approach can assess organelle-specific lipid peroxidation. |
| n | Name | Description |
|---|---|---|
| 1 | Proteomics | One can collaborate with specialist labs or access services to identify and quantify specific types of oxidised amino acid, such as carbonylated proteins (see Figure 6), on a proteome-wide scale by using bottom-up MS [182]. Sophisticated modification-specific workflows are available [183,184,185,186,187,188]. |
| 2 | ELISA | Simple and user-friendly ELISA kits can quantify total protein carbonylation [189]. |
| 3 | Immunoblot | Pan-proteome immunoblots with modification-specific antibodies or derivatisation reagents can be performed [190]. OxyBlot™ for protein carbonylation represents an enduringly popular approach [191]. |
| 4 | Fluorescence | Derivatising protein carbonyl groups with fluorophores, such as rhodamine-B hydrazine, allows for quantifying their levels via SDS-PAGE [192,193], especially when protein content can be normalised with a spectrally distinct amine-reactive probe like AlexaFluor™647-N-hydroxysuccinimide (F-NHS). Novel N-terminal reactive reagents, such as 2-pyridinecarboxyaldehyde, may also be used [194,195]. |
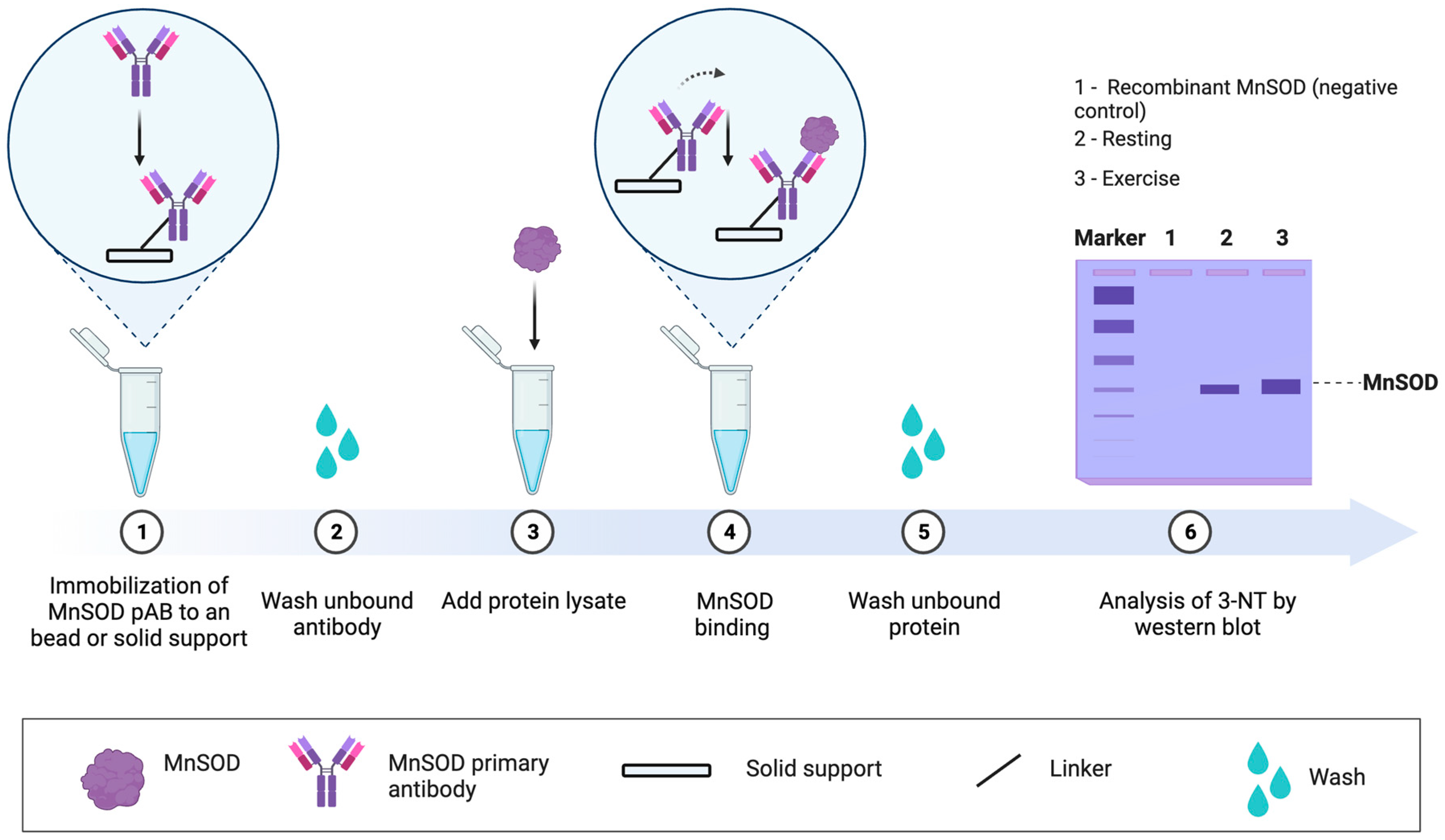
| 5 | Targeted approach | Specific proteins can be analysed by 1–4, such as MS for residue level analysis [196], when a protein is immunopurified. Targeted approaches can address specific questions [197,198], especially when the functional impact of the oxidation event is known (see Figure 7). For example, tyrosine 34 nitration impairs manganese SOD activity by electrically repelling superoxide [199]. Electrostatic repulsion helps explain why the rate of superoxide dismutation via O2·− + O2·− colliding to form hydrogen peroxide and oxygen is near zero [200]. Approach 4 combined with an ELISA assay may support protein-specific oxidative damage analysis in a microplate. |
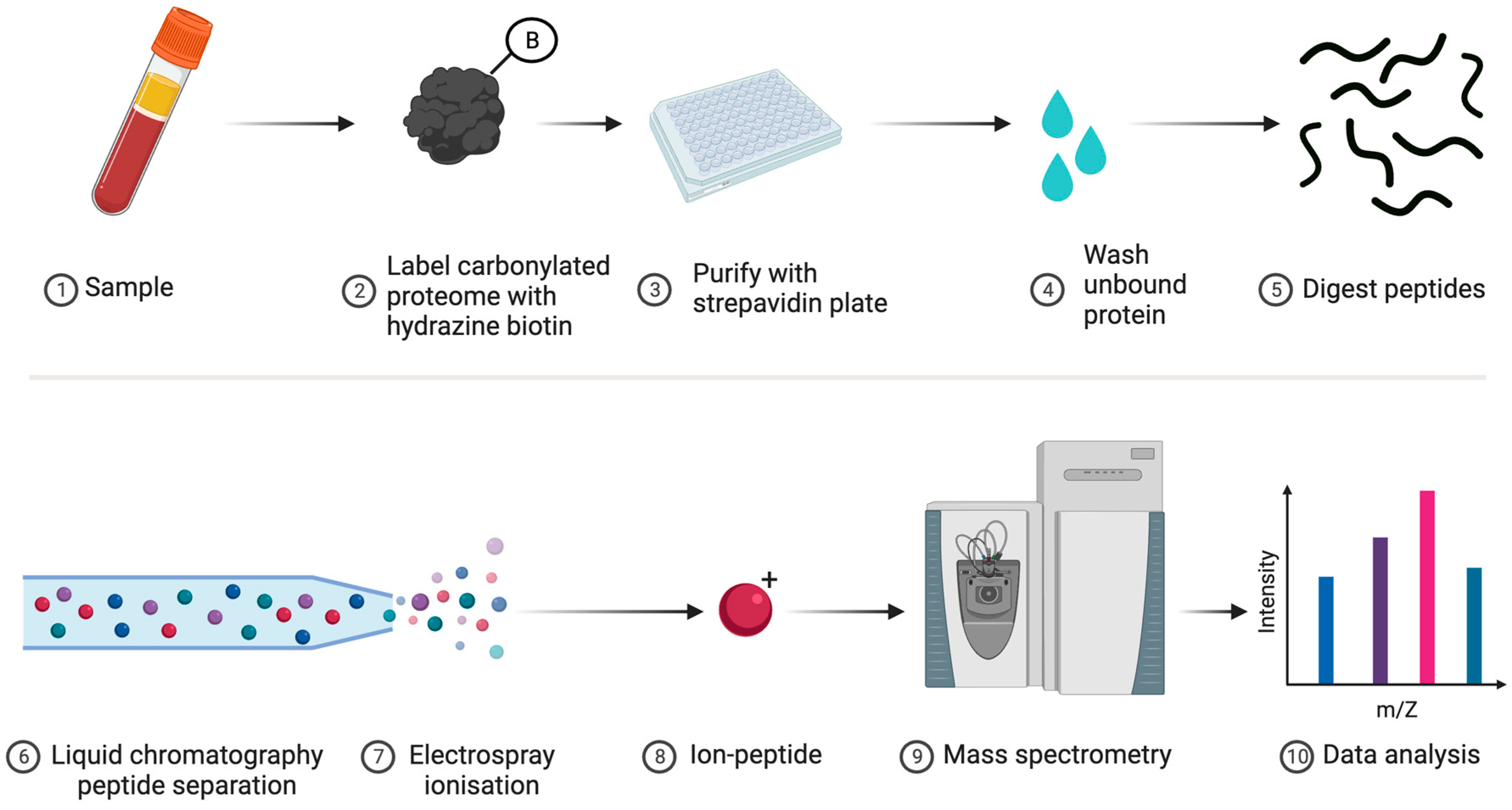
- Assay techniques: The validated analytical tools relating to DNA oxidation are applicable to RNA oxidation, as the latter is quantified by using ELISA, PCR-based technology, and chromatograph procedures, such as HPLC with electrochemical potential detection [216] and liquid chromatography/mass spectrometry or gas chromatography/mass spectrometry [217,218]. Note that most ELISA kits cannot discriminate between RNA and DNA oxidation products.
- Sample type: RNA oxidation can be quantified in urine, blood, and/or tissue (cells). The detection of 8-oxoGuo urinary excretion is possible but must be corrected for urine dilution (via urine volume, creatinine, or density). Blood plasma is an acceptable material to measure RNA oxidation with, although the data should be carefully interpreted with appropriate physiological modelling [217,219]. Tissue quantification has the advantage of tissue-specific interpretation, unlike plasma and urine collection.
| n | Name | Description |
|---|---|---|
| 1 | Proteomics | Sophisticated MS approaches, such as cysteine-reactive phosphate tag technology [242], can identify and quantify cysteine oxidation with residue resolution on a proteome-wide scale [243,244,245,246,247]. Modern methods allow for deep coverage of the cysteine proteome, such as ~34,000 residues across ~9000 proteins [242]. For reference, the full human proteome contains over 200,000 cysteine residues distributed across over 18,000 proteins [248]. New advances provide deep coverage and better quantification [249,250]. Protein-targeted methods are also possible [251,252]. Proteomic services, with some including data analytical packages, are available. |
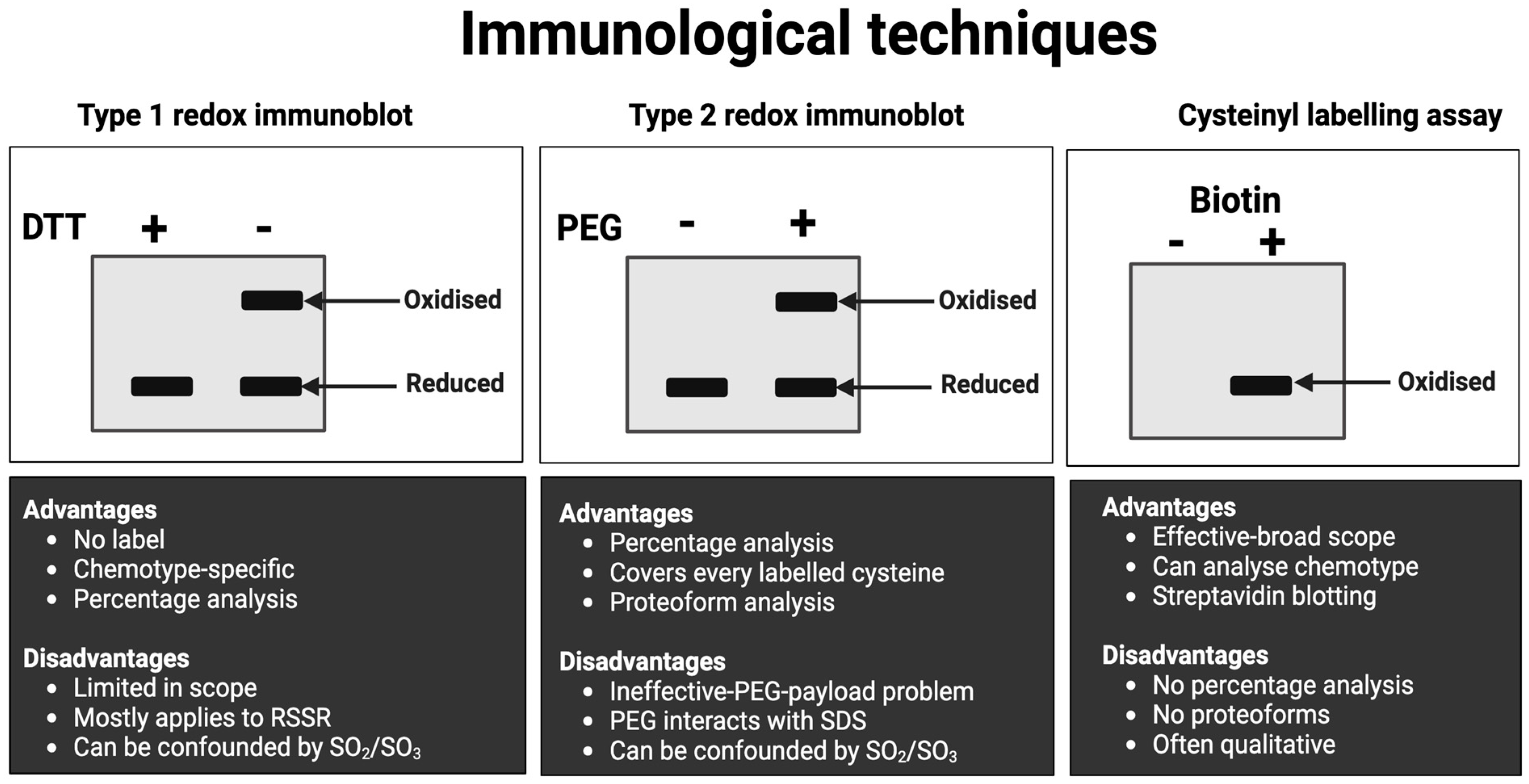
| 2 | Immunological approach | Approaches (see Figure 8) include immunocapture before the streptavidin immunoblotting of biotin-conjugated oxidised cysteines for target-specific cysteine oxidation [253]. Non-reducing immunoblotting quantifies the oxidation of some proteins, such as protein kinase G [254]. For the many proteins that fail to exhibit endogenous oxidation-induced mobility shifts, cysteines can be derivatised with mobility-shifting polyethylene glycol (PEG) payloads [255,256,257,258,259,260]. These assays quantify cysteine redox proteoforms [261]. |
| 3 | Outcomes | Some assays can infer the outcomes of redox regulation without measuring cysteine oxidation [262,263,264]. Transcriptional approaches, such as qPCR analysis, can infer the activation of redox-sensitive gene expression programmes, notably Nrf2 * activity. Immunoblot approaches include (1) degradation of KEAP1 * to infer Nrf2 activity; (2) monitoring a reporter, such as the phosphorylation of a signalling protein; and (3) quantifying protein content, such as heat shock proteins [265]. Changes in antioxidant enzyme activity, glutathione, or oxidative damage may also be instructive. |
- Do bear in mind that not every protein and every cysteine are yet measurable in one run by using MS technology [271].
- Do consider that different workflows measure different forms of cysteine oxidation, the so-called chemotypes. For example, a chemotype-specific proteomic approach demonstrated that fatiguing exercise increased S-glutathionylation, i.e., cysteine covalently attached to glutathione via a disulphide bond, in mice [272].
- Do consider that many techniques do not measure “over-oxidised” chemotypes, such as sulphinic acids.
- Do not assume that a technique will necessarily work! Mobility-shift immunoblots usually fail to detect the protein because the bulky PEG payloads sterically block primary antibody binding.
- Do not assume cysteine oxidation is functional without evidence.
- Do not assume the cysteine oxidation is necessarily oxidative eustress without evidence.
- Do not assume an outcome assay result is caused by cysteine oxidation without evidence.
- Standard operating procedures are available [276]. The cysteine-labelling procedures can be adapted to suit specific experimental needs. For example, to omit some costly preparatory steps, reduced cysteines can be labelled with an F-MAL in ALISA. In this case, increased cysteine oxidation would decrease the observed F-MAL signal.
- The assays can operate in different modes, from global (i.e., all proteins/no antibodies) to multiparametric array mode, in microscale and macroscale (e.g., slab-gel format).
- In some cases, the assays provide information on protein function, such as the difference in transcription factor cysteine redox states in the cytosol vs. the nucleus.
- Interpretationally, a change reflects a difference in the rate of ROS-sensitive cysteine oxidation and antioxidant-sensitive reduction across the entire protein. The summed weighted mean of all individual residues responding to both ROS and AOX inputs is useful.
- Whether the redox approach is general or targeted, where the general “catch-all” approach analyses focus on as many distinct oxidative stress processes as possible and the targeted ones focus on a specific process, such as lipid peroxidation. In both cases, multiple process-specific analytical indices are preferred. Still, the depth of the analyses depends on whether oxidative stress is a primary, secondary, or tertiary biochemical outcome variable.
- The type of biological material acquired, usually blood and/or tissue samples, and the number of samples dictate what can be measured and how. For example, performing MS-based proteomic analysis on 100 samples is unlikely to be financially viable. Relatedly, the relevant equipment and expertise to undertake the analysis must be available.
- Cheat code 4 to verify increased NAC loading via HPLC-based analysis of plasma NAC and assay GSH and GPX activity in erythrocytes or tissue lysates to infer whether NAC supports the glutathione redox system [330]. This might be expected to alter peroxide metabolism and hence oxidative damage to proteins via lipid peroxidation products, such as 4-HNE [168].
- Cheat code 5 to measure lipid peroxidation. In plasma, one might measure LOOH and 4-HNE via the FOX assay and immunoblotting, respectively. In tissue samples, one might measure 4-HNE via immunoblotting. Equally, one might implement a F2-isoprostanes ELISA in plasma or tissue [331].
- Cheat code 6 to measure oxidative damage to contractile proteins by using targeted analysis and the immunocapture of a specific protein followed by immunoblot analysis for 4-HNE, 3-NT, or protein carbonyls [197]. Like how cutting fingernails yields thiyl radicals in alpha keratin [332], mechanical stress produces protein-based free radicals [333]. Hence, one could add a spin trap to “clamp” protein radicals for targeted immunoblot analysis with an anti-trap reagent [334]. If only circulating samples were available, the same approaches could be applied in these compartments to test the plausibility of NAC minimising oxidative damage to proteins (albeit non-contractile ones).
- Cheat codes 8–9 with chemotype analysis to determine whether NAC, by supporting hydrogen sulphide production, elicits beneficial effects by inducing contractile protein-specific persulphidation [335,336,337]. If fluorescent labels are used, then cheat codes 6, 8, and 9 could be implemented simultaneously [338], for example, gel-based detection of persulphidation before 4-HNE immunoblotting.
- Cheat code 4: GSH levels (systemic or tissue). Or cheat code 10 (see below).
- Cheat code 5: 4-HNE blot (systemic or tissue).
- Cheat code 6: myosin-specific 4-HNE levels (tissue).
- NAC enters the circulation before it or a metabolite thereof accumulates in skeletal muscle (checked via cheat code 4: HPLC analysis of NAC).
- NAC indirectly acts as an antioxidant by impacting a process that influences the oxidation of contractile proteins. The former can be checked via GSH-related lipid peroxidation analysis (cheat codes 4–5) and the latter by targeted oxidative damage analysis pursuant to cheat code 6 or hydrogen sulphide donor effect as per cheat code 8 or 9.
- By so doing, NAC impacts a whole-body marker of fatigue such as exercise performance.
2. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cobley, J.N.; Marrin, K. Vitamin E Supplementation Does Not Alter Physiological Performance at Fixed Blood Lactate Concentrations in Trained Runners. J. Sports Med. Phys. Fit. 2012, 52, 63–70. [Google Scholar]
- Forman, H.J.; Davies, K.J.A.; Ursini, F. How Do Nutritional Antioxidants Really Work: Nucleophilic Tone and Para-Hormesis versus Free Radical Scavenging in Vivo. Free Radic. Biol. Med. 2014, 66, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; McHardy, H.; Morton, J.P.; Nikolaidis, M.G.; Close, G.L. Influence of Vitamin C and Vitamin E on Redox Signaling: Implications for Exercise Adaptations. Free Radic. Biol. Med. 2015, 84, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Close, G.L.; Bailey, D.M.; Davison, G.W. Exercise Redox Biochemistry: Conceptual, Methodological and Technical Recommendations. Redox Biol. 2017, 12, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Holmgren, A.; Larsson, N.-G.; Halliwell, B.; Chang, C.J.; Kalyanaraman, B.; Rhee, S.G.; Thornalley, P.J.; Partridge, L.; Gems, D.; et al. Unraveling the Biological Roles of Reactive Oxygen Species. Cell Metab. 2011, 13, 361–366. [Google Scholar] [CrossRef]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Gache, S.A.M.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653.e6. [Google Scholar] [CrossRef]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically Encoded Fluorescent Indicator for Intracellular Hydrogen Peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef]
- Forman, H.J.; Augusto, O.; Brigelius-Flohe, R.; Dennery, P.A.; Kalyanaraman, B.; Ischiropoulos, H.; Mann, G.E.; Radi, R.; Roberts, L.J.; Vina, J.; et al. Even Free Radicals Should Follow Some Rules: A Guide to Free Radical Research Terminology and Methodology. Free Radic. Biol. Med. 2015, 78, 233–235. [Google Scholar] [CrossRef]
- Halliwell, B.; Whiteman, M. Measuring Reactive Species and Oxidative Damage in Vivo and in Cell Culture: How Should You Do It and What Do the Results Mean? Br. J. Pharmacol. 2004, 142, 231–255. [Google Scholar] [CrossRef]
- Fridovich, I. The Biology of Oxygen Radicals. Science 1978, 201, 875–880. [Google Scholar] [CrossRef]
- Cobley, J.N.; Fiorello, M.L.; Bailey, D.M. 13 Reasons Why the Brain Is Susceptible to Oxidative Stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Oxygen Toxicity: A Radical Explanation. J. Exp. Biol. 1998, 201, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Ogilby, P.R. Singlet Oxygen: There Is Indeed Something New under the Sun. Chem. Soc. Rev. 2010, 39, 3181–3209. [Google Scholar] [CrossRef]
- Sies, H.; Menck, C.F.M. Singlet Oxygen Induced DNA Damage. Mutat. Res./DNAging 1992, 275, 367–375. [Google Scholar] [CrossRef]
- Meo, S.D.; Venditti, P. Evolution of the Knowledge of Free Radicals and Other Oxidants. Oxidative Med. Cell. Longev. 2020, 2020, 9829176. [Google Scholar] [CrossRef]
- Weiss, S.J.; King, G.W.; LoBuglio, A.F. Evidence for Hydroxyl Radical Generation by Human Monocytes. J. Clin. Investig. 1977, 60, 370–373. [Google Scholar] [CrossRef]
- Misra, H.P. Generation of Superoxide Free Radical during the Autoxidation of Thiols. J. Biol. Chem. 1974, 249, 2151–2155. [Google Scholar] [CrossRef]
- Flohé, L. Looking Back at the Early Stages of Redox Biology. Antioxidants 2020, 9, 1254. [Google Scholar] [CrossRef]
- Neuman, E.W. Potassium Superoxide and the Three-Electron Bond. J. Chem. Phys. 1934, 2, 31–33. [Google Scholar] [CrossRef]
- Sawyer, D.T.; Valentine, J.S. How Super Is Superoxide? Acc. Chem. Res. 1981, 14, 393–400. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. The Reduction of Cytochrome c by Milk Xanthine Oxidase. J. Biol. Chem. 1968, 243, 5753–5760. [Google Scholar] [CrossRef] [PubMed]
- McCord, J.M.; Fridovich, I. Superoxide Dismutase an Enzymic Function for Erythrocuprein (Hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.; Pattison, D.I.; Davies, J.B.; Anderson, R.F.; Lopez-Alarcon, C.; Davies, M.J. Superoxide Radicals React with Peptide-Derived Tryptophan Radicals with Very High Rate Constants to Give Hydroperoxides as Major Products. Free Radic. Biol. Med. 2018, 118, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Biochemistry of Oxidative Stress. Biochem. Soc. Trans. 2007, 35, 1147–1150. [Google Scholar] [CrossRef]
- Boveris, A.; Chance, B. The Mitochondrial Generation of Hydrogen Peroxide. General Properties and Effect of Hyperbaric Oxygen. Biochem. J. 1973, 134, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Sies, H.; Boveris, A. Hydroperoxide Metabolism in Mammalian Organs. Physiol. Rev. 1979, 59, 527–605. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, R.L.S.; Quinlan, C.L.; Perevoshchikova, I.V.; Hey-Mogensen, M.; Brand, M.D. Sites of Superoxide and Hydrogen Peroxide Production by Muscle Mitochondria Assessed Ex Vivo under Conditions Mimicking Rest and Exercise. J. Biol. Chem. 2015, 290, 209–227. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. Synapse Pruning: Mitochondrial ROS with Their Hands on the Shears. Bioessays 2018, 40, 1800031. [Google Scholar] [CrossRef]
- Sidlauskaite, E.; Gibson, J.W.; Megson, I.L.; Whitfield, P.D.; Tovmasyan, A.; Batinic-Haberle, I.; Murphy, M.P.; Moult, P.R.; Cobley, J.N. Mitochondrial ROS Cause Motor Deficits Induced by Synaptic Inactivity: Implications for Synapse Pruning. Redox Biol. 2018, 16, 344–351. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive Oxygen Species (ROS) as Pleiotropic Physiological Signalling Agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J. Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Gutteridge, J.M.C.; Halliwell, B. Antioxidants: Molecules, Medicines, and Myths. Biochem. Biophys. Res. Commun. 2010, 393, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Taverne, Y.J.; Merkus, D.; Bogers, A.J.; Halliwell, B.; Duncker, D.J.; Lyons, T.W. Reactive Oxygen Species: Radical Factors in the Evolution of Animal Life. Bioessays 2018, 40, 1700158. [Google Scholar] [CrossRef] [PubMed]
- Echtay, K.S.; Roussel, D.; St-Pierre, J.; Jekabsons, M.B.; Cadenas, S.; Stuart, J.A.; Harper, J.A.; Roebuck, S.J.; Morrison, A.; Pickering, S.; et al. Superoxide Activates Mitochondrial Uncoupling Proteins. Nature 2002, 415, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. Mechanisms of Mitochondrial ROS Production in Assisted Reproduction: The Known, the Unknown, and the Intriguing. Antioxidants 2020, 9, 933. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Understanding Mechanisms of Antioxidant Action in Health and Disease. Nat. Rev. Mol. Cell Biol. 2024, 25, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.A.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for Measuring Reactive Oxygen Species and Oxidative Damage in Cells and in Vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Superoxide as an Intracellular Radical Sink. Free Radic. Biol. Med. 1993, 14, 85–90. [Google Scholar] [CrossRef]
- Ganini, D.; Santos, J.H.; Bonini, M.G.; Mason, R.P. Switch of Mitochondrial Superoxide Dismutase into a Prooxidant Peroxidase in Manganese-Deficient Cells and Mice. Cell Chem. Biol. 2018, 25, 413–425.e6. [Google Scholar] [CrossRef]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide Dismutase 1 Acts as a Nuclear Transcription Factor to Regulate Oxidative Stress Resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Peskin, A.V.; Parsons-Mair, H.N. Thiol Oxidase Activity of Copper, Zinc Superoxide Dismutase. J. Biol. Chem. 2002, 277, 1906–1911. [Google Scholar] [CrossRef] [PubMed]
- Kettle, A.J.; Ashby, L.V.; Winterbourn, C.C.; Dickerhof, N. Superoxide: The Enigmatic Chemical Chameleon in Neutrophil Biology. Immunol. Rev. 2023, 314, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.; Fridovich, I. Inactivation of Glutathione Peroxidase by Superoxide Radical. Arch. Biochem. Biophys. 1985, 240, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Fridovich, I. Superoxide Radical Inhibits Catalase. J. Biol. Chem. 1982, 257, 5751–5754. [Google Scholar] [CrossRef]
- Switzer, C.H.; Kasamatsu, S.; Ihara, H.; Eaton, P. SOD1 Is an Essential H2S Detoxifying Enzyme. Proc. Natl. Acad. Sci. USA 2023, 120, e2205044120. [Google Scholar] [CrossRef] [PubMed]
- Liochev, S.I.; Fridovich, I. Copper, Zinc Superoxide Dismutase and H2O2 Effects of Bicarbonate on Inactivation and Oxidations of NADPH and Urate, and on Consumption of H2O2. J. Biol. Chem. 2002, 277, 34674–34678. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the Chemistry and Biology of Reactive Oxygen Species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef]
- Dickinson, B.C.; Chang, C.J. Chemistry and Biology of Reactive Oxygen Species in Signaling or Stress Responses. Nat. Chem. Biol. 2011, 7, 504–511. [Google Scholar] [CrossRef]
- Niki, E. Lipid Peroxidation: Physiological Levels and Dual Biological Effects. Free Radic. Biol. Med. 2009, 47, 469–484. [Google Scholar] [CrossRef]
- Yin, H.; Xu, L.; Porter, N.A. Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chem. Rev. 2011, 111, 5944–5972. [Google Scholar] [CrossRef] [PubMed]
- Melo, T.; Montero-Bullón, J.-F.; Domingues, P.; Domingues, M.R. Discovery of Bioactive Nitrated Lipids and Nitro-Lipid-Protein Adducts Using Mass Spectrometry-Based Approaches. Redox Biol. 2019, 23, 101106. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Zhao, K.; Whiteman, M. The Gastrointestinal Tract: A Major Site of Antioxidant Action? Free Radic. Res. 2000, 33, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.J.; Twist, C.; Cobley, J.N.; Howatson, G.; Close, G.L. Exercise-Induced Muscle Damage: What Is It, What Causes It and What Are the Nutritional Solutions? Eur. J. Sport Sci. 2019, 19, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Are Polyphenols Antioxidants or Pro-Oxidants? What Do We Learn from Cell Culture and in Vivo Studies? Arch. Biochem. Biophys. 2008, 476, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. The Wanderings of a Free Radical. Free Radic. Biol. Med. 2009, 46, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. 1—Oxidative Stress: Introductory Remarks. In Oxidative Stress; Academic Press: Cambridge, MA, USA, 1985; pp. 1–8. [Google Scholar] [CrossRef]
- Davies, M.J. Protein Oxidation and Peroxidation. Biochem. J. 2016, 473, 805–825. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Zhang, H. Targeting Oxidative Stress in Disease: Promise and Limitations of Antioxidant Therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidants and Human Disease: Some New Concepts1. FASEB J. 1987, 1, 358–364. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative Stress and Cancer: Have We Moved Forward? Biochem. J. 2006, 401, 1–11. [Google Scholar] [CrossRef] [PubMed]
- D’Autréaux, B.; Toledano, M.B. ROS as Signalling Molecules: Mechanisms That Generate Specificity in ROS Homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Holmström, K.M.; Finkel, T. Cellular Mechanisms and Physiological Consequences of Redox-Dependent Signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Jones, D.P. Redefining Oxidative Stress. Antioxid. Redox Signal. 2006, 8, 1865–1879. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: A Concept in Redox Biology and Medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative Stress: Concept and Some Practical Aspects. Antioxidants 2020, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Hydrogen Peroxide as a Central Redox Signaling Molecule in Physiological Oxidative Stress: Oxidative Eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Cobley, J.N. 50 Shades of Oxidative Stress: A State-Specific Cysteine Redox Pattern Hypothesis. Redox Biol. 2023, 67, 102936. [Google Scholar] [CrossRef]
- Cobley, J.N.; Margaritelis, N.V.; Morton, J.P.; Close, G.L.; Nikolaidis, M.G.; Malone, J.K. The Basic Chemistry of Exercise-Induced DNA Oxidation: Oxidative Damage, Redox Signaling, and Their Interplay. Front. Physiol. 2015, 6, 182. [Google Scholar] [CrossRef]
- Koppenol, W.H. Ferryl for Real. The Fenton Reaction near Neutral PH. Dalton Trans. 2022, 51, 17496–17502. [Google Scholar] [CrossRef]
- Halliwell, B.; Adhikary, A.; Dingfelder, M.; Dizdaroglu, M. Hydroxyl Radical Is a Significant Player in Oxidative DNA Damage in Vivo. Chem. Soc. Rev. 2021, 50, 8355–8360. [Google Scholar] [CrossRef] [PubMed]
- Illés, E.; Mizrahi, A.; Marks, V.; Meyerstein, D. Carbonate-Radical-Anions, and Not Hydroxyl Radicals, Are the Products of the Fenton Reaction in Neutral Solutions Containing Bicarbonate. Free Radic. Biol. Med. 2019, 131, 1–6. [Google Scholar] [CrossRef]
- Nikolaidis, M.G.; Margaritelis, N.V. Same Redox Evidence But Different Physiological “Stories”: The Rashomon Effect in Biology. Bioessays 2018, 40, 1800041. [Google Scholar] [CrossRef]
- Nikolaidis, M.G.; Kerksick, C.M.; Lamprecht, M.; McAnulty, S.R. Does Vitamin C and E Supplementation Impair the Favorable Adaptations of Regular Exercise? Oxidative Med. Cell. Longev. 2012, 2012, 707941. [Google Scholar] [CrossRef]
- Sies, H.; Ursini, F. Homeostatic Control of Redox Status and Health. IUBMB Life 2022, 74, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative Eustress: On Constant Alert for Redox Homeostasis. Redox Biol. 2021, 41, 101867. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. (Ed.) Oxidative Stress; Academic Press: Cambridge, MA, USA, 2019; ISBN 9780128186060. [Google Scholar]
- Ursini, F.; Maiorino, M.; Forman, H.J. Redox Homeostasis: The Golden Mean of Healthy Living. Redox Biol. 2016, 8, 205–215. [Google Scholar] [CrossRef]
- Gutteridge, J.M.C.; Halliwell, B. Mini-Review: Oxidative Stress, Redox Stress or Redox Success? Biochem. Biophys. Res. Commun. 2018, 502, 183–186. [Google Scholar] [CrossRef]
- Manford, A.G.; Rodríguez-Pérez, F.; Shih, K.Y.; Shi, Z.; Berdan, C.A.; Choe, M.; Titov, D.V.; Nomura, D.K.; Rape, M. A Cellular Mechanism to Detect and Alleviate Reductive Stress. Cell 2020, 183, 46–61.e21. [Google Scholar] [CrossRef] [PubMed]
- Manford, A.G.; Mena, E.L.; Shih, K.Y.; Gee, C.L.; McMinimy, R.; Martínez-González, B.; Sherriff, R.; Lew, B.; Zoltek, M.; Rodríguez-Pérez, F.; et al. Structural Basis and Regulation of the Reductive Stress Response. Cell 2021, 184, 5375–5390.e16. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How Mitochondria Produce Reactive Oxygen Species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Mootha, V.K. Oxygen and Mammalian Cell Culture: Are We Repeating the Experiment of Dr. Ox? Nat. Metab. 2019, 1, 858–860. [Google Scholar] [CrossRef] [PubMed]
- Keeley, T.P.; Mann, G.E. Defining Physiological Normoxia for Improved Translation of Cell Physiology to Animal Models and Humans. Physiol. Rev. 2019, 99, 161–234. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Kalyanaraman, B. Small-Molecule Luminescent Probes for the Detection of Cellular Oxidizing and Nitrating Species. Free Radic. Biol. Med. 2018, 128, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Eid, M.; Barayeu, U.; Sulková, K.; Aranda-Vallejo, C.; Dick, T.P. Using the Heme Peroxidase APEX2 to Probe Intracellular H2O2 Flux and Diffusion. Nat. Commun. 2024, 15, 1239. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, Y.C.; Altun, H.Y.; Secilmis, M.; Ata, B.N.; Sevimli, G.; Cokluk, Z.; Zaki, A.G.; Sezen, S.; Caglar, T.A.; Sevgen, İ.; et al. Complexities of the Chemogenetic Toolkit: Differential MDAAO Activation by d-Amino Substrates and Subcellular Targeting. Free Radic. Biol. Med. 2021, 177, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J.A.; Quintanilha, A.T.; Brooks, G.A.; Packer, L. Free Radicals and Tissue Damage Produced by Exercise. Biochem. Biophys. Res. Commun. 1982, 107, 1198–1205. [Google Scholar] [CrossRef]
- Bonini, M.G.; Rota, C.; Tomasi, A.; Mason, R.P. The Oxidation of 2′,7′-Dichlorofluorescin to Reactive Oxygen Species: A Self-Fulfilling Prophesy? Free Radic. Biol. Med. 2006, 40, 968–975. [Google Scholar] [CrossRef]
- Wardman, P. Fluorescent and Luminescent Probes for Measurement of Oxidative and Nitrosative Species in Cells and Tissues: Progress, Pitfalls, and Prospects. Free Radic. Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.M.; Davies, B.; Young, I.S.; Jackson, M.J.; Davison, G.W.; Isaacson, R.; Richardson, R.S. EPR Spectroscopic Detection of Free Radical Outflow from an Isolated Muscle Bed in Exercising Humans. J. Appl. Physiol. 2003, 94, 1714–1718. [Google Scholar] [CrossRef] [PubMed]
- Shchepinova, M.M.; Cairns, A.G.; Prime, T.A.; Logan, A.; James, A.M.; Hall, A.R.; Vidoni, S.; Arndt, S.; Caldwell, S.T.; Prag, H.A.; et al. MitoNeoD: A Mitochondria-Targeted Superoxide Probe. Cell Chem. Biol. 2017, 24, 1285–1298.e12. [Google Scholar] [CrossRef] [PubMed]
- Gatin-Fraudet, B.; Ottenwelter, R.; Saux, T.L.; Norsikian, S.; Pucher, M.; Lombès, T.; Baron, A.; Durand, P.; Doisneau, G.; Bourdreux, Y.; et al. Evaluation of Borinic Acids as New, Fast Hydrogen Peroxide–Responsive Triggers. Proc. Natl. Acad. Sci. USA 2021, 118, e2107503118. [Google Scholar] [CrossRef]
- Cheng, G.; Zielonka, M.; Dranka, B.; Kumar, S.N.; Myers, C.R.; Bennett, B.; Garces, A.M.; Machado, L.G.D.D.; Thiebaut, D.; Ouari, O.; et al. Detection of Mitochondria-Generated Reactive Oxygen Species in Cells Using Multiple Probes and Methods: Potentials, Pitfalls, and the Future. J. Biol. Chem. 2018, 293, 10363–10380. [Google Scholar] [CrossRef]
- Hausladen, A.; Fridovich, I. Superoxide and Peroxynitrite Inactivate Aconitases, but Nitric Oxide Does Not. J. Biol. Chem. 1994, 269, 29405–29408. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Oxygen Radicals, Nitric Oxide, and Peroxynitrite: Redox Pathways in Molecular Medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef]
- Gardner, P.R.; Nguyen, D.D.; White, C.W. Aconitase Is a Sensitive and Critical Target of Oxygen Poisoning in Cultured Mammalian Cells and in Rat Lungs. Proc. Natl. Acad. Sci. USA 1994, 91, 12248–12252. [Google Scholar] [CrossRef]
- Gardner, P.R. Aconitase: Sensitive Target and Measure of Superoxide. Methods Enzym. 2002, 349, 9–23. [Google Scholar] [CrossRef]
- Vásquez-Vivar, J.; Kalyanaraman, B.; Kennedy, M.C. Mitochondrial Aconitase Is a Source of Hydroxyl Radical an electron spin resonance investigation. J. Biol. Chem. 2000, 275, 14064–14069. [Google Scholar] [CrossRef]
- Roelofs, B.A.; Ge, S.X.; Studlack, P.E.; Polster, B.M. Low Micromolar Concentrations of the Superoxide Probe MitoSOX Uncouple Neural Mitochondria and Inhibit Complex IV. Free Radic. Biol. Med. 2015, 86, 250–258. [Google Scholar] [CrossRef]
- Cardozo, G.; Mastrogiovanni, M.; Zeida, A.; Viera, N.; Radi, R.; Reyes, A.M.; Trujillo, M. Mitochondrial Peroxiredoxin 3 Is Rapidly Oxidized and Hyperoxidized by Fatty Acid Hydroperoxides. Antioxidants 2023, 12, 408. [Google Scholar] [CrossRef]
- Karplus, P.A. A Primer on Peroxiredoxin Biochemistry. Free Radic. Biol. Med. 2015, 80, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Armas, M.I.D.; Esteves, R.; Viera, N.; Reyes, A.M.; Mastrogiovanni, M.; Alegria, T.G.P.; Netto, L.E.S.; Tórtora, V.; Radi, R.; Trujillo, M. Rapid Peroxynitrite Reduction by Human Peroxiredoxin 3: Implications for the Fate of Oxidants in Mitochondria. Free Radic. Biol. Med. 2019, 130, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Bryk, R.; Griffin, P.; Nathan, C. Peroxynitrite Reductase Activity of Bacterial Peroxiredoxins. Nature 2000, 407, 211–215. [Google Scholar] [CrossRef]
- Wood, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin Evolution and the Regulation of Hydrogen Peroxide Signaling. Science 2003, 300, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Peskin, A.V.; Low, F.M.; Paton, L.N.; Maghzal, G.J.; Hampton, M.B.; Winterbourn, C.C. The High Reactivity of Peroxiredoxin 2 with H2O2 Is Not Reflected in Its Reaction with Other Oxidants and Thiol Reagents. J. Biol. Chem. 2007, 282, 11885–11892. [Google Scholar] [CrossRef]
- Peskin, A.V.; Dickerhof, N.; Poynton, R.A.; Paton, L.N.; Pace, P.E.; Hampton, M.B.; Winterbourn, C.C. Hyperoxidation of Peroxiredoxins 2 and 3 Rate Constants for the Reactions of the Sulfenic Acid of the Peroxidatic Cysteine. J. Biol. Chem. 2013, 288, 14170–14177. [Google Scholar] [CrossRef] [PubMed]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-Dependent Reduction of Cysteine–Sulphinic Acid by S. Cerevisiae Sulphiredoxin. Nature 2003, 425, 980–984. [Google Scholar] [CrossRef]
- Akter, S.; Fu, L.; Jung, Y.; Conte, M.L.; Lawson, J.R.; Lowther, W.T.; Sun, R.; Liu, K.; Yang, J.; Carroll, K.S. Chemical Proteomics Reveals New Targets of Cysteine Sulfinic Acid Reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [Google Scholar] [CrossRef]
- Cobley, J.N.; Husi, H. Immunological Techniques to Assess Protein Thiol Redox State: Opportunities, Challenges and Solutions. Antioxidants 2020, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Stretton, C.; Pugh, J.N.; McDonagh, B.; McArdle, A.; Close, G.L.; Jackson, M.J. 2-Cys Peroxiredoxin Oxidation in Response to Hydrogen Peroxide and Contractile Activity in Skeletal Muscle: A Novel Insight into Exercise-Induced Redox Signalling? Free Radic. Biol. Med. 2020, 160, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Pugh, J.N.; Stretton, C.; McDonagh, B.; Brownridge, P.; McArdle, A.; Jackson, M.J.; Close, G.L. Exercise Stress Leads to an Acute Loss of Mitochondrial Proteins and Disruption of Redox Control in Skeletal Muscle of Older Subjects: An Underlying Decrease in Resilience with Aging? Free Radic. Biol. Med. 2021, 177, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Bersani, N.A.; Merwin, J.R.; Lopez, N.I.; Pearson, G.D.; Merrill, G.F. [29] Protein Electrophoretic Mobility Shift Assay to Monitor Redox State of Thioredoxin in Cells. Methods Enzym. 2002, 347, 317–326. [Google Scholar] [CrossRef]
- Low, F.M.; Hampton, M.B.; Peskin, A.V.; Winterbourn, C.C. Peroxiredoxin 2 Functions as a Noncatalytic Scavenger of Low-Level Hydrogen Peroxide in the Erythrocyte. Blood 2006, 109, 2611–2617. [Google Scholar] [CrossRef]
- Low, F.M.; Hampton, M.B.; Winterbourn, C.C. Peroxiredoxin 2 and Peroxide Metabolism in the Erythrocyte. Antioxid. Redox Signal. 2008, 10, 1621–1630. [Google Scholar] [CrossRef]
- Wayner, D.D.M.; Burton, G.W.; Ingold, K.U.; Locke, S. Quantitative Measurement of the Total, Peroxyl Radical-trapping Antioxidant Capability of Human Blood Plasma by Controlled Peroxidation. FEBS Lett. 1985, 187, 33–37. [Google Scholar] [CrossRef]
- Bartosz, G. Non-Enzymatic Antioxidant Capacity Assays: Limitations of Use in Biomedicine. Free Radic. Res. 2010, 44, 711–720. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Metodiewa, D. The Reaction of Superoxide with Reduced Glutathione. Arch. Biochem. Biophys. 1994, 314, 284–290. [Google Scholar] [CrossRef]
- Sies, H. Total Antioxidant Capacity: Appraisal of a Concept 1, 2. J. Nutr. 2007, 137, 1493–1495. [Google Scholar] [CrossRef]
- Powers, S.K.; Goldstein, E.; Schrager, M.; Ji, L.L. Exercise Training and Skeletal Muscle Antioxidant Enzymes: An Update. Antioxidants 2022, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.K.; Deminice, R.; Ozdemir, M.; Yoshihara, T.; Bomkamp, M.P.; Hyatt, H. Exercise-Induced Oxidative Stress: Friend or Foe? J. Sport Health Sci. 2020, 9, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.K.; Radak, Z.; Ji, L.L. Exercise-induced Oxidative Stress: Past, Present and Future. J. Physiol. 2016, 594, 5081–5092. [Google Scholar] [CrossRef] [PubMed]
- Peskin, A.V.; Winterbourn, C.C. Assay of Superoxide Dismutase Activity in a Plate Assay Using WST-1. Free Radic. Biol. Med. 2017, 103, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Crapo, J.D.; McCord, J.M.; Fridovich, I. [41] Preparation and Assay of Superioxide Dismutases. Methods Enzym. 1978, 53, 382–393. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide Dismutase: Improved Assays and an Assay Applicable to Acrylamide Gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Misra, H.P.; Fridovich, I. The Role of Superoxide Anion in the Autoxidation of Epinephrine and a Simple Assay for Superoxide Dismutase. J. Biol. Chem. 1972, 247, 3170–3175. [Google Scholar] [CrossRef] [PubMed]
- Stolwijk, J.M.; Falls-Hubert, K.C.; Searby, C.C.; Wagner, B.A.; Buettner, G.R. Simultaneous Detection of the Enzyme Activities of GPx1 and GPx4 Guide Optimization of Selenium in Cell Biological Experiments. Redox Biol. 2020, 32, 101518. [Google Scholar] [CrossRef] [PubMed]
- Dagnell, M.; Pace, P.E.; Cheng, Q.; Frijhoff, J.; Östman, A.; Arnér, E.S.J.; Hampton, M.B.; Winterbourn, C.C. Thioredoxin Reductase 1 and NADPH Directly Protect Protein Tyrosine Phosphatase 1B from Inactivation during H2O2 Exposure. J. Biol. Chem. 2017, 292, 14371–14380. [Google Scholar] [CrossRef]
- Arnér, E.S.J. Selenoproteins, Methods and Protocols. Methods Mol. Biol. 2017, 1661, 301–309. [Google Scholar] [CrossRef]
- Woo, H.A.; Yim, S.H.; Shin, D.H.; Kang, D.; Yu, D.-Y.; Rhee, S.G. Inactivation of Peroxiredoxin I by Phosphorylation Allows Localized H2O2 Accumulation for Cell Signaling. Cell 2010, 140, 517–528. [Google Scholar] [CrossRef]
- Kerins, M.J.; Milligan, J.; Wohlschlegel, J.A.; Ooi, A. Fumarate Hydratase Inactivation in Hereditary Leiomyomatosis and Renal Cell Cancer Is Synthetic Lethal with Ferroptosis Induction. Cancer Sci. 2018, 109, 2757–2766. [Google Scholar] [CrossRef]
- Gomez-Cabrera, M.C.; Carretero, A.; Millan-Domingo, F.; Garcia-Dominguez, E.; Correas, A.G.; Olaso-Gonzalez, G.; Viña, J. Redox-Related Biomarkers in Physical Exercise. Redox Biol. 2021, 42, 101956. [Google Scholar] [CrossRef] [PubMed]
- Giustarini, D.; Dalle-Donne, I.; Colombo, R.; Milzani, A.; Rossi, R. An Improved HPLC Measurement for GSH and GSSG in Human Blood. Free Radic. Biol. Med. 2003, 35, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.B.; Furdui, C.M.; King, S.B. Introduction to Approaches and Tools for the Evaluation of Protein Cysteine Oxidation. Essays Biochem. 2020, 64, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Margaritelis, N.V.; Nastos, G.G.; Vasileiadou, O.; Chatzinikolaou, P.N.; Theodorou, A.A.; Paschalis, V.; Vrabas, I.S.; Kyparos, A.; Fatouros, I.G.; Nikolaidis, M.G. Inter-individual Variability in Redox and Performance Responses after Antioxidant Supplementation: A Randomized Double Blind Crossover Study. Acta Physiol. 2023, 238, e14017. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.M.; Culcasi, M.; Filipponi, T.; Brugniaux, J.V.; Stacey, B.S.; Marley, C.J.; Soria, R.; Rimoldi, S.F.; Cerny, D.; Rexhaj, E.; et al. EPR Spectroscopic Evidence of Iron-Catalysed Free Radical Formation in Chronic Mountain Sickness: Dietary Causes and Vascular Consequences. Free Radic. Biol. Med. 2022, 184, 99–113. [Google Scholar] [CrossRef]
- Beyer, W.F.; Fridovich, I. Assaying for Superoxide Dismutase Activity: Some Large Consequences of Minor Changes in Conditions. Anal. Biochem. 1987, 161, 559–566. [Google Scholar] [CrossRef]
- Salzano, S.; Checconi, P.; Hanschmann, E.-M.; Lillig, C.H.; Bowler, L.D.; Chan, P.; Vaudry, D.; Mengozzi, M.; Coppo, L.; Sacre, S.; et al. Linkage of Inflammation and Oxidative Stress via Release of Glutathionylated Peroxiredoxin-2, Which Acts as a Danger Signal. Proc. Natl. Acad. Sci. USA 2014, 111, 12157–12162. [Google Scholar] [CrossRef]
- Lisi, V.; Moulton, C.; Fantini, C.; Grazioli, E.; Guidotti, F.; Sgrò, P.; Dimauro, I.; Capranica, L.; Parisi, A.; Luigi, L.D.; et al. Steady-State Redox Status in Circulating Extracellular Vesicles: A Proof-of-Principle Study on the Role of Fitness Level and Short-Term Aerobic Training in Healthy Young Males. Free Radic. Biol. Med. 2023, 204, 266–275. [Google Scholar] [CrossRef]
- Hansen, R.E.; Winther, J.R. An Introduction to Methods for Analyzing Thiols and Disulfides: Reactions, Reagents, and Practical Considerations. Anal. Biochem. 2009, 394, 147–158. [Google Scholar] [CrossRef]
- Flohé, L. The Fairytale of the GSSG/GSH Redox Potential. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3139–3142. [Google Scholar] [CrossRef]
- Marx, V. Finding the Right Antibody for the Job. Nat. Methods 2013, 10, 703–707. [Google Scholar] [CrossRef]
- Janes, K.A. An Analysis of Critical Factors for Quantitative Immunoblotting. Sci. Signal. 2015, 8, rs2. [Google Scholar] [CrossRef]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Signal. 2015, 23, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Dillard, C.J.; Litov, R.E.; Savin, W.M.; Dumelin, E.E.; Tappel, A.L. Effects of Exercise, Vitamin E, and Ozone on Pulmonary Function and Lipid Peroxidation. J. Appl. Physiol. 1978, 45, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Bielski, B.H.J.; Cabelli, D.E.; Arudi, R.L.; Ross, A.B. Reactivity of HO2/O−2 Radicals in Aqueous Solution. J. Phys. Chem. Ref. Data 1985, 14, 1041–1100. [Google Scholar] [CrossRef]
- Barayeu, U.; Schilling, D.; Eid, M.; da Silva, T.N.X.; Schlicker, L.; Mitreska, N.; Zapp, C.; Gräter, F.; Miller, A.K.; Kappl, R.; et al. Hydropersulfides Inhibit Lipid Peroxidation and Ferroptosis by Scavenging Radicals. Nat. Chem. Biol. 2023, 19, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, Biology and Role in Disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Bayır, H.; Anthonymuthu, T.S.; Tyurina, Y.Y.; Patel, S.J.; Amoscato, A.A.; Lamade, A.M.; Yang, Q.; Vladimirov, G.K.; Philpott, C.C.; Kagan, V.E. Achieving Life through Death: Redox Biology of Lipid Peroxidation in Ferroptosis. Cell Chem. Biol. 2020, 27, 387–408. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Mao, G.; Qu, F.; Angeli, J.P.F.; Doll, S.; Croix, C.S.; Dar, H.H.; Liu, B.; Tyurin, V.A.; Ritov, V.B.; et al. Oxidized Arachidonic and Adrenic PEs Navigate Cells to Ferroptosis. Nat. Chem. Biol. 2017, 13, 81–90. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Erve, T.J.; Lih, F.B.; Kadiiska, M.B.; Deterding, L.J.; Eling, T.E.; Mason, R.P. Reinterpreting the Best Biomarker of Oxidative Stress: The 8-Iso-PGF2α/PGF2α Ratio Distinguishes Chemical from Enzymatic Lipid Peroxidation. Free Radic. Biol. Med. 2015, 83, 245–251. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Erve, T.J.; Kadiiska, M.B.; London, S.J.; Mason, R.P. Classifying Oxidative Stress by F2-Isoprostane Levels across Human Diseases: A Meta-Analysis. Redox Biol. 2017, 12, 582–599. [Google Scholar] [CrossRef] [PubMed]
- Spirlandeli, A.; Deminice, R.; Jordao, A. Plasma Malondialdehyde as Biomarker of Lipid Peroxidation: Effects of Acute Exercise. Int. J. Sports Med. 2013, 35, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Lee, C.Y.J. Using Isoprostanes as Biomarkers of Oxidative Stress: Some Rarely Considered Issues. Antioxid. Redox Signal. 2010, 13, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.G.; Kyparos, A.; Vrabas, I.S. F2-Isoprostane Formation, Measurement and Interpretation: The Role of Exercise. Prog. Lipid Res. 2011, 50, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Margaritelis, N.V.; Theodorou, A.A.; Paschalis, V.; Veskoukis, A.S.; Dipla, K.; Zafeiridis, A.; Panayiotou, G.; Vrabas, I.S.; Kyparos, A.; Nikolaidis, M.G. Adaptations to Endurance Training Depend on Exercise-induced Oxidative Stress: Exploiting Redox Interindividual Variability. Acta Physiol. 2018, 222, e12898. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Cobley, J.N.; Paschalis, V.; Veskoukis, A.S.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Going Retro: Oxidative Stress Biomarkers in Modern Redox Biology. Free Radic. Biol. Med. 2016, 98, 2–12. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Kyparos, A.; Paschalis, V.; Theodorou, A.A.; Panayiotou, G.; Zafeiridis, A.; Dipla, K.; Nikolaidis, M.G.; Vrabas, I.S. Reductive Stress after Exercise: The Issue of Redox Individuality. Redox Biol. 2014, 2, 520–528. [Google Scholar] [CrossRef]
- Wolff, S.P. [18] Ferrous Ion Oxidation in Presence of Ferric Ion Indicator Xylenol Orange for Measurement of Hydroperoxides. Methods Enzym. 1994, 233, 182–189. [Google Scholar] [CrossRef]
- Williamson, J.; Hughes, C.M.; Cobley, J.N.; Davison, G.W. The Mitochondria-Targeted Antioxidant MitoQ, Attenuates Exercise-Induced Mitochondrial DNA Damage. Redox Biol. 2020, 36, 101673. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Porter, N.A. Specificity of the Ferrous Oxidation of Xylenol Orange Assay: Analysis of Autoxidation Products of Cholesteryl Arachidonate. Anal. Biochem. 2003, 313, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forman, H.J. 4-Hydroxynonenal-Mediated Signaling and Aging. Free Radic. Biol. Med. 2017, 111, 219–225. [Google Scholar] [CrossRef]
- Waeg, G.; Dimsity, G.; Esterbauer, H. Monoclonal Antibodies for Detection of 4-Hydroxynonenal Modified Proteins. Free Radic. Res. 1996, 25, 149–159. [Google Scholar] [CrossRef]
- Cobley, J.N.; Davison, G.W. Measuring Oxidative Damage and Redox Signalling. In Oxidative Eustress in Exercise Physiology; CRC Press: Boca Raton, FL, USA, 2022; pp. 11–22. [Google Scholar] [CrossRef]
- Cobley, J.N.; Davison, G.W. Oxidative Eustress in Exercise Physiology; CRC Press: Boca Raton, FL, USA, 2022; ISBN 9781003051619. [Google Scholar]
- Slavov, N. Counting Protein Molecules for Single-Cell Proteomics. Cell 2022, 185, 232–234. [Google Scholar] [CrossRef]
- Slavov, N. Unpicking the Proteome in Single Cells. Science 2020, 367, 512–513. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Kitamura, N.; Galligan, J.J. A Global View of the Human Post-Translational Modification Landscape. Biochem. J. 2023, 480, 1241–1265. [Google Scholar] [CrossRef]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Baker, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.F.; Fenselau, C.; Garcia, B.A.; et al. How Many Human Proteoforms Are There? Nat. Chem. Biol. 2018, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L.; Linial, M.; Goodlett, D.; Langridge-Smith, P.; Goo, Y.A.; Safford, G.; Bonilla, L.; Kruppa, G.; Zubarev, R.; et al. Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next Proteomics Currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.M.; Meyer, A.S. Cataloguing the Proteome: Current Developments in Single-Molecule Protein Sequencing. Biophys. Rev. 2022, 3, 011304. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.L.; Davies, M.J. Detection, Identification, and Quantification of Oxidative Protein Modifications. J. Biol. Chem. 2019, 294, 19683–19708. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, C.L.; Morgan, P.E.; Davies, M.J. Quantification of Protein Modification by Oxidants. Free Radic. Biol. Med. 2009, 46, 965–988. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass Spectrometry-Based Proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Batthyány, C.; Bartesaghi, S.; Mastrogiovanni, M.; Lima, A.; Demicheli, V.; Radi, R. Tyrosine-Nitrated Proteins: Proteomic and Bioanalytical Aspects. Antioxid. Redox Signal. 2017, 26, 313–328. [Google Scholar] [CrossRef]
- Madian, A.G.; Diaz-Maldonado, N.; Gao, Q.; Regnier, F.E. Oxidative Stress Induced Carbonylation in Human Plasma. J. Proteom. 2011, 74, 2395–2416. [Google Scholar] [CrossRef]
- Madian, A.G.; Regnier, F.E. Profiling Carbonylated Proteins in Human Plasma. J. Proteome Res. 2010, 9, 1330–1343. [Google Scholar] [CrossRef]
- Fedorova, M.; Bollineni, R.C.; Hoffmann, R. Protein Carbonylation as a Major Hallmark of Oxidative Damage: Update of Analytical Strategies. Mass Spectrom. Rev. 2014, 33, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Aldini, G.; Domingues, M.R.; Spickett, C.M.; Domingues, P.; Altomare, A.; Sánchez-Gómez, F.J.; Oeste, C.L.; Pérez-Sala, D. Protein Lipoxidation: Detection Strategies and Challenges. Redox Biol. 2015, 5, 253–266. [Google Scholar] [CrossRef]
- Bollineni, R.C.; Hoffmann, R.; Fedorova, M. Proteome-Wide Profiling of Carbonylated Proteins and Carbonylation Sites in HeLa Cells under Mild Oxidative Stress Conditions. Free Radic. Biol. Med. 2014, 68, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Buss, H.; Chan, T.P.; Sluis, K.B.; Domigan, N.M.; Winterbourn, C.C. Protein Carbonyl Measurement by a Sensitive ELISA Method. Free Radic. Biol. Med. 1997, 23, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Sakellariou, G.K.; Owens, D.J.; Murray, S.; Waldron, S.; Gregson, W.; Fraser, W.D.; Burniston, J.G.; Iwanejko, L.A.; McArdle, A.; et al. Lifelong Training Preserves Some Redox-Regulated Adaptive Responses after an Acute Exercise Stimulus in Aged Human Skeletal Muscle. Free Radic. Biol. Med. 2014, 70, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Frijhoff, J.; Winyard, P.G.; Zarkovic, N.; Davies, S.S.; Stocker, R.; Cheng, D.; Knight, A.R.; Taylor, E.L.; Oettrich, J.; Ruskovska, T.; et al. Clinical Relevance of Biomarkers of Oxidative Stress. Antioxid. Redox Signal. 2015, 23, 1144–1170. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Davies, M.J.; Grune, T. Determination of Protein Carbonyls in Plasma, Cell Extracts, Tissue Homogenates, Isolated Proteins: Focus on Sample Preparation and Derivatization Conditions. Redox Biol. 2015, 5, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, C.D.; Zisimopoulos, D.; Argyropoulou, V.; Kalaitzopoulou, E.; Ioannou, P.V.; Salachas, G.; Grune, T. Protein Carbonyl Determination by a Rhodamine B Hydrazide-Based Fluorometric Assay. Redox Biol. 2018, 17, 236–245. [Google Scholar] [CrossRef]
- MacDonald, J.I.; Munch, H.K.; Moore, T.; Francis, M.B. One-Step Site-Specific Modification of Native Proteins with 2-Pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Bridge, H.N.; Leiter, W.; Frazier, C.L.; Weeks, A.M. An N Terminomics Toolbox Combining 2-Pyridinecarboxaldehyde Probes and Click Chemistry for Profiling Protease Specificity. Cell Chem. Biol. 2024, 31, 534–549.e8. [Google Scholar] [CrossRef]
- Cobley, J.N.; Sakellariou, G.K.; Husi, H.; McDonagh, B. Proteomic Strategies to Unravel Age-Related Redox Signalling Defects in Skeletal Muscle. Free Radic. Biol. Med. 2019, 132, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Place, N.; Ivarsson, N.; Venckunas, T.; Neyroud, D.; Brazaitis, M.; Cheng, A.J.; Ochala, J.; Kamandulis, S.; Girard, S.; Volungevičius, G.; et al. Ryanodine Receptor Fragmentation and Sarcoplasmic Reticulum Ca2+ Leak after One Session of High-Intensity Interval Exercise. Proc. Natl. Acad. Sci. USA 2015, 112, 15492–15497. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Hamadeh, M.J.; Kaczor, J.J.; Raha, S.; deBeer, J.; Tarnopolsky, M.A. Aberrant Mitochondrial Homeostasis in the Skeletal Muscle of Sedentary Older Adults. PLoS ONE 2010, 5, e10778. [Google Scholar] [CrossRef]
- MacMillan-Crow, L.A.; Crow, J.P.; Kerby, J.D.; Beckman, J.S.; Thompson, J.A. Nitration and Inactivation of Manganese Superoxide Dismutase in Chronic Rejection of Human Renal Allografts. Proc. Natl. Acad. Sci. USA 1996, 93, 11853–11858. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide Dismutases An Adaptation to a Paramagnetic Gas. J. Biol. Chem. 1989, 264, 7761–7764. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Moustafa, A.-E.A.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Davison, G.W. Exercise and Oxidative Damage in Nucleoid DNA Quantified Using Single Cell Gel Electrophoresis: Present and Future Application. Front. Physiol. 2016, 7, 249. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R.; Azqueta, A. DNA Repair as a Biomarker in Human Biomonitoring Studies; Further Applications of the Comet Assay. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2012, 736, 122–129. [Google Scholar] [CrossRef]
- Sykora, P.; Witt, K.L.; Revanna, P.; Smith-Roe, S.L.; Dismukes, J.; Lloyd, D.G.; Engelward, B.P.; Sobol, R.W. Next Generation High Throughput DNA Damage Detection Platform for Genotoxic Compound Screening. Sci. Rep. 2018, 8, 2771. [Google Scholar] [CrossRef]
- Li, W.; Sancar, A. Methodologies for Detecting Environmentally Induced DNA Damage and Repair. Environ. Mol. Mutagen. 2020, 61, 664–679. [Google Scholar] [CrossRef]
- Figueroa-González, G.; Pérez-Plasencia, C. Strategies for the Evaluation of DNA Damage and Repair Mechanisms in Cancer. Oncol. Lett. 2017, 13, 3982–3988. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.; Meyer, J.N.; Mandavilli, B.S.; Houten, B.V. DNA Repair Protocols, Mammalian Systems. Methods Mol. Biol. 2006, 314, 183–199. [Google Scholar] [CrossRef]
- Williamson, J.; Hughes, C.M.; Burke, G.; Davison, G.W. A Combined γ-H2AX and 53BP1 Approach to Determine the DNA Damage-Repair Response to Exercise in Hypoxia. Free Radic. Biol. Med. 2020, 154, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Ravanat, J.-L.; Guicherd, P.; Tuce, Z.; Cadet, J. Simultaneous Determination of Five Oxidative DNA Lesions in Human Urine. Chem. Res. Toxicol. 1999, 12, 802–808. [Google Scholar] [CrossRef]
- Taghizadeh, K.; McFaline, J.L.; Pang, B.; Sullivan, M.; Dong, M.; Plummer, E.; Dedon, P.C. Quantification of DNA Damage Products Resulting from Deamination, Oxidation and Reaction with Products of Lipid Peroxidation by Liquid Chromatography Isotope Dilution Tandem Mass Spectrometry. Nat. Protoc. 2008, 3, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Zatopek, K.M.; Potapov, V.; Maduzia, L.L.; Alpaslan, E.; Chen, L.; Evans, T.C.; Ong, J.L.; Ettwiller, L.M.; Gardner, A.F. RADAR-Seq: A RAre DAmage and Repair Sequencing Method for Detecting DNA Damage on a Genome-Wide Scale. DNA Repair 2019, 80, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Biernacka, A.; Pardo, B.; Dojer, N.; Forey, R.; Skrzypczak, M.; Fongang, B.; Nde, J.; Yousefi, R.; Pasero, P.; et al. QDSB-Seq Is a General Method for Genome-Wide Quantification of DNA Double-Strand Breaks Using Sequencing. Nat. Commun. 2019, 10, 2313. [Google Scholar] [CrossRef]
- Poetsch, A.R.; Boulton, S.J.; Luscombe, N.M. Genomic Landscape of Oxidative DNA Damage and Repair Reveals Regioselective Protection from Mutagenesis. Genome Biol. 2018, 19, 215. [Google Scholar] [CrossRef]
- Rodriguez, R.; Krishnan, Y. The Chemistry of Next-Generation Sequencing. Nat. Biotechnol. 2023, 41, 1709–1715. [Google Scholar] [CrossRef]
- Larsen, E.L.; Karstoft, K.; Poulsen, H.E. Exercise and RNA Oxidation. In Oxidative Eustress in Exercise Physiology; Cobley, J.N., Davison, G.W., Eds.; CRC Press: Boca Raton, FL, USA, 2022; ISBN 9781003051619. [Google Scholar]
- Floyd, R.A.; Watson, J.J.; Harris, J.; West, M.; Wong, P.K. Formation of 8-Hydroxydeoxyguanosine, Hydroxyl Free Radical Adduct of DNA in Granulocytes Exposed to the Tumor Promoter, Tetradeconylphorbolacetate. Biochem. Biophys. Res. Commun. 1986, 137, 841–846. [Google Scholar] [CrossRef]
- Larsen, E.L.; Weimann, A.; Poulsen, H.E. Interventions Targeted at Oxidatively Generated Modifications of Nucleic Acids Focused on Urine and Plasma Markers. Free Radic. Biol. Med. 2019, 145, 256–283. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Jaruga, P.; Rodriguez, H. Identification and Quantification of 8,5′-Cyclo-2′-Deoxy-Adenosine in DNA by Liquid Chromatography/Mass Spectrometry. Free Radic. Biol. Med. 2001, 30, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, H.E.; Weimann, A.; Henriksen, T.; Kjær, L.K.; Larsen, E.L.; Carlsson, E.R.; Christensen, C.K.; Brandslund, I.; Fenger, M. Oxidatively Generated Modifications to Nucleic Acids in Vivo: Measurement in Urine and Plasma. Free Radic. Biol. Med. 2019, 145, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Jacob, H.S.; Jandl, J.H. Effects of Sulfhydryl Inhibition on Red Blood Cells III. Glutathione in the Regulation of the Hexose Monophosphate Pathway. J. Biol. Chem. 1966, 241, 4243–4250. [Google Scholar] [CrossRef] [PubMed]
- Saran, M.; Bors, W. Oxygen Radicals Acting as Chemical Messengers: A Hypothesis. Free Radic. Res. Commun. 1989, 7, 213–220. [Google Scholar] [CrossRef] [PubMed]
- GUTTERIDGE, J.M.C.; HALLIWELL, B. Free Radicals and Antioxidants in the Year 2000: A Historical Look to the Future. Ann. N. Y. Acad. Sci. 2000, 899, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.S.; Donato, A.J.; Uberoi, A.; Wray, D.W.; Lawrenson, L.; Nishiyama, S.; Bailey, D.M. Exercise-Induced Brachial Artery Vasodilation: Role of Free Radicals. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1516–H1522. [Google Scholar] [CrossRef] [PubMed]
- Viña, J.; Gimeno, A.; Sastre, J.; Desco, C.; Asensi, M.; Pallardó, F.V.; Cuesta, A.; Ferrero, J.A.; Terada, L.S.; Repine, J.E. Mechanism of Free Radical Production in Exhaustive Exercise in Humans and Rats; Role of Xanthine Oxidase and Protection by Allopurinol. IUBMB Life 2000, 49, 539–544. [Google Scholar] [CrossRef]
- Gómez-Cabrera, M.-C.; Pallardó, F.V.; Sastre, J.; Viña, J.; García-del-Moral, L. Allopurinol and Markers of Muscle Damage Among Participants in the Tour de France. JAMA 2003, 289, 2503–2504. [Google Scholar] [CrossRef]
- Khassaf, M.; McArdle, A.; Esanu, C.; Vasilaki, A.; McArdle, F.; Griffiths, R.D.; Brodie, D.A.; Jackson, M.J. Effect of Vitamin C Supplements on Antioxidant Defence and Stress Proteins in Human Lymphocytes and Skeletal Muscle. J. Physiol. 2003, 549, 645–652. [Google Scholar] [CrossRef]
- Ristow, M.; Zarse, K.; Oberbach, A.; Klöting, N.; Birringer, M.; Kiehntopf, M.; Stumvoll, M.; Kahn, C.R.; Blüher, M. Antioxidants Prevent Health-Promoting Effects of Physical Exercise in Humans. Proc. Natl. Acad. Sci. USA 2009, 106, 8665–8670. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cabrera, M.; Borrás, C.; Pallardó, F.V.; Sastre, J.; Ji, L.L.; Viña, J. Decreasing Xanthine Oxidase-mediated Oxidative Stress Prevents Useful Cellular Adaptations to Exercise in Rats. J. Physiol. 2005, 567, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cabrera, M.-C.; Domenech, E.; Romagnoli, M.; Arduini, A.; Borras, C.; Pallardo, F.V.; Sastre, J.; Viña, J. Oral Administration of Vitamin C Decreases Muscle Mitochondrial Biogenesis and Hampers Training-Induced Adaptations in Endurance Performance. Am. J. Clin. Nutr. 2008, 87, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Redox Basis of Exercise Physiology. Redox Biol. 2020, 35, 101499. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.A.; Trewin, A.J.; Parker, L.; Wadley, G.D. Antioxidant Supplements and Endurance Exercise: Current Evidence and Mechanistic Insights. Redox Biol. 2020, 35, 101471. [Google Scholar] [CrossRef] [PubMed]
- Henriquez-Olguin, C.; Meneses-Valdes, R.; Jensen, T.E. Compartmentalized Muscle Redox Signals Controlling Exercise Metabolism—Current State, Future Challenges. Redox Biol. 2020, 35, 101473. [Google Scholar] [CrossRef]
- Pregel, M.J.; Storer, A.C. Active Site Titration of the Tyrosine Phosphatases SHP-1 and PTP1B Using Aromatic Disulfides Reaction with the Essential Cysteine Residue in the Active Site. J. Biol. Chem. 1997, 272, 23552–23558. [Google Scholar] [CrossRef]
- Salmeen, A.; Andersen, J.N.; Myers, M.P.; Meng, T.-C.; Hinks, J.A.; Tonks, N.K.; Barford, D. Redox Regulation of Protein Tyrosine Phosphatase 1B Involves a Sulphenyl-Amide Intermediate. Nature 2003, 423, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Allison, W.S. Formation and Reactions of Sulfenic Acids in Proteins. Acc. Chem. Res. 1976, 9, 293–299. [Google Scholar] [CrossRef]
- Lennicke, C.; Cochemé, H.M. Redox Metabolism: ROS as Specific Molecular Regulators of Cell Signaling and Function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Parvez, S.; Long, M.J.C.; Poganik, J.R.; Aye, Y. Redox Signaling by Reactive Electrophiles and Oxidants. Chem. Rev. 2018, 118, 8798–8888. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Hampton, M.B. Thiol Chemistry and Specificity in Redox Signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohé, R.; Flohé, L. Basic Principles and Emerging Concepts in the Redox Control of Transcription Factors. Antioxid. Redox Signal. 2011, 15, 2335–2381. [Google Scholar] [CrossRef] [PubMed]
- Antunes, F.; Brito, P.M. Quantitative Biology of Hydrogen Peroxide Signaling. Redox Biol. 2017, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen Peroxide Sensing, Signaling and Regulation of Transcription Factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging. Cell 2020, 180, 968–983.e24. [Google Scholar] [CrossRef]
- Leichert, L.I.; Gehrke, F.; Gudiseva, H.V.; Blackwell, T.; Ilbert, M.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Jakob, U. Quantifying Changes in the Thiol Redox Proteome upon Oxidative Stress in Vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 8197–8202. [Google Scholar] [CrossRef]
- Day, N.J.; Gaffrey, M.J.; Qian, W.-J. Stoichiometric Thiol Redox Proteomics for Quantifying Cellular Responses to Perturbations. Antioxidants 2021, 10, 499. [Google Scholar] [CrossRef]
- Li, X.; Gluth, A.; Zhang, T.; Qian, W. Thiol Redox Proteomics: Characterization of Thiol-based Post-translational Modifications. Proteomics 2023, 23, e2200194. [Google Scholar] [CrossRef]
- Kim, H.; Ha, S.; Lee, H.Y.; Lee, K. ROSics: Chemistry and Proteomics of Cysteine Modifications in Redox Biology. Mass Spectrom. Rev. 2015, 34, 184–208. [Google Scholar] [CrossRef]
- Yang, J.; Carroll, K.S.; Liebler, D.C. The Expanding Landscape of the Thiol Redox Proteome. Mol. Cell. Proteom. 2016, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The Cysteine Proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Derks, J.; Leduc, A.; Wallmann, G.; Huffman, R.G.; Willetts, M.; Khan, S.; Specht, H.; Ralser, M.; Demichev, V.; Slavov, N. Increasing the Throughput of Sensitive Proteomics by PlexDIA. Nat. Biotechnol. 2023, 41, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Sinitcyn, P.; Richards, A.L.; Weatheritt, R.J.; Brademan, D.R.; Marx, H.; Shishkova, E.; Meyer, J.G.; Hebert, A.S.; Westphall, M.S.; Blencowe, B.J.; et al. Global Detection of Human Variants and Isoforms by Deep Proteome Sequencing. Nat. Biotechnol. 2023, 41, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Held, J.M.; Danielson, S.R.; Behring, J.B.; Atsriku, C.; Britton, D.J.; Puckett, R.L.; Schilling, B.; Campisi, J.; Benz, C.C.; Gibson, B.W. Targeted Quantitation of Site-Specific Cysteine Oxidation in Endogenous Proteins Using a Differential Alkylation and Multiple Reaction Monitoring Mass Spectrometry Approach. Mol. Cell. Proteom. 2010, 9, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.A.; Cobley, J.N.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Koch, L.G.; Britton, S.L.; Burniston, J.G. Label-Free LC-MS Profiling of Skeletal Muscle Reveals Heart-Type Fatty Acid Binding Protein as a Candidate Biomarker of Aerobic Capacity. Proteomes 2013, 1, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Boivin, B.; Zhang, S.; Arbiser, J.L.; Zhang, Z.-Y.; Tonks, N.K. A Modified Cysteinyl-Labeling Assay Reveals Reversible Oxidation of Protein Tyrosine Phosphatases in Angiomyolipoma Cells. Proc Natl. Acad. Sci. USA 2008, 105, 9959–9964. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Madhani, M.; Cuello, F.; Charles, R.L.; Brennan, J.P.; Schröder, E.; Browning, D.D.; Eaton, P. Cysteine Redox Sensor in PKGIa Enables Oxidant-Induced Activation. Science 2007, 317, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Makmura, L.; Hamann, M.; Areopagita, A.; Furuta, S.; Muoz, A.; Momand, J. Development of a Sensitive Assay to Detect Reversibly Oxidized Protein Cysteine Sulfhydryl Groups. Antioxid. Redox Signal. 2001, 3, 1105–1118. [Google Scholar] [CrossRef]
- van Leeuwen, L.A.G.; Hinchy, E.C.; Murphy, M.P.; Robb, E.L.; Cochemé, H.M. Click-PEGylation—A Mobility Shift Approach to Assess the Redox State of Cysteines in Candidate Proteins. Free Radic. Biol. Med. 2017, 108, 374–382. [Google Scholar] [CrossRef]
- Cobley, J.N.; Noble, A.; Jimenez-Fernandez, E.; Moya, M.-T.V.; Guille, M.; Husi, H. Catalyst-Free Click PEGylation Reveals Substantial Mitochondrial ATP Synthase Sub-Unit Alpha Oxidation before and after Fertilisation. Redox Biol. 2019, 26, 101258. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Oviosu, O.; Eaton, P. The PEG-Switch Assay: A Fast Semi-Quantitative Method to Determine Protein Reversible Cysteine Oxidation. J. Pharmacol. Toxicol. 2013, 68, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Chang, G. Quantitative Display of the Redox Status of Proteins with Maleimide-polyethylene Glycol Tagging. Electrophoresis 2019, 40, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.; Noble, A.; Bessell, R.; Guille, M.; Husi, H. Reversible Thiol Oxidation Inhibits the Mitochondrial ATP Synthase in Xenopus Laevis Oocytes. Antioxidants 2020, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N. Oxiforms: Unique Cysteine Residue- and Chemotype-specified Chemical Combinations Can Produce Functionally-distinct Proteoforms. Bioessays 2023, 45, 2200248. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, E.L.; Traustadóttir, T. Aerobic Exercise Training Partially Reverses the Impairment of Nrf2 Activation in Older Humans. Free Radic. Biol. Med. 2020, 160, 418–432. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, E.L.; Berry, S.R.; Traustadóttir, T. Effects of Exercise Training on Redox Stress Resilience in Young and Older Adults. Adv. Redox Res. 2021, 2, 100007. [Google Scholar] [CrossRef]
- Ostrom, E.L.; Valencia, A.P.; Marcinek, D.J.; Traustadóttir, T. High Intensity Muscle Stimulation Activates a Systemic Nrf2-Mediated Redox Stress Response. Free Radic. Biol. Med. 2021, 172, 82–89. [Google Scholar] [CrossRef]
- Cobley, J.N.; Moult, P.R.; Burniston, J.G.; Morton, J.P.; Close, G.L. Exercise Improves Mitochondrial and Redox-Regulated Stress Responses in the Elderly: Better Late than Never! Biogerontology 2015, 16, 249–264. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-Related Factor 2 (Nrf2), a NF-E2-like Basic Leucine Zipper Transcriptional Activator That Binds to the Tandem NF-E2/AP1 Repeat of the Beta-Globin Locus Control Region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct Evidence That Sulfhydryl Groups of Keap1 Are the Sensors Regulating Induction of Phase 2 Enzymes That Protect against Carcinogens and Oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef] [PubMed]
- Reisz, J.A.; Bechtold, E.; King, S.B.; Poole, L.B.; Furdui, C.M. Thiol-blocking Electrophiles Interfere with Labeling and Detection of Protein Sulfenic Acids. FEBS J. 2013, 280, 6150–6161. [Google Scholar] [CrossRef] [PubMed]
- Schilling, D.; Barayeu, U.; Steimbach, R.R.; Talwar, D.; Miller, A.K.; Dick, T.P. Commonly Used Alkylating Agents Limit Persulfide Detection by Converting Protein Persulfides into Thioethers. Angew. Chem. Int. Ed. 2022, 61, e202203684. [Google Scholar] [CrossRef] [PubMed]
- Timp, W.; Timp, G. Beyond Mass Spectrometry, the next Step in Proteomics. Sci. Adv. 2020, 6, eaax8978. [Google Scholar] [CrossRef] [PubMed]
- Kramer, P.A.; Duan, J.; Gaffrey, M.J.; Shukla, A.K.; Wang, L.; Bammler, T.K.; Qian, W.-J.; Marcinek, D.J. Fatiguing Contractions Increase Protein S-Glutathionylation Occupancy in Mouse Skeletal Muscle. Redox Biol. 2018, 17, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L. Tissue Sulfhydryl Groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Saurin, A.T.; Neubert, H.; Brennan, J.P.; Eaton, P. Widespread Sulfenic Acid Formation in Tissues in Response to Hydrogen Peroxide. Proc. Natl. Acad. Sci. USA 2004, 101, 17982–17987. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.; Guille, M.; Cobley, J.N. ALISA: A Microplate Assay to Measure Protein Thiol Redox State. Free Radic. Biol. Med. 2021, 174, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Tuncay, A.; Noble, A.; Guille, M.; Cobley, J.N. RedoxiFluor: A Microplate Technique to Quantify Target-Specific Protein Thiol Redox State in Relative Percentage and Molar Terms. Free Radic. Biol. Med. 2022, 181, 118–129. [Google Scholar] [CrossRef]
- Cobley, J.N. Chapter 23—How exercise induces oxidative eustress. In Oxidative Stress; Academic Press: Cambridge, MA, USA, 2020; pp. 447–462. [Google Scholar] [CrossRef]
- Muggeridge, D.J.; Crabtree, D.R.; Tuncay, A.; Megson, I.L.; Davison, G.; Cobley, J.N. Exercise Decreases PP2A-Specific Reversible Thiol Oxidation in Human Erythrocytes: Implications for Redox Biomarkers. Free Radic. Biol. Med. 2022, 182, 73–78. [Google Scholar] [CrossRef]
- Tuncay, A.; Crabtree, D.R.; Muggeridge, D.J.; Husi, H.; Cobley, J.N. Performance Benchmarking Microplate-Immunoassays for Quantifying Target-Specific Cysteine Oxidation Reveals Their Potential for Understanding Redox-Regulation and Oxidative Stress. Free Radic. Biol. Med. 2023, 204, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Talwar, D.; Miller, C.G.; Grossmann, J.; Szyrwiel, L.; Schwecke, T.; Demichev, V.; Drazic, A.-M.M.; Mayakonda, A.; Lutsik, P.; Veith, C.; et al. The GAPDH Redox Switch Safeguards Reductive Capacity and Enables Survival of Stressed Tumour Cells. Nat. Metab. 2023, 5, 660–676. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, C.A.; Delfinis, L.J.; Hughes, M.C.; Turnbull, P.C.; Gandhi, S.; DiBenedetto, S.N.; Rahman, F.A.; Tadi, P.; Amaral, C.A.; Dehghani, A.; et al. Mitochondrial Creatine Sensitivity Is Lost in the D2.Mdx Model of Duchenne Muscular Dystrophy and Rescued by the Mitochondrial-Enhancing Compound Olesoxime. Am. J. Physiol. Cell Physiol. 2023, 324, C1141–C1157. [Google Scholar] [CrossRef]
- Alcock, L.J.; Perkins, M.V.; Chalker, J.M. Chemical Methods for Mapping Cysteine Oxidation. Chem. Soc. Rev. 2017, 47, 231–268. [Google Scholar] [CrossRef]
- Shi, Y.; Carroll, K.S. Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation. Acc. Chem. Res. 2020, 53, 20–31. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Fu, L.; Jung, Y.; Yang, J.; Carroll, K.S. Reaction-Based Fluorogenic Probes for Detecting Protein Cysteine Oxidation in Living Cells. Nat. Commun. 2022, 13, 5522. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Methner, C.; Nadtochiy, S.M.; Logan, A.; Pell, V.R.; Ding, S.; James, A.M.; Cochemé, H.M.; Reinhold, J.; Lilley, K.S.; et al. Cardioprotection by S-Nitrosation of a Cysteine Switch on Mitochondrial Complex I. Nat. Med. 2013, 19, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Burger, N.; James, A.M.; Mulvey, J.F.; Hoogewijs, K.; Ding, S.; Fearnley, I.M.; Loureiro-López, M.; Norman, A.A.I.; Arndt, S.; Mottahedin, A.; et al. ND3 Cys39 in Complex I Is Exposed during Mitochondrial Respiration. Cell Chem. Biol. 2022, 29, 636–649.e14. [Google Scholar] [CrossRef] [PubMed]
- Egan, B.; Sharples, A.P. Molecular Responses to Acute Exercise and Their Relevance for Adaptations in Skeletal Muscle to Exercise Training. Physiol. Rev. 2023, 103, 2057–2170. [Google Scholar] [CrossRef]
- Egan, B.; Zierath, J.R. Exercise Metabolism and the Molecular Regulation of Skeletal Muscle Adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Bartlett, J.D.; Kayani, A.; Murray, S.W.; Louhelainen, J.; Donovan, T.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; et al. PGC-1α Transcriptional Response and Mitochondrial Adaptation to Acute Exercise Is Maintained in Skeletal Muscle of Sedentary Elderly Males. Biogerontology 2012, 13, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, E.; Lang, L.; Zimmermann, J.; Luczak, M.; Kiefer, A.M.; Niedner-Schatteburg, G.; Manolikakes, G.; Morgan, B.; Deponte, M. Glutathione Kinetically Outcompetes Reactions between Dimedone and a Cyclic Sulfenamide or Physiological Sulfenic Acids. Free Radic. Biol. Med. 2023, 208, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Davies, M.J.; Krämer, A.C.; Miotto, G.; Zaccarin, M.; Zhang, H.; Ursini, F. Protein Cysteine Oxidation in Redox Signaling: Caveats on Sulfenic Acid Detection and Quantification. Arch. Biochem. Biophys. 2017, 617, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Stöcker, S.; Laer, K.V.; Mijuskovic, A.; Dick, T.P. The Conundrum of Hydrogen Peroxide Signaling and the Emerging Role of Peroxiredoxins as Redox Relay Hubs. Antioxid. Redox Signal. 2018, 28, 558–573. [Google Scholar] [CrossRef] [PubMed]
- Anschau, V.; Ferrer-Sueta, G.; Aleixo-Silva, R.L.; Fernandes, R.B.; Tairum, C.A.; Tonoli, C.C.C.; Murakami, M.T.; de Oliveira, M.A.; Netto, L.E.S. Reduction of Sulfenic Acids by Ascorbate in Proteins, Connecting Thiol-Dependent to Alternative Redox Pathways. Free Radic. Biol. Med. 2020, 156, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Computational Systems Biology. Nature 2002, 420, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Mogilner, A.; Wollman, R.; Marshall, W.F. Quantitative Modeling in Cell Biology: What Is It Good For? Dev. Cell 2006, 11, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J. Why Model? J. Artif. Soc. Soc. Simul. 2008, 11, 12. [Google Scholar]
- Wolkenhauer, O. Why Model? Front. Physiol. 2014, 5, 21. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Chatzinikolaou, P.N.; Chatzinikolaou, A.N.; Paschalis, V.; Theodorou, A.A.; Vrabas, I.S.; Kyparos, A.; Nikolaidis, M.G. The Redox Signal: A Physiological Perspective. IUBMB Life 2022, 74, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Buettner, G.R. Superoxide Dismutase in Redox Biology: The Roles of Superoxide and Hydrogen Peroxide. Anti-Cancer Agents Med. Chem. 2011, 11, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Salvador, A.; Antunes, A. How Gradients and Microdomains Determine H2O2 Redox Signalling. In Peroxiporins: Redox Signal Mediators in and between Cells, 1st ed.; Medrano-Fren, I., Bienert, G.P., Sitia, R., Eds.; CRC Press: Boca Raton, FL, USA, 2023; ISBN 9781003160649. [Google Scholar]
- Salvador, A. Pillars of Theoretical Biology: “Biochemical Systems Analysis, I, II and III”. J. Theor. Biol. 2024, 576, 111655. [Google Scholar] [CrossRef] [PubMed]
- Held, J.M. Redox Systems Biology: Harnessing the Sentinels of the Cysteine Redoxome. Antioxid. Redox Signal. 2020, 32, 659–676. [Google Scholar] [CrossRef] [PubMed]
- Pillay, C.S.; Hofmeyr, J.-H.; Mashamaite, L.N.; Rohwer, J.M. From Top-Down to Bottom-Up: Computational Modeling Approaches for Cellular Redoxin Networks. Antioxid. Redox Signal. 2013, 18, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.R. A Tutorial on the Diffusibility and Reactivity of Free Nitric Oxide. Nitric Oxide 1997, 1, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.B.; Langford, T.F.; Huang, B.K.; Deen, W.M.; Sikes, H.D. A Reaction-Diffusion Model of Cytosolic Hydrogen Peroxide. Free Radic. Biol. Med. 2016, 90, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.B.; Huang, B.K.; Deen, W.M.; Sikes, H.D. Analysis of the Lifetime and Spatial Localization of Hydrogen Peroxide Generated in the Cytosol Using a Reduced Kinetic Model. Free Radic. Biol. Med. 2015, 89, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.K.; Sikes, H.D. Quantifying Intracellular Hydrogen Peroxide Perturbations in Terms of Concentration. Redox Biol. 2014, 2, 955–962. [Google Scholar] [CrossRef]
- Langford, T.F.; Deen, W.M.; Sikes, H.D. A Mathematical Analysis of Prx2-STAT3 Disulfide Exchange Rate Constants for a Bimolecular Reaction Mechanism. Free Radic. Biol. Med. 2018, 120, 239–245. [Google Scholar] [CrossRef]
- Lim, C.H.; Dedon, P.C.; Deen, W.M. Kinetic Analysis of Intracellular Concentrations of Reactive Nitrogen Species. Chem. Res. Toxicol. 2008, 21, 2134–2147. [Google Scholar] [CrossRef] [PubMed]
- Travasso, R.D.M.; dos Aidos, F.S.; Bayani, A.; Abranches, P.; Salvador, A. Localized Redox Relays as a Privileged Mode of Cytoplasmic Hydrogen Peroxide Signaling. Redox Biol. 2017, 12, 233–245. [Google Scholar] [CrossRef] [PubMed]
- TSOUKIAS, N.M. Nitric Oxide Bioavailability in the Microcirculation: Insights from Mathematical Models. Microcirculation 2008, 15, 813–834. [Google Scholar] [CrossRef] [PubMed]
- Orrico, F.; Möller, M.N.; Cassina, A.; Denicola, A.; Thomson, L. Kinetic and Stoichiometric Constraints Determine the Pathway of H2O2 Consumption by Red Blood Cells. Free Radic. Biol. Med. 2018, 121, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Brito, P.M.; Antunes, F. Estimation of Kinetic Parameters Related to Biochemical Interactions between Hydrogen Peroxide and Signal Transduction Proteins. Front. Chem. 2014, 2, 82. [Google Scholar] [CrossRef] [PubMed]
- Pillay, C.S.; Eagling, B.D.; Driscoll, S.R.E.; Rohwer, J.M. Quantitative Measures for Redox Signaling. Free Radic. Biol. Med. 2016, 96, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Sousa, T.; Gouveia, M.; Travasso, R.D.M.; Salvador, A. How Abundant Are Superoxide and Hydrogen Peroxide in the Vasculature Lumen, How Far Can They Reach? Redox Biol. 2022, 58, 102527. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Hampton, M.B.; Livesey, J.H.; Kettle, A.J. Modeling the Reactions of Superoxide and Myeloperoxidase in the Neutrophil Phagosome Implications For Microbial Killing. J. Biol. Chem. 2006, 281, 39860–39869. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Phillips, R. Cell Biology by the Numbers, 2nd ed.; Garland Science: New York, NY, USA, 2006. [Google Scholar]
- Phillips, R.; Kondev, J.; Theriot, J.; Garcia, H. Physical Biology of the Cell, 2nd ed.; Garland Science: New York, NY, USA, 2012. [Google Scholar]
- Nikolaidis, M.; Margaritelis, N.; Matsakas, A. Quantitative Redox Biology of Exercise. Int. J. Sports Med. 2020, 41, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Chatzinikolaou, P.N.; Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Vrabas, I.S.; Kyparos, A.; D’Alessandro, A.; Nikolaidis, M.G. Erythrocyte Metabolism. Acta Physiol. 2024, 240, e14081. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Cobley, J.N.; Paschalis, V.; Veskoukis, A.S.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Principles for Integrating Reactive Species into in Vivo Biological Processes: Examples from Exercise Physiology. Cell. Signal. 2016, 28, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Michailidis, Y.; Jamurtas, A.Z.; Nikolaidis, M.G.; Fatouros, I.G.; Koutedakis, Y.; Papassotiriou, I.; Kouretas, D. Sampling Time Is Crucial for Measurement of Aerobic Exercise-Induced Oxidative Stress. Med. Sci. Sports Exerc. 2007, 39, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Jordan, A.C.; Perry, C.G.R.; Cheng, A.J. Promoting a Pro-Oxidant State in Skeletal Muscle: Potential Dietary, Environmental, and Exercise Interventions for Enhancing Endurance-Training Adaptations. Free Radic. Biol. Med. 2021, 176, 189–202. [Google Scholar] [CrossRef]
- Cobley, J.N.; Sakellariou, G.K.; Murray, S.; Waldron, S.; Gregson, W.; Burniston, J.G.; Morton, J.P.; Iwanejko, L.A.; Close, G.L. Lifelong Endurance Training Attenuates Age-Related Genotoxic Stress in Human Skeletal Muscle. Longev. Heal. 2013, 2, 11. [Google Scholar] [CrossRef]
- Murphy, M.P.; Hartley, R.C. Mitochondria as a Therapeutic Target for Common Pathologies. Nat. Rev. Drug Discov. 2018, 17, 865–886. [Google Scholar] [CrossRef]
- Murphy, M.P. Antioxidants as Therapies: Can We Improve on Nature? Free Radic. Biol. Med. 2014, 66, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.J.; Porteous, C.M.; Gane, A.M.; Murphy, M.P. Delivery of Bioactive Molecules to Mitochondria in Vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 5407–5412. [Google Scholar] [CrossRef]
- Cobley, J.N.; McGlory, C.; Morton, J.P.; Close, G.L. N-Acetylcysteine’s Attenuation of Fatigue After Repeated Bouts of Intermittent Exercise: Practical Implications for Tournament Situations. Int. J. Sport Nutr. Exerc. Metab. 2011, 21, 451–461. [Google Scholar] [CrossRef]
- Reid, M.B. Free Radicals and Muscle Fatigue: Of ROS, Canaries, and the IOC. Free Radic. Biol. Med. 2008, 44, 169–179. [Google Scholar] [CrossRef]
- Giustarini, D.; Milzani, A.; Dalle-Donne, I.; Rossi, R. How to Increase Cellular Glutathione. Antioxidants 2023, 12, 1094. [Google Scholar] [CrossRef]
- Paschalis, V.; Theodorou, A.A.; Margaritelis, N.V.; Kyparos, A.; Nikolaidis, M.G. N-Acetylcysteine Supplementation Increases Exercise Performance and Reduces Oxidative Stress Only in Individuals with Low Levels of Glutathione. Free Radic. Biol. Med. 2018, 115, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.; Symons, M.C.R. Sulphur Radicals Formed by Cutting α-Keratin. Nature 1987, 328, 833–834. [Google Scholar] [CrossRef] [PubMed]
- Zapp, C.; Obarska-Kosinska, A.; Rennekamp, B.; Kurth, M.; Hudson, D.M.; Mercadante, D.; Barayeu, U.; Dick, T.P.; Denysenkov, V.; Prisner, T.; et al. Mechanoradicals in Tensed Tendon Collagen as a Source of Oxidative Stress. Nat. Commun. 2020, 11, 2315. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.P.; Ganini, D. Immuno-Spin Trapping of Macromolecules Free Radicals in Vitro and in Vivo—One Stop Shopping for Free Radical Detection. Free Radic. Biol. Med. 2019, 131, 318–331. [Google Scholar] [CrossRef]
- Ezeriņa, D.; Takano, Y.; Hanaoka, K.; Urano, Y.; Dick, T.P. N-Acetyl Cysteine Functions as a Fast-Acting Antioxidant by Triggering Intracellular H2S and Sulfane Sulfur Production. Cell Chem. Biol. 2018, 25, 447–459.e4. [Google Scholar] [CrossRef] [PubMed]
- Pedre, B.; Barayeu, U.; Ezeriņa, D.; Dick, T.P. The Mechanism of Action of N-Acetylcysteine (NAC): The Emerging Role of H2S and Sulfane Sulfur Species. Pharmacol. Ther. 2021, 228, 107916. [Google Scholar] [CrossRef]
- Kalyanaraman, B. NAC, NAC, Knockin’ on Heaven’s Door: Interpreting the Mechanism of Action of N-Acetylcysteine in Tumor and Immune Cells. Redox Biol. 2022, 57, 102497. [Google Scholar] [CrossRef]
- Zivanovic, J.; Kouroussis, E.; Kohl, J.B.; Adhikari, B.; Bursac, B.; Schott-Roux, S.; Petrovic, D.; Miljkovic, J.L.; Thomas-Lopez, D.; Jung, Y.; et al. Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration. Cell Metab. 2019, 30, 1152–1170.e13. [Google Scholar] [CrossRef]
- Huang, J.; Co, H.K.; Lee, Y.; Wu, C.; Chen, S. Multistability Maintains Redox Homeostasis in Human Cells. Mol. Syst. Biol. 2021, 17, e10480. [Google Scholar] [CrossRef]
- Raftos, J.E.; Whillier, S.; Kuchel, P.W. Glutathione Synthesis and Turnover in the Human Erythrocyte Alignment of a Model Based on Detailed Enzyme Kinetics with Experimental Data. J. Biol. Chem. 2010, 285, 23557–23567. [Google Scholar] [CrossRef]
- Lyons, J.; Rauh-Pfeiffer, A.; Yu, Y.M.; Lu, X.-M.; Zurakowski, D.; Tompkins, R.G.; Ajami, A.M.; Young, V.R.; Castillo, L. Blood Glutathione Synthesis Rates in Healthy Adults Receiving a Sulfur Amino Acid-Free Diet. Proc. Natl. Acad. Sci. USA 2000, 97, 5071–5076. [Google Scholar] [CrossRef] [PubMed]
- Burgunder, J.M.; Varriale, A.; Lauterburg, B.H. Effect of N-Acetylcysteine on Plasma Cysteine and Glutathione Following Paracetamol Administration. Eur. J. Clin. Pharmacol. 1989, 36, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.G.; Margaritelis, N.V. Free Radicals and Antioxidants: Appealing to Magic. Trends Endocrinol. Metab. 2023, 34, 503–504. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Peskin, A.V.; Kleffmann, T.; Radi, R.; Pace, P.E. Carbon Dioxide/Bicarbonate Is Required for Sensitive Inactivation of Mammalian Glyceraldehyde-3-Phosphate Dehydrogenase by Hydrogen Peroxide. Proc. Natl. Acad. Sci. 2023, 120, e2221047120. [Google Scholar] [CrossRef] [PubMed]
- Radi, R. Interplay of Carbon Dioxide and Peroxide Metabolism in Mammalian Cells. J. Biol. Chem. 2022, 298, 102358. [Google Scholar] [CrossRef] [PubMed]
- Dagnell, M.; Cheng, Q.; Rizvi, S.H.M.; Pace, P.E.; Boivin, B.; Winterbourn, C.C.; Arnér, E.S.J. Bicarbonate Is Essential for Protein-Tyrosine Phosphatase 1B (PTP1B) Oxidation and Cellular Signaling through EGF-Triggered Phosphorylation Cascades. J. Biol. Chem. 2019, 294, 12330–12338. [Google Scholar] [CrossRef] [PubMed]
- Cobley, J.N.; Ab. Malik, Z.; Morton, J.P.; Close, G.L.; Edwards, B.J.; Burniston, J.G. Age- and Activity-Related Differences in the Abundance of Myosin Essential and Regulatory Light Chains in Human Muscle. Proteomes 2016, 4, 15. [Google Scholar] [CrossRef]
- Vasileiadou, O.; Nastos, G.G.; Chatzinikolaou, P.N.; Papoutsis, D.; Vrampa, D.I.; Methenitis, S.; Margaritelis, N.V. Redox Profile of Skeletal Muscles: Implications for Research Design and Interpretation. Antioxidants 2023, 12, 1738. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, F.A.; Thielert, M.; Mann, M. Making Single-Cell Proteomics Biologically Relevant. Nat. Methods 2023, 20, 320–323. [Google Scholar] [CrossRef]
- Glover, M.R.; Davies, M.J.; Fuentes-Lemus, E. Oxidation of the Active Site Cysteine Residue of Glyceraldehyde-3-Phosphate Dehydrogenase to the Hyper-Oxidized Sulfonic Acid Form Is Favored under Crowded Conditions. Free Radic. Biol. Med. 2024, 212, 1–9. [Google Scholar] [CrossRef]
- Henriquez-Olguin, C.; Meneses-Valdes, R.; Kritsiligkou, P.; Fuentes-Lemus, E. From Workout to Molecular Switches: How Does Skeletal Muscle Produce, Sense, and Transduce Subcellular Redox Signals? Free Radic. Biol. Med. 2023, 209, 355–365. [Google Scholar] [CrossRef]
- Fuentes-Lemus, E.; Davies, M.J. Effect of Crowding, Compartmentalization and Nanodomains on Protein Modification and Redox Signaling—Current State and Future Challenges. Free Radic. Biol. Med. 2023, 196, 81–92. [Google Scholar] [CrossRef]
- Dai, Y.; Chamberlayne, C.F.; Messina, M.S.; Chang, C.J.; Zare, R.N.; You, L.; Chilkoti, A. Interface of Biomolecular Condensates Modulates Redox Reactions. Chem 2023, 9, 1594–1609. [Google Scholar] [CrossRef] [PubMed]
- Kritsiligkou, P.; Bosch, K.; Shen, T.K.; Meurer, M.; Knop, M.; Dick, T.P. Proteome-Wide Tagging with an H2O2 Biosensor Reveals Highly Localized and Dynamic Redox Microenvironments. Proc. Natl. Acad. Sci. USA 2023, 120, e2314043120. [Google Scholar] [CrossRef]
- Margaritelis, N.V. Personalized Redox Biology: Designs and Concepts. Free Radic. Biol. Med. 2023, 208, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J. An Update on Methods and Approaches for Interrogating Mitochondrial Reactive Oxygen Species Production. Redox Biol. 2021, 45, 102044. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Hsieh, C.-J.; Li, S.; Lee, H.; Graham, T.J.; Xu, K.; Weng, C.-C.; Doot, R.K.; Chu, W.; Chakraborty, S.K.; et al. Development of a Positron Emission Tomography Radiotracer for Imaging Elevated Levels of Superoxide in Neuroinflammation. ACS Chem. Neurosci. 2018, 9, 578–586. [Google Scholar] [CrossRef]
- Zielonka, J.; Kalyanaraman, B. Hydroethidine- and MitoSOX-Derived Red Fluorescence Is Not a Reliable Indicator of Intracellular Superoxide Formation: Another Inconvenient Truth. Free Radic. Biol. Med. 2010, 48, 983–1001. [Google Scholar] [CrossRef]
- Milne, G.L.; Nogueira, M.S.; Gao, B.; Sanchez, S.C.; Amin, W.; Thomas, S.; Oger, C.; Galano, J.-M.; Murff, H.J.; Yang, G.; et al. Identification of Novel F2-Isoprostane Metabolites by Specific UDP-Glucuronosyltransferases. Redox Biol. 2024, 70, 103020. [Google Scholar] [CrossRef]
- Hughes, A.J.; Spelke, D.P.; Xu, Z.; Kang, C.-C.; Schaffer, D.V.; Herr, A.E. Single-Cell Western Blotting. Nat. Methods 2014, 11, 749–755. [Google Scholar] [CrossRef]
- Liu, Y.; Herr, A.E. DropBlot: Single-Cell Western Blotting of Chemically Fixed Cancer Cells. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Hie, B.L.; Shanker, V.R.; Xu, D.; Bruun, T.U.J.; Weidenbacher, P.A.; Tang, S.; Wu, W.; Pak, J.E.; Kim, P.S. Efficient Evolution of Human Antibodies from General Protein Language Models. Nat. Biotechnol. 2024, 42, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Krause, B.; Greene, E.R.; Subramanian, S.; Mohr, B.P.; Holton, J.M.; Olmos, J.L.; Xiong, C.; Sun, Z.Z.; Socher, R.; et al. Large Language Models Generate Functional Protein Sequences across Diverse Families. Nat. Biotechnol. 2023, 41, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Biochemistry of Oxidative Stress. Angew. Chem. Int. Ed. Engl. 1986, 25, 1058–1071. [Google Scholar] [CrossRef]
- Adhikari, S.; Nice, E.C.; Deutsch, E.W.; Lane, L.; Omenn, G.S.; Pennington, S.R.; Paik, Y.-K.; Overall, C.M.; Corrales, F.J.; Cristea, I.M.; et al. A High-Stringency Blueprint of the Human Proteome. Nat. Commun. 2020, 11, 5301. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.S.; Ahn, S.B.; Mohamedali, A.; Islam, M.T.; Cantor, D.; Verhaert, P.D.; Fanayan, S.; Sharma, S.; Nice, E.C.; Connor, M.; et al. Accelerating the Search for the Missing Proteins in the Human Proteome. Nat. Commun. 2017, 8, 14271. [Google Scholar] [CrossRef] [PubMed]
- Carbonara, K.; Andonovski, M.; Coorssen, J.R. Proteomes Are of Proteoforms: Embracing the Complexity. Proteomes 2021, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Martin-Baniandres, P.; Lan, W.-H.; Board, S.; Romero-Ruiz, M.; Garcia-Manyes, S.; Qing, Y.; Bayley, H. Enzyme-Less Nanopore Detection of Post-Translational Modifications within Long Polypeptides. Nat. Nanotechnol. 2023, 18, 1335–1340. [Google Scholar] [CrossRef]
- Yu, L.; Kang, X.; Li, F.; Mehrafrooz, B.; Makhamreh, A.; Fallahi, A.; Foster, J.C.; Aksimentiev, A.; Chen, M.; Wanunu, M. Unidirectional Single-File Transport of Full-Length Proteins through a Nanopore. Nat. Biotechnol. 2023, 41, 1130–1139. [Google Scholar] [CrossRef]
- Brinkerhoff, H.; Kang, A.S.W.; Liu, J.; Aksimentiev, A.; Dekker, C. Multiple Rereads of Single Proteins at Single–Amino Acid Resolution Using Nanopores. Science 2021, 374, 1509–1513. [Google Scholar] [CrossRef]
- Bleier, L.; Wittig, I.; Heide, H.; Steger, M.; Brandt, U.; Dröse, S. Generator-Specific Targets of Mitochondrial Reactive Oxygen Species. Free Radic. Biol. Med. 2015, 78, 1–10. [Google Scholar] [CrossRef] [PubMed]

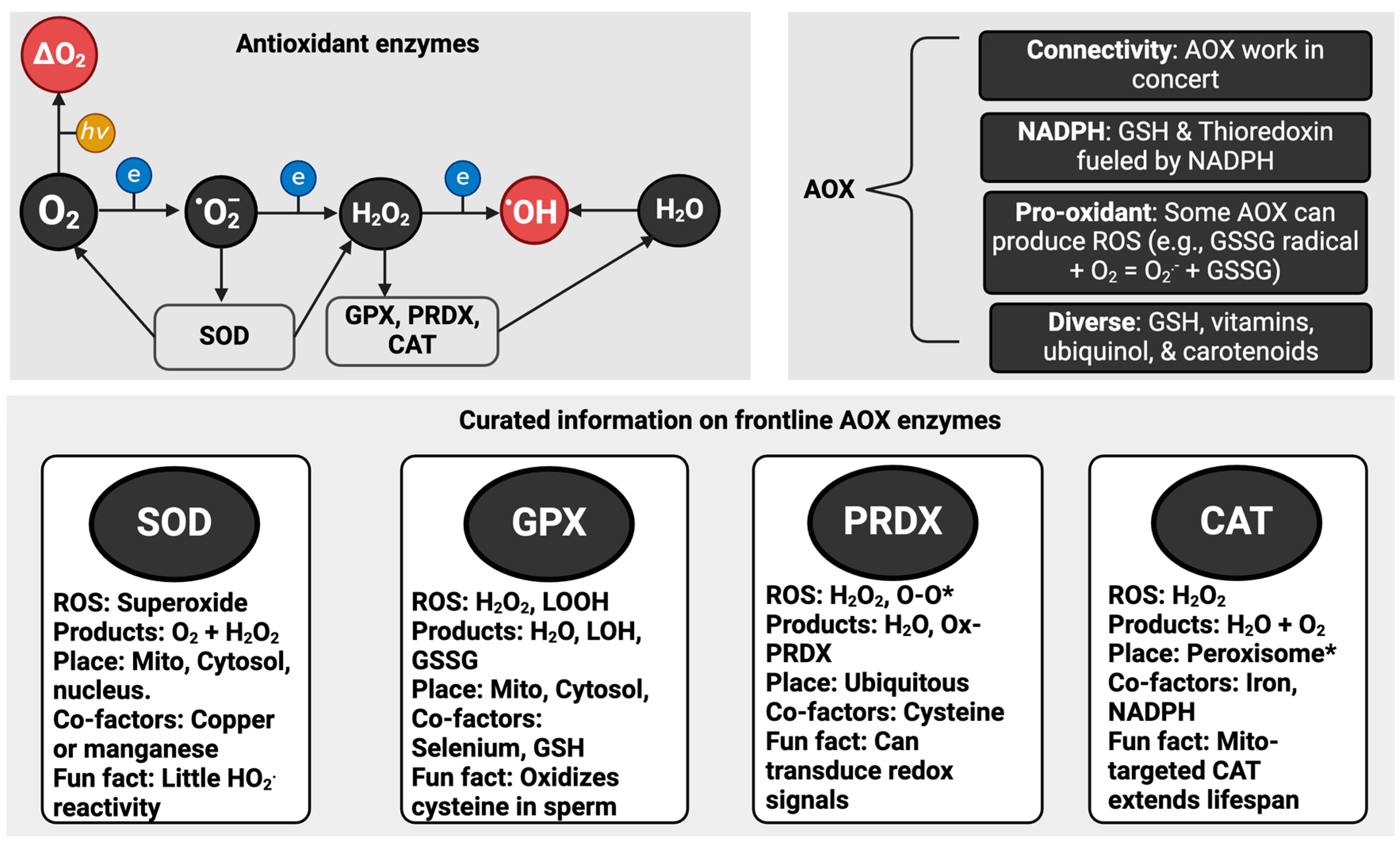
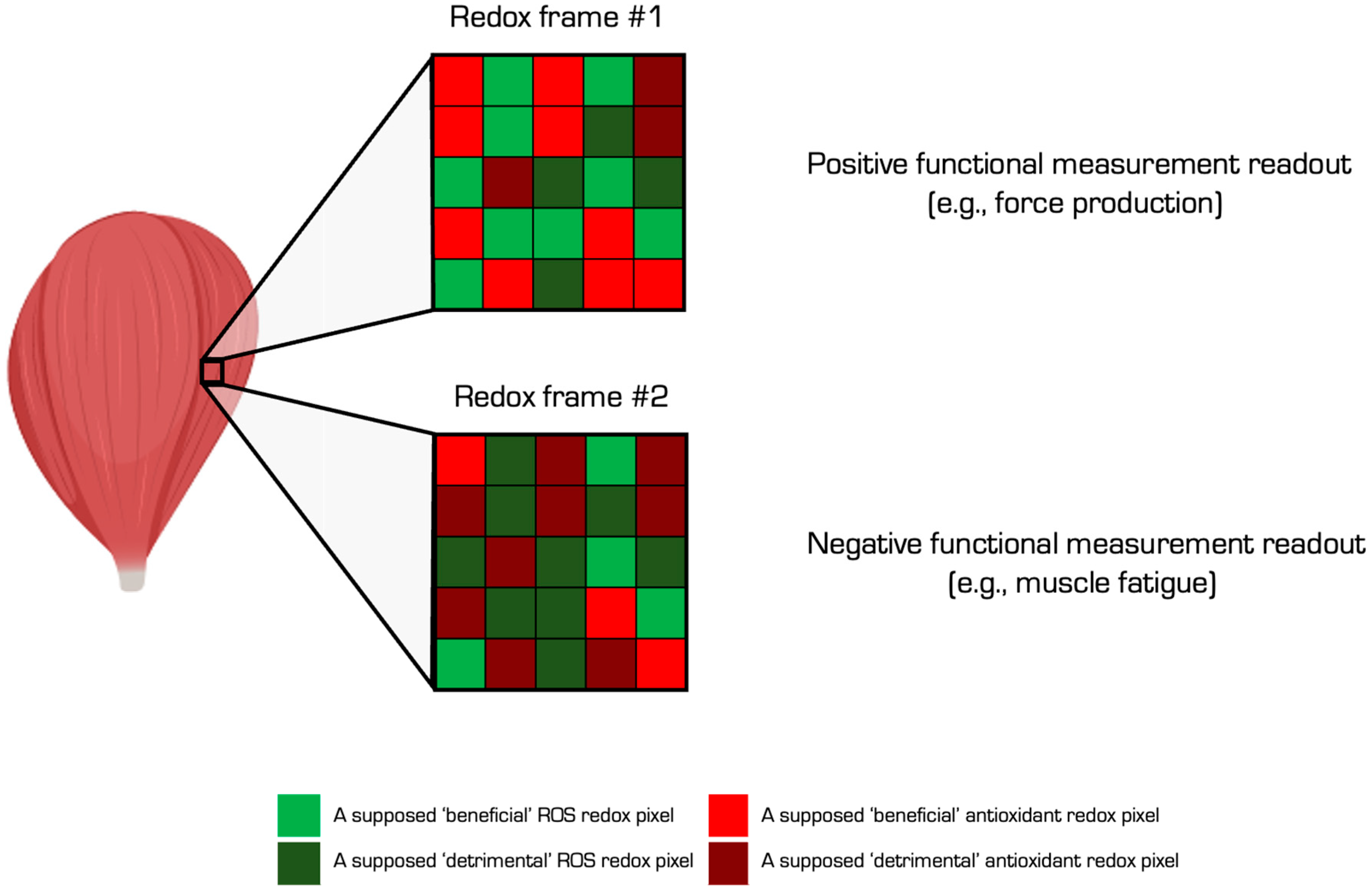

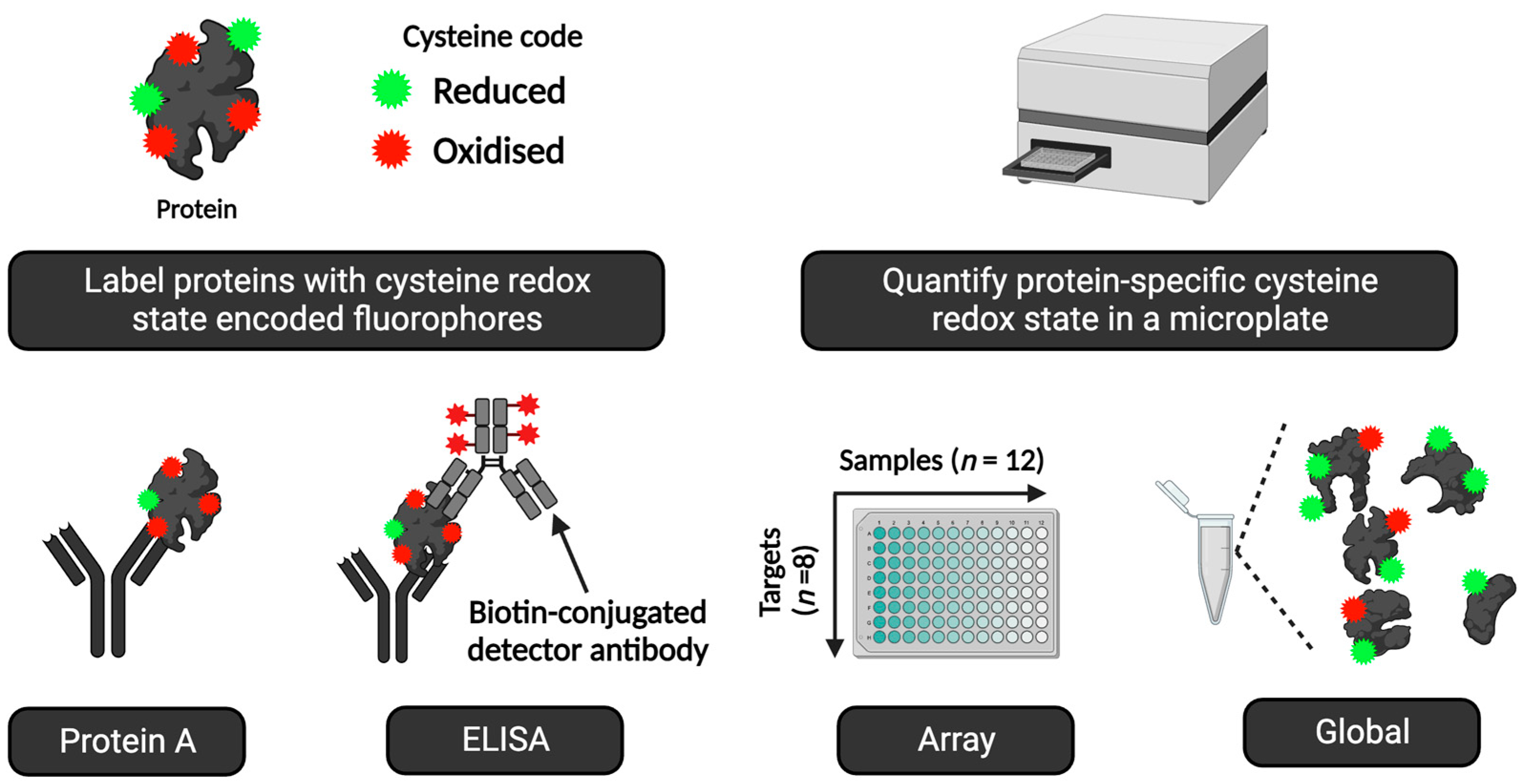

| Term | Definition | Example |
|---|---|---|
| Free radical | A molecule with one or more unpaired valence electrons that is capable of independent existence. | Hydroxyl radical |
| Non-radical | A spin-paired molecule devoid of unpaired electrons. | Hydrogen peroxide |
| ROS | Free radical and non-radical oxygen-derived reactive species. | Superoxide |
| Reactive species | A catch-all term for all free radical and non-radical reactive species that includes oxygen-, sulphur-, nitrogen-, and carbon-atom-centred species. | Peroxynitrite (sum of the protonated and deprotonated forms) |
| Reductant | A molecule that donates one or more electrons to a redox reaction partner. | NADPH donating a hydride to reduce oxidise glutathione reductase |
| Oxidant | A molecule that takes one or more electrons from a redox reaction partner. | Hydroxyl radical oxidising L-cysteine to a thiyl radical |
| Antioxidant | Any substance that delays, prevents, or removes oxidative damage to a target molecule. A substance that reacts with an oxidant to regulate its reactions with other targets, thus influencing redox-dependent biological signalling pathways and/or oxidative damage. | Vitamin E preventing lipid peroxidation by reducing an alkoxyl radical to a hydroperoxide CAT metabolising hydrogen peroxide to prevent reaction with other targets, such as a transition metal ion |
| Oxidative stress | An imbalance between ROS and antioxidants leading to disrupted redox regulation and/or oxidative damage. | See below |
| Oxidative eustress | A beneficial imbalance between ROS and antioxidants leading to disrupted redox regulation and/or oxidative damage. | Specific cysteine oxidation patterns and oxidative macromolecule damage in an acute-exercise context |
| Oxidative distress | A deleterious imbalance between ROS and antioxidants leading to disrupted redox regulation and/or oxidative damage. | Aberrant and potentially random patterns of cysteine oxidation and oxidative damage in a musculoskeletal ageing context |
| n | Name | Description |
|---|---|---|
| 1 | Enzyme activity | The basis of modern SOD assays [127]—intercepting xanthine oxidase-induced superoxide before it reduces a reporter molecule like cytochrome C—was in place before SOD was discovered! [21]. Many valid microplate and gel-based SOD assays exist. They can be used to quantify SOD activity and, with appropriate controls, discern between isoforms [128,129,130]. Glutathione peroxidase (GPX) activity and CAT activity can be assessed by using established assays [31,131]. Meanwhile, PRDX and thioredoxin activities can be inferred via non-reducing immunoblotting. Assays to quantify thioredoxin reductase or glutathione reductase activity are available [132,133]. |
| 2 | Surrogate markers | Antioxidant enzyme activity can be inferred by using surrogate markers [4]. For example, PRDX1 activity can reflect the phosphorylation state of the enzyme [134]. Other useful post-translational surrogates include manganese SOD acetylation state, disulphide bond formation in copper zinc SOD, and succinylation of GPX4 [135]. Measuring the concentration of a protein by immunoblotting or ELISA, in combination with other markers, can be used to infer a change in antioxidant activity [136]. |
| 3 | Glutathione | High-performance liquid chromatography (HPLC)-based methods are available for measuring reduced (GSH), oxidised (GSSG), and total (GSH + GSSG) glutathione [37,137]. Although HPLC is preferred, plate- and kit-based assays are available. Controls (see cheat code 8) can prevent artificial cysteine oxidation [138]. |
| 4 | Low-molecular-weight molecules | HPLC-based methods can measure the concentration of low-molecular-weight antioxidants like vitamin C [139]. EPR spectrometry can measure vitamin C and E radicals to provide information on their redox activity in human samples [140]. |
| n | Name | Description |
|---|---|---|
| 1 | Fluorescence | Single-cell gel electrophoresis (SCGE), or the comet assay, is a technically simple and sensitive method for quantifying single- and double-stranded DNA breaks in mononuclear cells. The SCGE assay embeds cells on agarose gel, and if supercoiling is relaxed due to a single or double break, the loop of the DNA is free to migrate during electrophoresis and thus form a comet tail [202]. The SCGE assay can resolve damage up to ~3 breaks per 109 Da [203]. Several variations in the SCGE assay exist. As oxidation of DNA can occur at a similar rate to strand breaks, base oxidation can be examined via a simple SCGE modification using lesion-specific enzymes (e.g., formamidopyrimidine DNA glycosylase) to accurately capture oxidised purines (guanine/adenine) and/or pyrimidines (thymine/cytosine). The assay may be combined with fluorescent in situ hybridisation (FISH) to detect whole-genome, telomeric and centrometric DNA, and gene region-specific DNA damage [202]. CometChip technology is now used to minimise sample-to-sample variation [204]. |
| 2 | ELISA | ELISA application is a popular immunological method to measure DNA damage, mainly in the form of 8-oxodG. There are several commercially available kit-based options; however, caution is paramount regarding assay specificity and reliability. |
| 3 | Molecular approach | Polymerase chain reaction (PCR) or quantitative PCR (qPCR) can map nuclear and mitochondrial DNA damage at nucleotide resolution [205]. In PCR, DNA amplification is stalled at the damaged site via the blocking of the progression of Taq polymerase, which ultimately reduces the number of DNA templates that are devoid of any Taq-blocked lesions. qPCR quantifies DNA damage on both duplex strands. It is possible to quantitatively detect and analyse gene-specific DNA damage (and repair) by using qPCR and with only 1–2 ng of total genomic DNA. qPCR has caveats: it depends on high-molecular-weight DNA, well-defined qPCR conditions, and the intricate calculation of lesion frequencies [206,207]. Long-Amplicon Quantitative PCR (LA-qPCR) can also provide an overview of total genomic mitochondrial DNA damage [166]. Following double-stranded DNA damage, a repair response is usually initiated, where the subsequent phosphorylation of Serine-139 of histone H2AX ensues [208]. The γH2AX assay is relatively simple to execute and is based on immunofluorescence using a specific Serine-139-γH2AX antibody to show the location in the chromatin foci at the sites of DNA damage. |
| 4 | Analytical approach | HPLC coupled with tandem MS is reliable in detecting oxidative DNA damage (e.g., 8-hydroxy-2′-deoxyguanosine) with excellent separation of nucleosides. HPLC-MS yields robust information on the location of DNA damage, but high assay cost and required extensive experience can preclude assay use. Gas chromatography coupled with mass spectrometry (GC/MS) is highly sensitive to the detection of several forms of DNA damage, including those of the sugar moiety and four heterocyclic bases (e.g., 8-oxodG, 5-HMUra, 8-oxoAde, 5-OHUra, and 8-oxoGua) [209]. Although the technique provides impressive structural data in complex samples (such as the detection of a single DNA lesion in DNA with several lesions), the quantification of nucleoside forms of base damage is not as robust compared with liquid chromatography-based methods [210]. |
| 5 | Sequencing | Innovative next-generation sequencing technology now exists, providing high-throughput and high accuracy DNA sequence data. RADAR-Seq [211], qDSB-Seq [212], and AP-Seq [213] are typical examples of sequencing-based technologies designed to quantify and map DNA damage on a genome-wide scale. This approach determines the precise gene locations of DNA damage. Interestingly, the quenching of fluorophores on account of the low redox potential of guanine had to be addressed before next-generation sequencing technologies could be developed [214]. |
| Type | Benefits | Description (Useful Properties as Applicable) |
|---|---|---|
| ELISA | Throughput Multiplexed Sensitive Rapid | High-sample n-plex analysis (adds statistical power) Parallel assessment of multiple 2–10 proteins (enables screening) Picomole sensitivity (supports human biomarker studies) Performed in 1 day with minimal hands-on time (benefits screens) |
| Redox | Cysteine holistic Percentages Moles Context Chemotype Process sensitive | Agnostic of any one cysteine residue (adds coverage of the entire molecule) Quantifies cysteine redox state in percentages (interpretationally useful) Quantifies cysteine redox state in moles (interpretationally useful) Provides cysteine proteome context (interpretationally useful) Supports chemotype-specific analysis (supports mechanistic studies) Results are sensitive to oxidative and antioxidative processes scaled across every cysteine residue on the target protein (interpretationally useful) |
| Performance | Valid Effective Accurate Reliable Reproducible Range | Draws on highly principled redox and immunological methods (robust) They work (e.g., compare to Click-PEG) (means to study the specific protein) Data correspond to ground-truth standards (adds percentage analysis) High consistency between samples (adds robustness) Delivers consistent results (adds robustness) Operates across a large dynamic range (useful for human applications) |
| Microplate | Simple Easy to perform Off the shelf Automated | Simple to understand, interpret, and operate (supports accessibility) Little technical skill required to deliver actionable results (accessibility) Compatible with commercial ELISA kits (accessibility) Delivers rapid and automated data within seconds (time efficient) |
| Code | Name | Analytical Approaches |
|---|---|---|
| 1 | Avoid the minefield of measuring ROS directly in humans (at least for now) | n/a |
| 2 | How to infer ROS production in human samples by using endogenous reporter molecules | Aconitase assay Peroxiredoxin dimer assay |
| 3 | How to hack “TAC” in human samples | TAC |
| 4 | How to measure antioxidants in human samples | Enzyme assays Surrogate markers Glutathione Low-molecular-weight compounds |
| 5 | How to measure lipid peroxidation in human samples | Lipidomics HPLC-MDA ELISA—F2-isoprostanes Fox assay—LOOH Immunoblot—4-HNE Targeted-protein-specific approach—4-HNE |
| 6 | How to measure protein oxidation in human samples | Proteomics Fluorescent-in-gel carbonylation assay ELISA—protein carbonylation Immunoblot—OxyBlot™ Targeted-protein-specific nitration |
| 7 | How to measure DNA and RNA oxidation in human samples | Fluorescence—comet assay Sequencing—RADAR-Seq Analytical approach—HPLC ELISA—8-oxo-G Molecular approach—QPCR |
| 8 | How to measure redox regulation in human samples | Redox proteomics Immunological assays, such as Click-PEG Outcomes, such as keap1 degradation |
| 9 | How to use redox ELISA technology to measure protein cysteine oxidation in humans | ALISA RedoxiFluor |
| 10 | How to exploit mathematical modelling and computational analyses in redox biology | Mathematical modelling and bioinformatics using appropriate software packages |
| Are You Interested in… | Answer | Refinement | Selection Outcomes | Assay(s) |
|---|---|---|---|---|
| Oxidative stress input or output? | Output | n/a | Consider cheat codes 5–9 Disregard cheat codes 1 and 2 | n/a |
| Reactive species or antioxidant input? | Yes | Interested in an antioxidant (yes) | Consider cheat code 4 Disregard cheat codes 1–3 | n/a |
| Oxidative damage or redox regulation output? | Both | n/a | n/a | n/a |
| A specific oxidative damage output? | Yes | Yes | Consider cheat codes 5–6 Disregard cheat code 7 (based on mechanism) | Global 4-HNE immunoblot Contractile protein immunocapture for targeted 4-HNE analysis |
| A specific cysteine oxidation event? | Yes | Yes | Consider cheat codes 8 and 9 | Gel-based analysis of persulphides using a fluorescent probe |
| Using a specific antioxidant? | Yes | n/a | Consider cheat code 4 | n/a |
| Using an antioxidant with a known mode of action? | Yes | Use mechanism-directed assay | Consider cheat code 4 | HPLC of [NAC] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cobley, J.N.; Margaritelis, N.V.; Chatzinikolaou, P.N.; Nikolaidis, M.G.; Davison, G.W. Ten “Cheat Codes” for Measuring Oxidative Stress in Humans. Antioxidants 2024, 13, 877. https://doi.org/10.3390/antiox13070877
Cobley JN, Margaritelis NV, Chatzinikolaou PN, Nikolaidis MG, Davison GW. Ten “Cheat Codes” for Measuring Oxidative Stress in Humans. Antioxidants. 2024; 13(7):877. https://doi.org/10.3390/antiox13070877
Chicago/Turabian StyleCobley, James N., Nikos V. Margaritelis, Panagiotis N. Chatzinikolaou, Michalis G. Nikolaidis, and Gareth W. Davison. 2024. "Ten “Cheat Codes” for Measuring Oxidative Stress in Humans" Antioxidants 13, no. 7: 877. https://doi.org/10.3390/antiox13070877
APA StyleCobley, J. N., Margaritelis, N. V., Chatzinikolaou, P. N., Nikolaidis, M. G., & Davison, G. W. (2024). Ten “Cheat Codes” for Measuring Oxidative Stress in Humans. Antioxidants, 13(7), 877. https://doi.org/10.3390/antiox13070877








