Extraction Optimization of Quercus cerris L. Wood Chips: A Comparative Study between Full Factorial Design (FFD) and Artificial Neural Network (ANN)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Wood Samples
2.3. Experimental Design
2.4. Extraction Procedure and Yield
2.5. 2,2-Diphenyl-1-Picrylhydrazyl (DPPH) Scavenging Activity Assay
2.6. Ferric Reducing Antioxidant Power (FRAP) Assay
2.7. Measurement of Total Phenol Content (TPC)
2.8. Measurement of Total Flavonoid Content (TFC)
2.9. Measurement of Condensed Tannin Content (CTC)
2.10. Measurement of Hydrolyzable Tannin Content (HTC)
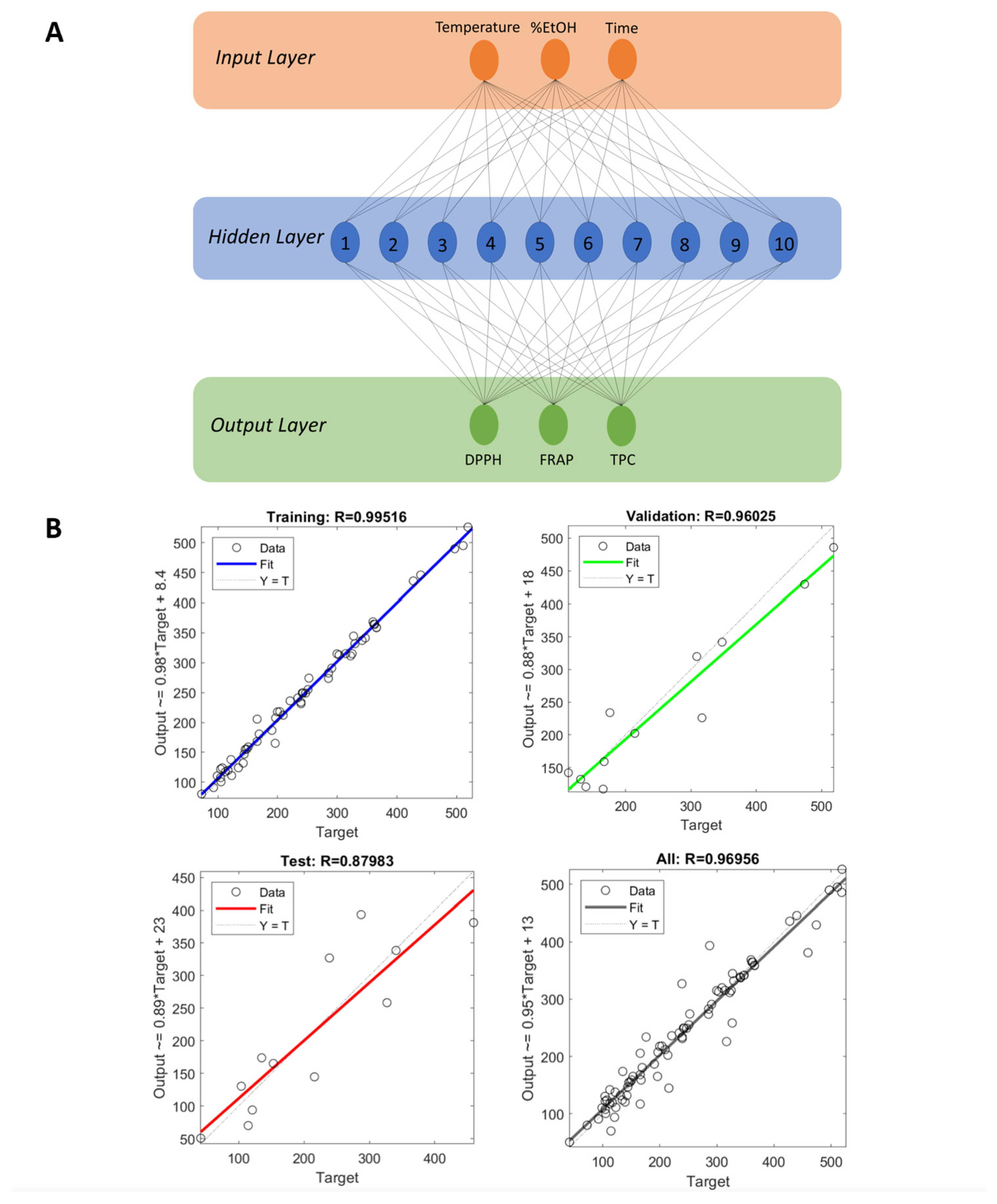
2.11. Artificial Neural Network (ANN) Modeling
2.12. Comparison of FFD and ANN Models Prediction Ability
2.13. Optimization of the Process
2.14. Statistical Analysis
3. Results and Discussion
3.1. Model Adequacy
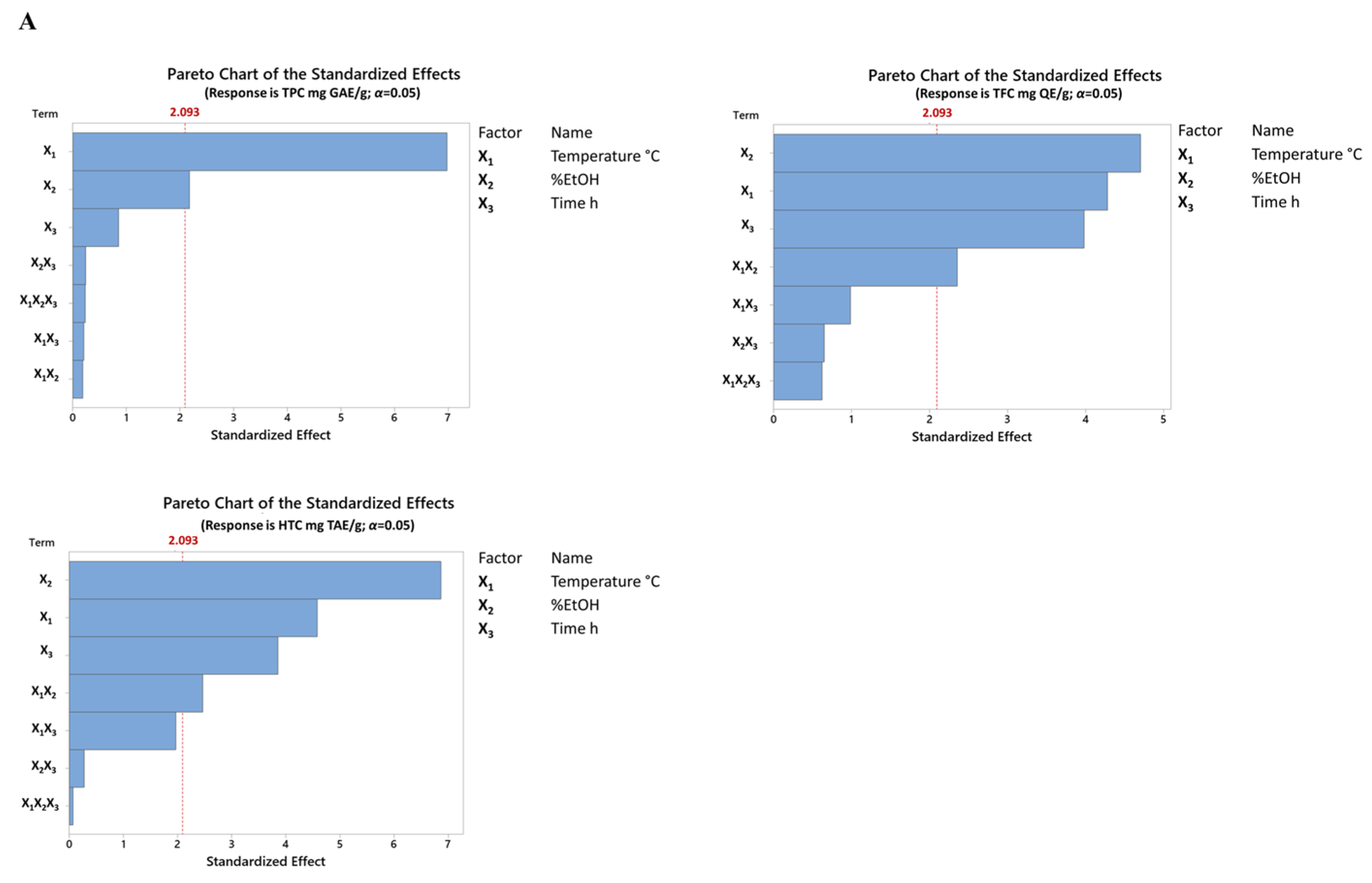
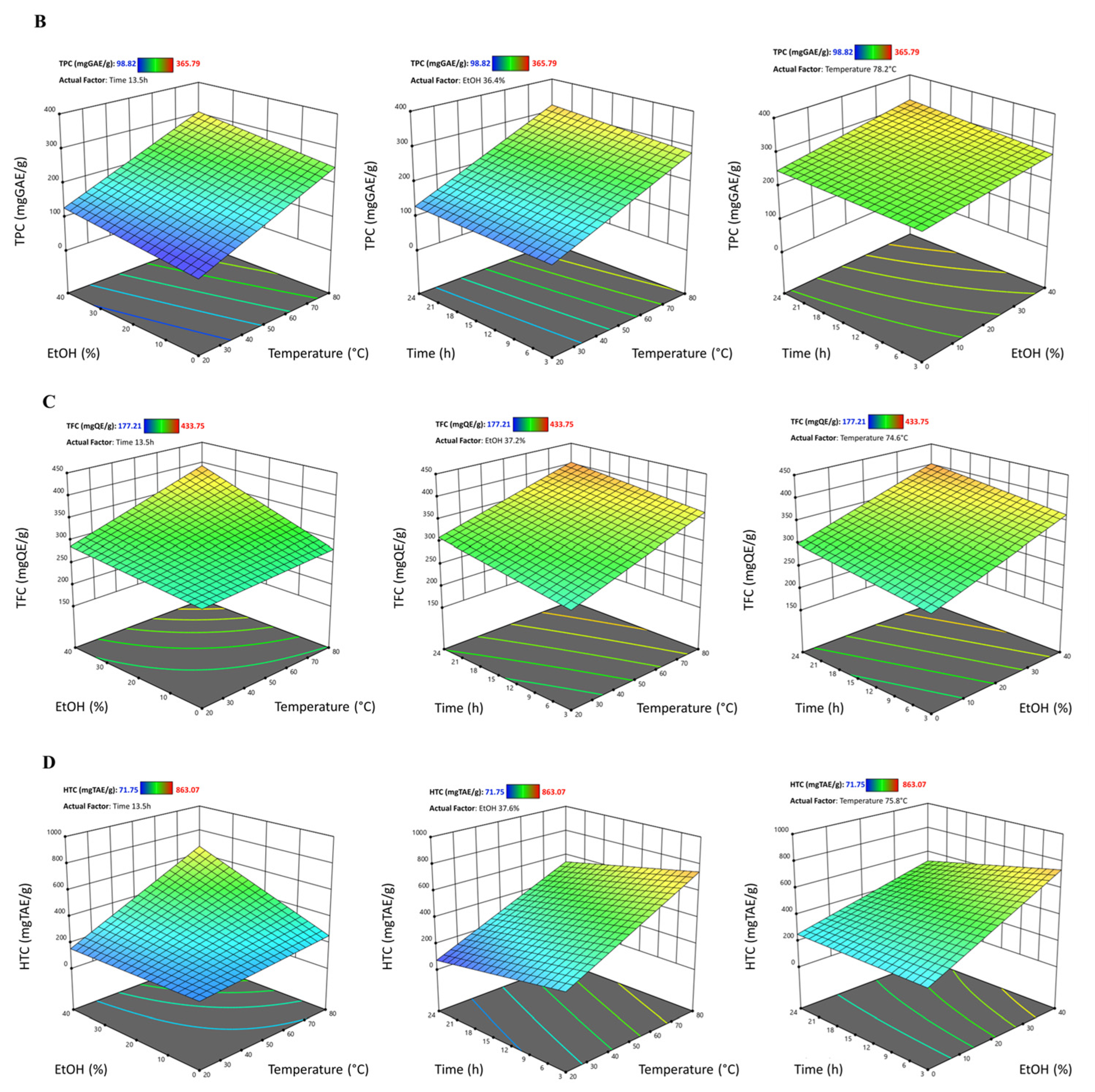
3.2. Effect of Extraction Parameters on Yield
3.3. Effect of Extraction Conditions on the Antioxidant Activity Based FFD
3.4. Effect of Extraction Conditions on Specialized Metabolites Based FFD
3.4.1. Effect on Total Phenolic Content (TPC)
3.4.2. Effect on Total Flavonoid Content (TFC)
3.4.3. Effect Hydrolyzable Tannin Content (HTC)
3.5. Multiple Response Prediction
3.6. Pearson Correlation Existing between the Dependent Variables
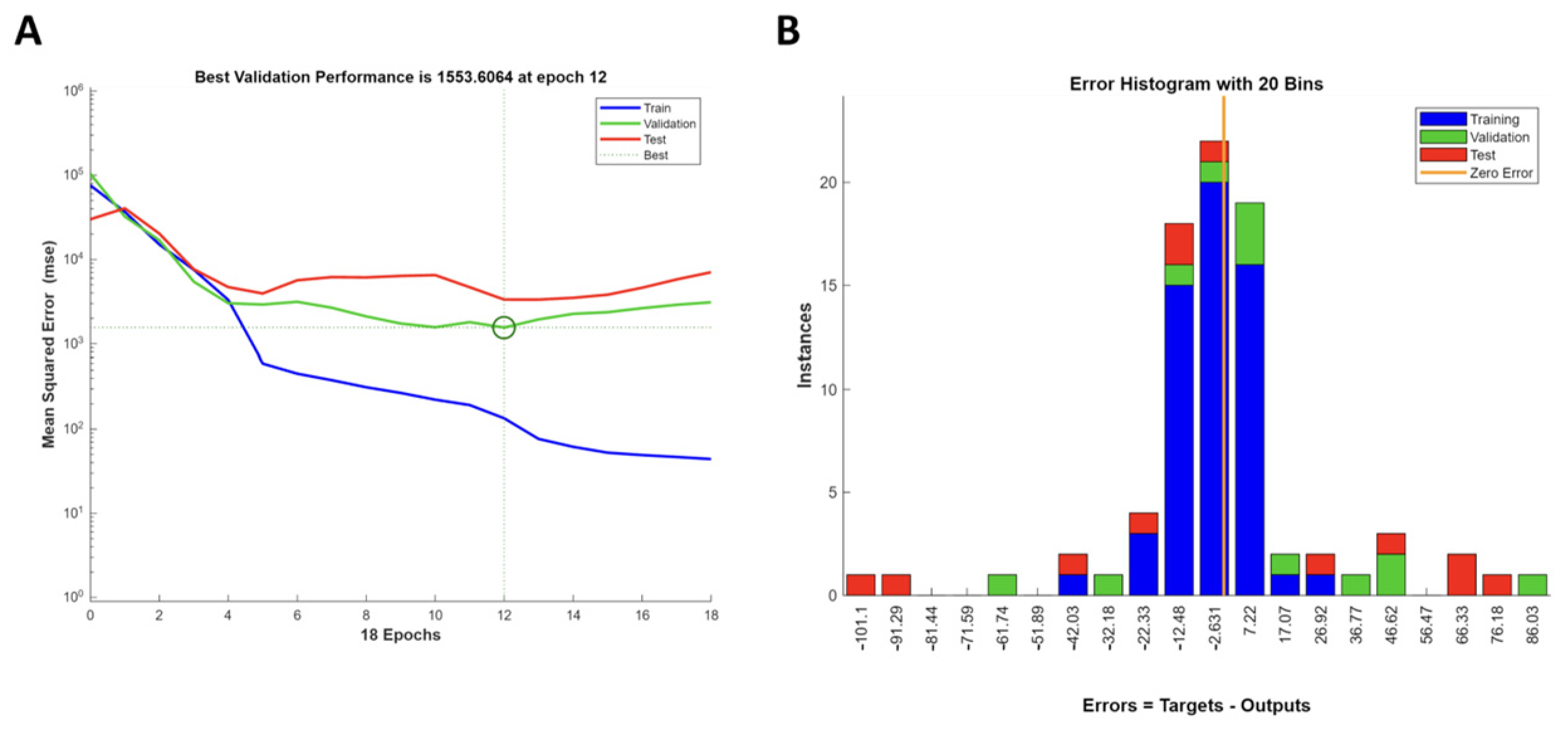
3.7. Artificial Neural Network Model
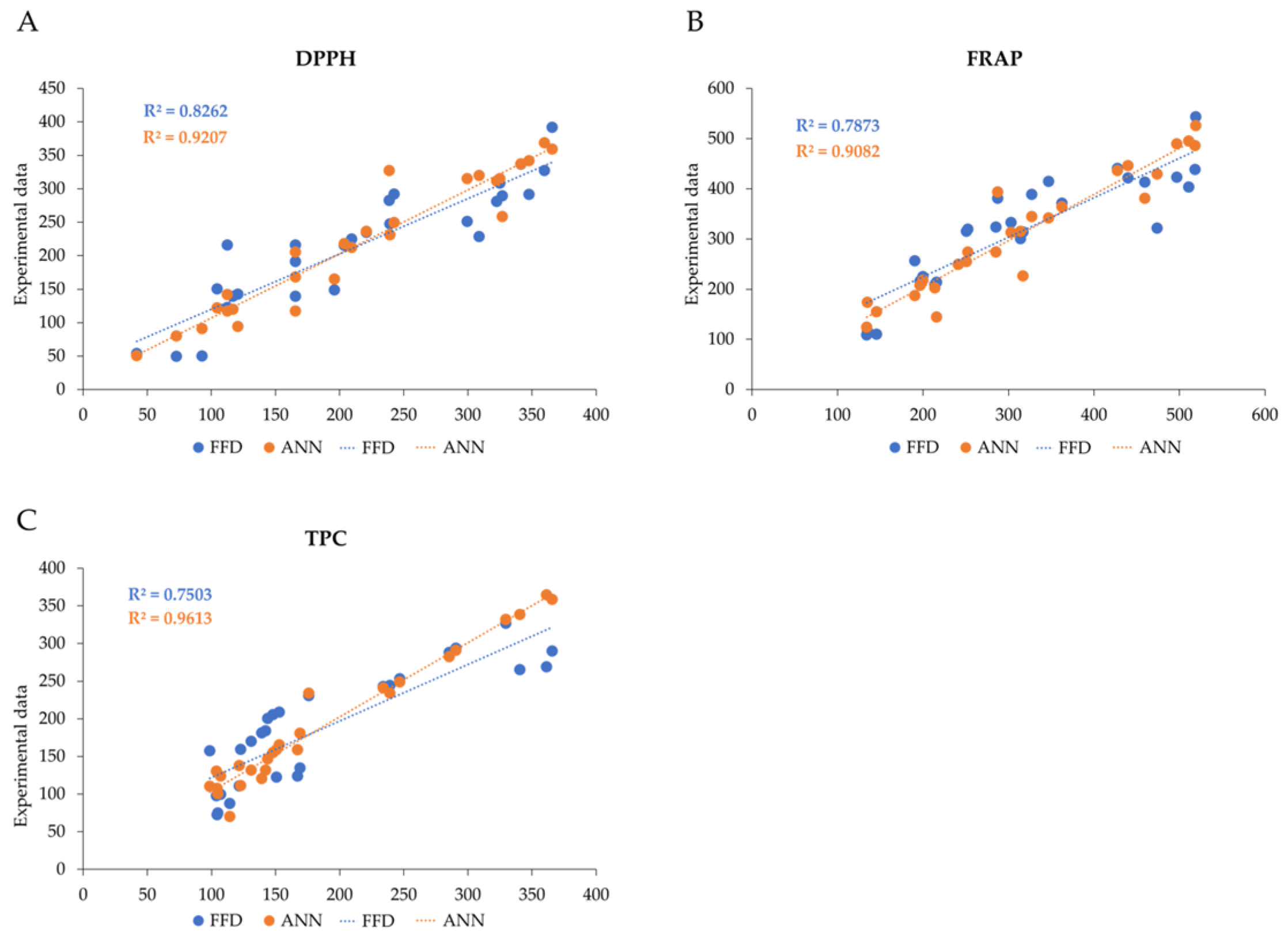
3.8. Comparison of FFD and ANN Models
3.9. Process Optimization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braghiroli, F.L.; Passarini, L. Valorization of biomass residues from forest operations and wood manufacturing presents a wide range of sustainable and innovative possibilities. Curr. For. Rep. 2020, 6, 172–183. [Google Scholar] [CrossRef]
- Bergamasco, S.; Zikeli, F.; Vinciguerra, V.; Sobolev, A.P.; Scarnati, L.; Tofani, G.; Scarascia Mugnozza, G.; Romagnoli, M. Extraction and Characterization of Acidolysis Lignin from Turkey Oak (Quercus cerris L.) and Eucalypt (Eucalyptus camaldulensis Dehnh.) Wood from Population Stands in Italy. Polymers 2023, 15, 3591. [Google Scholar] [CrossRef]
- Ianni, F.; Segoloni, E.; Blasi, F.; Di Maria, F. Low-Molecular-Weight Phenols Recovery by Eco-Friendly Extraction from Quercus Spp. Wastes: An Analytical and Biomass-Sustainability Evaluation. Processes 2020, 8, 387. [Google Scholar] [CrossRef]
- Lavisci, P.; Masson, D.; Deglise, X. Quality of turkey oak (Quercus cerris L.) wood. II. Analysis of some physico-chemical parameters related to its gluability. J. Holzforsch. 1991, 45, 415–418. [Google Scholar] [CrossRef]
- Bajraktari, A. Wood Quality of Quercus cerris from Kosovo; Universidade de Lisboa: Lisbon, Portugal, 2018. [Google Scholar]
- Bernetti, I. Il mercato delle biomasse forestali per scopi energetici: Un modello di offerta. Riv. Di Econ. Agrar. 1998, 53, 1–36. [Google Scholar]
- Bernetti, I.; Fagarazzi, C.; Romano, S. Biomass production as an energy source in coppices of the province of Florence, Italy. In Proceedings of the Atti della Sixth Joint Conference on Food, Agriculture and the Environment, Minneapolis, MN, USA, 31 August–2 September 1998. [Google Scholar]
- Tálos-Nebehaj, E.; Hofmann, T.; Albert, L. Seasonal changes of natural antioxidant content in the leaves of Hungarian forest trees. Ind. Crops Prod. 2017, 98, 53–59. [Google Scholar] [CrossRef]
- Cetera, P.; Russo, D.; Milella, L.; Todaro, L. Thermo-treatment affects Quercus cerris L. wood properties and the antioxidant activity and chemical composition of its by-product extracts. Ind. Crops Prod. 2019, 130, 380–388. [Google Scholar] [CrossRef]
- Yang, C.S.; Sang, S.; Lambert, J.D.; Lee, M.J. Bioavailability issues in studying the health effects of plant polyphenolic compounds. Mol. Nutr. Food Res. 2008, 52, S139–S151. [Google Scholar] [CrossRef]
- Yang, G.; Jaakkola, P. Wood chemistry and isolation of extractives from wood. Lit. Study BIOTULI Proj.-Saimaa Univ. Appl. Sci. 2011, 10, 22. [Google Scholar]
- Nisca, A.; Ștefănescu, R.; Moldovan, C.; Mocan, A.; Mare, A.D.; Ciurea, C.N.; Man, A.; Muntean, D.-L.; Tanase, C. Optimization of Microwave Assisted Extraction Conditions to Improve Phenolic Content and In Vitro Antioxidant and Anti-Microbial Activity in Quercus cerris Bark Extracts. Plants 2022, 11, 240. [Google Scholar] [CrossRef]
- Rakić, S.; Petrović, S.; Kukić, J.; Jadranin, M.; Tešević, V.; Povrenović, D.; Šiler-Marinković, S. Influence of thermal treatment on phenolic compounds and antioxidant properties of oak acorns from Serbia. Food Chem. 2007, 104, 830–834. [Google Scholar] [CrossRef]
- Şen, U.; Viegas, C.; Duarte, M.P.; Maurício, E.M.; Nobre, C.; Correia, R.; Pereira, H.; Gonçalves, M. Maceration of Waste Cork in Binary Hydrophilic Solvents for the Production of Functional Extracts. Environments 2023, 10, 142. [Google Scholar] [CrossRef]
- Luís, Â.; Gil, N.; Amaral, M.E.; Domingues, F.; Duartea, A.P. Ailanthus altissima (Miller) Swingle: A source of bioactive compounds with antioxidant activity. BioResources 2012, 7, 2105–2120. [Google Scholar] [CrossRef]
- Anastas, P.; Eghbali, N. Green chemistry: Principles and practice. Chem. Soc. Rev. 2010, 39, 301–312. [Google Scholar] [CrossRef] [PubMed]
- ISO 1985; Wood–Determination of Density for Physical Mechanical Tests. UNI Ente Nazionale Italiano di Unificazione: Milano, Italy, 1985.
- Mitić, M.; Tošić, S.; Pavlović, A.; Mašković, P.; Kostić, D.; Mitić, J.; Stevanović, V. Optimization of the extraction process of minerals from Salvia officinalis L. using factorial design methodology. Microchem. J. 2019, 145, 1224–1230. [Google Scholar] [CrossRef]
- Faraone, I.; Rai, D.K.; Chiummiento, L.; Fernandez, E.; Choudhary, A.; Prinzo, F.; Milella, L. Antioxidant Activity and Phytochemical Characterization of Senecio clivicolus Wedd. Molecules 2018, 23, 2497. [Google Scholar] [CrossRef]
- Libutti, A.; Russo, D.; Lela, L.; Ponticelli, M.; Milella, L.; Rivelli, A.R. Enhancement of Yield, Phytochemical Content and Biological Activity of a Leafy Vegetable (Beta vulgaris L. var. cycla) by Using Organic Amendments as an Alternative to Chemical Fertilizer. Plants 2023, 12, 569. [Google Scholar] [CrossRef]
- Ahajji, A.; Diouf, P.; Aloui, F.; Elbakali, I.; Perrin, D.; Merlin, A.; George, B. Influence of heat treatment on antioxidant properties and colour stability of beech and spruce wood and their extractives. Wood Sci. Technol. 2009, 43, 69–83. [Google Scholar] [CrossRef]
- Gironi, F.; Piemonte, V. Temperature and solvent effects on polyphenol extraction process from chestnut tree wood. Chem. Eng. Res. Des. 2011, 89, 857–862. [Google Scholar] [CrossRef]
- Gortzi, O.; Metaxa, X.; Mantanis, G.; Lalas, S. Effect of artificial ageing using different wood chips on the antioxidant activity, resveratrol and catechin concentration, sensory properties and colour of two Greek red wines. Food Chem. 2013, 141, 2887–2895. [Google Scholar] [CrossRef]
- Sen, A.; Miranda, I.; Esteves, B.; Pereira, H. Chemical characterization, bioactive and fuel properties of waste cork and phloem fractions from Quercus cerris L. bark. Ind. Crops Prod. 2020, 157, 112909. [Google Scholar] [CrossRef]
- Yang, L.; Jiang, J.G.; Li, W.F.; Chen, J.; Wang, D.Y.; Zhu, L. Optimum extraction process of polyphenols from the bark of Phyllanthus emblica L. based on the response surface methodology. J. Sep. Sci. 2009, 32, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Ștefănescu, R.; Ciurea, C.N.; Mare, A.D.; Man, A.; Nisca, A.; Nicolescu, A.; Mocan, A.; Babotă, M.; Coman, N.-A.; Tanase, C. Quercus Robur Older Bark—A Source of Polyphenolic Extracts with Biological Activities. Appl. Sci. 2022, 12, 11738. [Google Scholar] [CrossRef]
- Ferreira, J.; Santos, S.; Pereira, H. In Vitro Screening for Acetylcholinesterase Inhibition and Antioxidant Activity of Quercus suber Cork and Corkback Extracts. Evid.-Based Complement. Altern. Med. 2020, 2020, 3825629. [Google Scholar] [CrossRef] [PubMed]
- Bajraktari, A.; Nunes, L.; Knapic, S.; Pimenta, R.; Pinto, T.; Duarte, S.; Miranda, I.; Pereira, H. Chemical characterization, hardness and termite resistance of Quercus cerris heartwood from Kosovo. Maderas. Cienc. Y Tecnol. 2018, 20, 305–314. [Google Scholar] [CrossRef]
- Todaro, L.; Russo, D.; Cetera, P.; Milella, L. Effects of thermo-vacuum treatment on secondary metabolite content and antioxidant activity of poplar (Populus nigra L.) wood extracts. Ind. Crops Prod. 2017, 109, 384–390. [Google Scholar] [CrossRef]
- Khanbabaee, K.; Van Ree, T. Tannins: Classification and definition. Nat. Prod. Rep. 2001, 18, 641–649. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Gammoudi, N.; Mabrouk, M.; Bouhemda, T.; Nagaz, K.; Ferchichi, A. Modeling and optimization of capsaicin extraction from Capsicum annuum L. using response surface methodology (RSM), artificial neural network (ANN), and Simulink simulation. Ind. Crops Prod. 2021, 171, 113869. [Google Scholar] [CrossRef]
- Youssefi, S.; Emam-Djomeh, Z.; Mousavi, S.M. Comparison of artificial neural network (ANN) and response surface methodology (RSM) in the prediction of quality parameters of spray-dried pomegranate juice. Dry. Technol. 2009, 27, 910–917. [Google Scholar] [CrossRef]


| Symbols | Coded Levels | |||
|---|---|---|---|---|
| Independent variables | −1 | 0 | 1 | |
| Temperature (°C) | X1 | 25 | 50 | 80 |
| Solvent (% EtOH/H2O) | X2 | 0 | 20 | 40 |
| Time (h) | X3 | 3 | 6 | 24 |
| Independent Variables | Dependent Variables | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Run | X1 | X2 | X3 | Yield% | 1 DPPH Scavenging Activity mg TE/g DW | 2 FRAP mg TE/g DW | 3 TPC mg GAE/g DW | 4 TFC mg QE/g DW | 5 CTC mg TAE/g DW | 6 HTC mg TAE/g DW |
| 1 | 20 (−1) | 0 (−1) | 3 (−1) | 0.61 ± 0.03 m | 72.69 ± 3.57 h, i | 134.32 ± 9.15 j | 104.45 ± 1.16 g | 177.21 ± 10.91 e | 13.09 ± 1.26 g, h, i, j | 161.82 ± 7.55 j, k |
| 2 | 20 (−1) | 0 (−1) | 6 (0) | 0.63 ± 0.01 m | 92.66 ± 3.55 h | 145.65 ± 7.79 j | 105.18 ± 17.61 g | 234.19 ± 20.83 d, e | 5.34 ± 0.46 j | 187.64 ± 9.32 i, j, k |
| 3 | 20 (−1) | 0 (−1) | 24 (1) | 0.78 ± 0.05 l, m | 41.88 ± 7.46 j | 135.10 ± 4.13 j | 114.46 ± 13.45 f, g | 299.03 ± 37.81 c, d, e | 5.70 ± 0.54 i, j | 202.88 ± 13.96 h, i, j |
| 4 | 20 (−1) | 20 (0) | 3 (−1) | 0.67 ± 0.02 l, m | 120.78 ± 2.96 h | 215.87 ± 8.01 h, i, j | 103.87 ± 1.39 g | 225.38 ± 10.75 d, e | 5.55 ± 0.63 j | 182.83 ± 17.77 i, j, k |
| 5 | 20 (−1) | 20 (0) | 6 (0) | 0.92 ± 0.08 j, k, l, m | 116.87 ± 5.94 h | 196.71 ± 11.69 i, j | 107.29 ± 45.76 g | 251.13 ± 10.17 d, e | 7.71 ± 0.88 h, i, j | 191.70 ± 15.54 i, j, k |
| 6 | 20 (−1) | 20 (0) | 24 (1) | 1.03 ± 0.08 i, j, k, l, m | 112.53 ± 6.47 h | 199.78 ± 19.26 i, j | 121.82 ± 3.01 e, f, g | 301.84 ± 23.50 c, d | 5.60 ± 0.46 j | 71.75 ± 0.96 l |
| 7 | 20 (−1) | 40 (1) | 3 (−1) | 0.76 ± 0.02 l, m | 220.89 ± 8.12 d, e, f | 252.48 ± 15.78 f, g, h, i | 150.76 ± 1.04 e, f, g | 277.22 ± 17.31 c, d, e | 19.51 ± 1.71 f, g | 301.72 ± 3.85 d, e, f |
| 8 | 20 (−1) | 40 (1) | 6 (0) | 1.09 ± 0.03 i, j, k, l, m | 308.96 ± 14.91 c | 474.09 ± 44.52 b | 167.19 ± 8.83 e, f | 280.49 ± 28.01 c, d, e | 36.95 ± 2.43 c, d | 284.59 ± 26.30 e, f, g |
| 9 | 20 (−1) | 40 (1) | 24 (1) | 1.46 ± 0.09 e, f, g, h, i | 165.69 ± 8.43 g | 303.11 ± 26.73 e, f, g | 169.16 ± 26.57 e, f | 324.47 ± 12.30 a, b, c, d | 36.57 ± 3.43 c, d | 118.97 ± 10.74 k, l |
| 10 | 50 (0) | 0 (−1) | 3 (−1) | 0.87 ± 0.07 k, l, m | 104.4 ± 6.85 h | 190.43 ± 6.28 i, j | 98.82 ± 9.49 g | 276.22 ± 23.26 c, d, e | 38.75 ± 3.64 b, c, d | 217.85 ± 10.30 g, h, i, j |
| 11 | 50 (0) | 0 (−1) | 6 (0) | 1.03 ± 0.04 i, j, k, l, m | 195.99 ± 15.98 f | 241.46 ± 13.35 g, h, i | 122.95 ± 4.07 e, f, g | 278.36 ± 25.93 c, d, e | 8.74 ± 0.77 h, i, j | 371.73 ± 32.03 c, d |
| 12 | 50 (0) | 0 (−1) | 24 (1) | 1.12 ± 0.08 i, j, k, l | 165.69 ± 8.43 g | 214.05 ± 16.02 h, i, j | 131.17 ± 0.12 e, f, g | 323.66 ± 25.68 a, b, c, d | 8.97 ± 0.16 h, i, j | 152.46 ± 12.57 j, k |
| 13 | 50 (0) | 20 (0) | 3 (−1) | 1.30 ± 0.05 h, i, j, k | 112.44 ± 6.05 h | 316.91 ± 17.35 e, f, g | 139.18 ± 13.21 e, f, g | 301.84 ± 11.23 c, d | 13.51 ± 4.33 g, h, i, j | 218.73 ± 8.00 g, h, i, j |
| 14 | 50 (0) | 20 (0) | 6 (0) | 1.62 ± 0.07 e, f, g, h | 165.69 ± 8.43 g | 250.94 ± 21.47 f, g, h, i | 142.38 ± 0.37 e, f, g | 312.25 ± 27.83 a, b, c, d | 14.50 ± 1.33 g, h, i | 162.55 ± 14.71 j, k |
| 15 | 50 (0) | 20 (0) | 24 (1) | 1.73 ± 0.08 e, f, g, h | 203.64 ± 5.75 f | 285.5 ± 11.90 e, f, g, h | 143.95 ± 4.09 e, f, g | 317.85 ± 31.11 a, b, c, d | 13.60 ± 0.48 g, h, i, j | 217.51 ± 20.11 g, h, i, j |
| 16 | 50 (0) | 40 (1) | 3 (−1) | 1.44 ± 0.05 f, g, h, i | 322.10 ± 9.75 b, c | 362.79 ± 3.36 c, d, e | 148.16 ± 17.27 e, f, g | 297.84 ± 9.02 c, d, e | 46.79 ± 3.06 b | 410.08 ± 35.66 c |
| 17 | 50 (0) | 40 (1) | 6 (0) | 1.69 ± 0.06 e, f, g, h | 238.77 ± 7.92 d, e | 287.31 ± 21.35 e, f, g, h | 152.97 ± 9.10 e, f, g | 297.04 ± 13.25 c, d, e | 26.74 ± 2.55 e, f | 521.61 ± 45.63 b |
| 18 | 50 (0) | 40 (1) | 24 (1) | 2.33 ± 0.14 b, c | 347.71 ± 25.97 a, b | 518.48 ± 51.93 a | 175.98 ± 17.70 d, e | 347.08 ± 24.71 a, b, c, d | 38.18 ± 2.98 b, c, d | 261.22 ± 23.77 f, g, h, i |
| 19 | 80 (1) | 0 (−1) | 3 (−1) | 1.39 ± 0.08 g, h, i, j | 299.60 ± 5.66 c | 511.04 ± 26.71 a, b | 234.04 ± 6.43 c, d | 263.61 ± 16.89 d, e | 34.10 ± 1.48 d, e | 203.48 ± 19.72 h, i, j |
| 20 | 80 (1) | 0 (−1) | 6 (0) | 1.43 ± 0.07 g, h, i | 239.29 ± 10.51 d, e | 327.34 ± 27.20 e, f | 239.19 ± 14.59 c | 279.16 ± 17.53 c, d, e | 16.38 ± 1.48 g, h | 359.12 ± 26.16 c, d, e |
| 21 | 80 (1) | 0 (−1) | 24 (1) | 1.95 ± 0.05 c, d, e | 209.33 ± 2.89 e, f | 314.25 ± 27.46 e, f, g | 246.83 ± 29.19 c | 304.24 ± 4.21 b, c, d | 8.95 ± 0.80 h, i, j | 276.43 ± 17.66 f, g, h |
| 22 | 80 (1) | 20 (0) | 3 (−1) | 1.83 ± 0.08 d, e, f, g | 326.85 ± 8.95 b, c | 459.44 ± 42.61 b | 340.69 ± 1.70 a, b | 263.81 ± 12.79 d, e | 36.69 ± 2.39 c, d | 530.85 ± 50.88 b |
| 23 | 80 (1) | 20 (0) | 6 (0) | 1.92 ± 0.10 c, d, e, f | 242.53 ± 12.83 d | 347.02 ± 26.92 d, e | 361.30 ± 36.53 a | 274.62 ± 6.79 c, d, e | 27.17 ± 2.53 e, f | 378.71 ± 3.62 c, d |
| 24 | 80 (1) | 20 (0) | 24 (1) | 2.67 ± 0.12 a, b | 324.98 ± 2.87 b, c | 439.91 ± 36.33 b, c | 365.79 ± 14.77 a | 329.46 ± 28.33 a, b, c, d | 74.82 ± 6.57 a | 436.89 ± 12.34 c |
| 25 | 80 (1) | 40 (1) | 3 (−1) | 2.28 ± 0.16 b, c, d | 359.87 ± 9.72 a | 496.85 ± 44.57 a, b | 285.46 ± 21.20 b, c | 394.45 ± 36.83 a, b, c | 40.87 ± 2.83 b, c, d | 863.07 ± 25.67 a |
| 26 | 80 (1) | 40 (1) | 6 (0) | 2.46 ± 0.10 a, b | 341.56 ± 7.11 b | 427.55 ± 36.92 b, c, d | 290.76 ± 11.12 b, c | 427.01 ± 12.12 a, b | 74.82 ± 6.57 a | 821.22 ± 46.95 a |
| 27 | 80 (1) | 40 (1) | 24 (1) | 2.85 ± 0.19 a | 365.78 ± 1.95 a | 519.11 ± 41.19 a | 329.59 ± 32.78 a, b | 433.75 ± 15.57 a | 44.67 ± 4.01 b, c | 591.50 ± 42.93 b |
| Variables | Std. Dev. | Mean | C.V.% | R2 | Adj. R2 | Pred. R2 | Adequate Precision |
|---|---|---|---|---|---|---|---|
| 1 DPPH scavenging activity | 48.41 | 215.54 | 22.46 | 0.8262 | 0.7622 | 0.6632 | 12.9832 |
| 2 FRAP | 67.25 | 317.31 | 21.20 | 0.7873 | 0.7089 | 0.5342 | 11.8708 |
| 3 TPC | 50.94 | 188.65 | 27.00 | 0.7503 | 0.6583 | 0.5260 | 9.1674 |
| 4 TFC | 30.05 | 299.75 | 10.03 | 0.7873 | 0.7090 | 0.5661 | 11.9922 |
| 5 CTC | 15.00 | 26.08 | 57.51 | 0.5777 | 0.4221 | −0.1252 | 7.4330 |
| 6 HTC | 50.94 | 188.65 | 27.00 | 0.7503 | 0.7906 | 0.7175 | 14.2945 |
| Response | Independent Variables | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 DPPH | YDPPH = | +215.49 | +83.26 X1 | +70.72 X2 | −0.22 X3 | −10.00 X1 X2 | +9.77 X1 X3 | +5.37 X2 X3 | +17.28 X1 X2 X3 |
| p-values | <0.0001 * | <0.0001 * | 0.9839 | 0.4981 | 0.4590 | 0.6822 | 0.2885 | ||
| 2 FRAP | YFRAP = | +318.45 | +99.05 X1 | +86.16 X2 | +4.82 X3 | −20.49 X1 X2 | −0.58 X X3 | +28.64 X2 X3 | +27.26 X1 X2 X3 |
| p-values | <0.0001 * | <0.0001 * | 0.7458 | 0.3210 | 0.9745 | 0.1271 | 0.2301 | ||
| 3 TPC | YTPC = | +190.90 | +86.75 X1 | +27.04 X2 | +9.47 X3 | +2.81 X1 X2 | +2.79 X1 X3 | +3.22 X2 X3 | +3.83 X1 X2 X3 |
| p-values | <0.0001 * | 0.0425 * | 0.4041 | 0.8558 | 0.8396 | 0.8152 | 0.8206 | ||
| 4 TFC | YTFC = | +305.96 | +31.41 X1 | +34.52 X2 | +26.07 X3 | +21.15 X1 X2 | −7.89 X1 X3 | −5.19 X2 X3 | +6.09 X1 X2 X3 |
| p-values | 0.0004 * | 0.0002 * | 0.0008 * | 0.0295 * | 0.3374 | 0.5253 | 0.5427 | ||
| 5 HTC | YHTC = | +310.52 | +152.80 X1 | +101.96 X2 | −48.99 X3 | +105.06 X1 X2 | −1.61 X1 X3 | −47.88 X2 X3 | −8.10 X1 X2 X3 |
| p-values | <0.0001 * | 0.0002 * | 0.0234 * | 0.0011 * | 0.9478 | 0.0629 | 0.7886 | ||
| Methods | Response Prediction | 95% PI a | SE Fit b |
|---|---|---|---|
| DPPH 1 scavenging activity | 378.7 mg TE/g | (257.2; 500.2) | 32.1 |
| FRAP 2 | 519.1 mg TE/g | (350.3; 688.0) | 44.5 |
| TPC 3 | 319.0 mg GAE/g | (191.2; 446.9) | 33.7 |
| TFC 4 | 404.4 mg QE/g | (329.0; 479.9) | 19.9 |
| HTC 5 | 606.6 mg TAE/g | (377.7; 835.6) | 60.4 |
| 3 TPC (mg GAE/g) | 4 TFC (mg QE/g) | 5 HTC (mg TAE/g) | FRAP (mg TE/g) | |
|---|---|---|---|---|
| TFC (mg QE/g) | 0.484 | |||
| HTC (mg TAE/g) | 0.661 | 0.621 | ||
| 2 FRAP (mg TE/g) | 0.733 | 0.576 | 0.601 | |
| 1 DPPH scavenging activity (mg TE/g) | 0.737 | 0.597 | 0.726 | 0.931 |
| 1 DPPH Scavenging Activity (mg TE/g) | 2 FRAP (mg TE/g) | 3 TPC (mg GAE/g) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Temperature | % EtOH | Time | Exp. | FFD | ANN | Exp. | FFD | ANN | Exp. | FFD | ANN |
| 20 | 0 | 3 | 72.69 | 49.58 | 80.03 | 134.32 | 108.73 | 124.03 | 104.45 | 72.63 | 107.60 |
| 20 | 0 | 6 | 92.66 | 50.13 | 90.91 | 145.65 | 109.88 | 154.68 | 105.18 | 74.71 | 100.55 |
| 20 | 0 | 24 | 41.88 | 53.43 | 50.56 | 135.10 | 116.78 | 173.86 | 114.46 | 87.21 | 70.00 |
| 20 | 20 | 3 | 120.78 | 142.21 | 93.88 | 215.87 | 214.00 | 144.68 | 103.87 | 97.47 | 130.31 |
| 20 | 20 | 6 | 116.87 | 139.36 | 119.72 | 196.71 | 215.54 | 207.41 | 107.29 | 99.38 | 123.91 |
| 20 | 20 | 24 | 112.53 | 122.25 | 117.40 | 199.78 | 224.81 | 217.71 | 121.82 | 110.84 | 137.75 |
| 20 | 40 | 3 | 220.89 | 234.83 | 236.11 | 252.48 | 319.27 | 274.15 | 150.76 | 122.32 | 158.90 |
| 20 | 40 | 6 | 308.96 | 228.58 | 319.80 | 474.09 | 321.21 | 429.46 | 167.19 | 124.05 | 158.94 |
| 20 | 40 | 24 | 165.69 | 191.06 | 205.53 | 303.11 | 332.83 | 313.00 | 169.16 | 134.47 | 180.55 |
| 50 | 0 | 3 | 104.49 | 150.35 | 121.93 | 190.43 | 256.11 | 186.74 | 98.82 | 157.61 | 110.35 |
| 50 | 0 | 6 | 195.99 | 148.75 | 165.07 | 241.46 | 249.31 | 249.51 | 122.95 | 159.39 | 111.00 |
| 50 | 0 | 24 | 165.69 | 139.18 | 116.96 | 214.05 | 208.48 | 202.20 | 131.17 | 170.11 | 131.99 |
| 50 | 20 | 3 | 112.44 | 215.70 | 141.92 | 316.91 | 313.63 | 225.96 | 139.18 | 181.43 | 120.49 |
| 50 | 20 | 6 | 165.69 | 215.64 | 168.19 | 250.94 | 315.01 | 255.13 | 142.38 | 184.14 | 131.82 |
| 50 | 20 | 24 | 203.64 | 215.27 | 218.12 | 285.25 | 323.27 | 273.73 | 143.95 | 200.37 | 146.86 |
| 50 | 40 | 3 | 322.41 | 281.06 | 311.50 | 362.79 | 371.15 | 364.13 | 148.16 | 205.25 | 155.19 |
| 50 | 40 | 6 | 238.77 | 282.53 | 326.93 | 287.31 | 380.71 | 393.38 | 152.97 | 208.88 | 165.17 |
| 50 | 40 | 24 | 347.71 | 291.37 | 341.79 | 518.48 | 438.07 | 485.69 | 175.98 | 230.63 | 233.86 |
| 80 | 0 | 3 | 299.60 | 251.12 | 314.86 | 511.04 | 403.49 | 494.92 | 234.04 | 242.59 | 241.04 |
| 80 | 0 | 6 | 239.29 | 247.38 | 231.34 | 327.34 | 388.73 | 344.60 | 239.19 | 244.08 | 233.94 |
| 80 | 0 | 24 | 209.33 | 224.92 | 212.08 | 314.25 | 300.18 | 315.31 | 246.83 | 253.01 | 249.10 |
| 80 | 20 | 3 | 326.85 | 289.20 | 258.35 | 459.44 | 413.26 | 381.11 | 340.69 | 265.39 | 338.50 |
| 80 | 20 | 6 | 242.53 | 291.93 | 249.25 | 347.02 | 414.48 | 341.47 | 361.30 | 268.89 | 364.56 |
| 80 | 20 | 24 | 324.98 | 308.30 | 315.08 | 439.91 | 421.74 | 445.88 | 365.79 | 289.91 | 358.56 |
| 80 | 40 | 3 | 359.87 | 327.28 | 368.56 | 496.85 | 423.04 | 489.75 | 285.46 | 288.18 | 282.63 |
| 80 | 40 | 6 | 341.56 | 336.48 | 337.29 | 427.55 | 440.22 | 435.94 | 290.76 | 293.70 | 290.92 |
| 80 | 40 | 24 | 365.78 | 391.67 | 359.10 | 519.11 | 543.31 | 526.07 | 329.59 | 338.99 | 340.89 |
| Model-predicted capability | |||||||||||
| 1 DPPH scavenging activity | 2 FRAP | 3 TPC | |||||||||
| FFD | ANN | FFD | ANN | FFD | ANN | ||||||
| 4 MAE | 33.92 | 18.43 | 43.22 | 24.12 | 34.63 | 11.30 | |||||
| 5 RMSE | 40.61 | 27.72 | 56.42 | 37.34 | 42.73 | 17.17 | |||||
| 6 R2 | 0.83 | 0.92 | 0.79 | 0.96 | 0.75 | 0.91 | |||||
| Temperature °C | Time h | % EtOH/H2O | 1 DPPH Scavenging Activity mg TE/g | 2 FRAP mg TE/g | 3 TPC mg GAE/g | Composite Desirability | |
|---|---|---|---|---|---|---|---|
| FFD | 80.00 | 24.00 | 40.00 | 391.67 | 543.31 | 326.80 | 0.95 |
| ANN | 80.00 | 23.47 | 39.32 | 380.22 | 526.22 | 357.24 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponticelli, M.; Carlucci, V.; Mecca, M.; Todaro, L.; Milella, L.; Russo, D. Extraction Optimization of Quercus cerris L. Wood Chips: A Comparative Study between Full Factorial Design (FFD) and Artificial Neural Network (ANN). Antioxidants 2024, 13, 1115. https://doi.org/10.3390/antiox13091115
Ponticelli M, Carlucci V, Mecca M, Todaro L, Milella L, Russo D. Extraction Optimization of Quercus cerris L. Wood Chips: A Comparative Study between Full Factorial Design (FFD) and Artificial Neural Network (ANN). Antioxidants. 2024; 13(9):1115. https://doi.org/10.3390/antiox13091115
Chicago/Turabian StylePonticelli, Maria, Vittorio Carlucci, Marisabel Mecca, Luigi Todaro, Luigi Milella, and Daniela Russo. 2024. "Extraction Optimization of Quercus cerris L. Wood Chips: A Comparative Study between Full Factorial Design (FFD) and Artificial Neural Network (ANN)" Antioxidants 13, no. 9: 1115. https://doi.org/10.3390/antiox13091115








