Influence of High Temperature and Ammonia and Nitrite Accumulation on the Physiological, Structural, and Genetic Aspects of the Biology of Largemouth Bass (Micropterus salmoides)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Experimental Fish
2.3. Fish Treatment and Sampling
2.4. Calculation of Growth Rate and Feed Conversion Ratio
2.5. Water Quality Measurement
2.6. Tisue Biochemical Assay
2.7. Histological Analysis and TUNEL Assay
2.8. Total RNA Isolation
2.9. RNA-Seq and Data Analysis
2.10. Quantitative Real-Time PCR
2.11. Statistics
3. Results
3.1. Feeding-Based Accumulation of Nitrogenous Pollutants
3.2. Growth Rate and Feed Conversion Ratio of the Fish
3.3. Ammonia Content and Oxidative Indices in the Liver
3.4. Tissue Damage and Cell Apoptosis
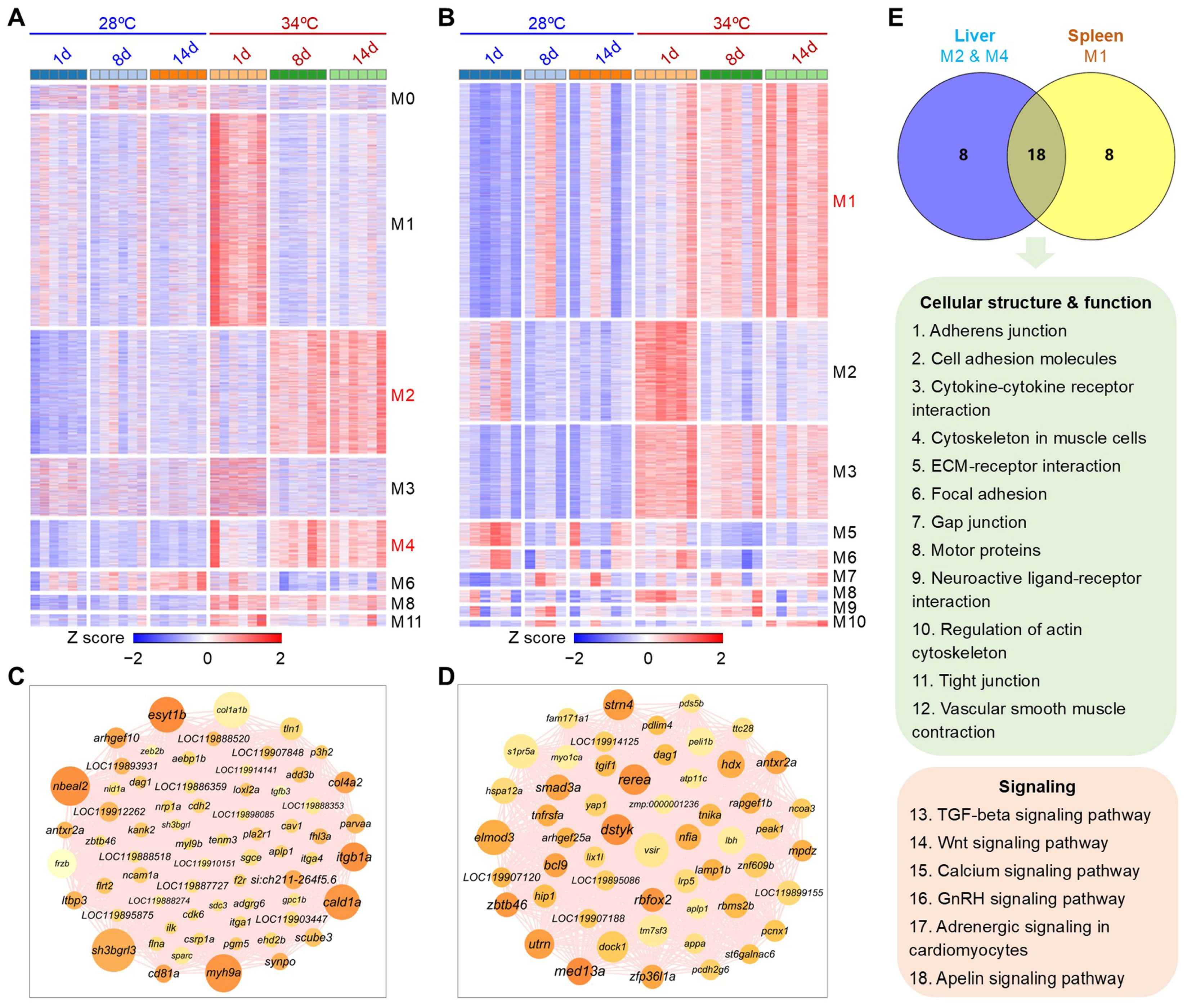
3.5. Overall Changes in Gene Expression
3.6. Biological Processes Affected by Hyperthermia and Ammonia/Nitrite Toxicity
3.7. Gene Modules Affected by Hyperthermia and Ammonia/Nitrite Toxicity
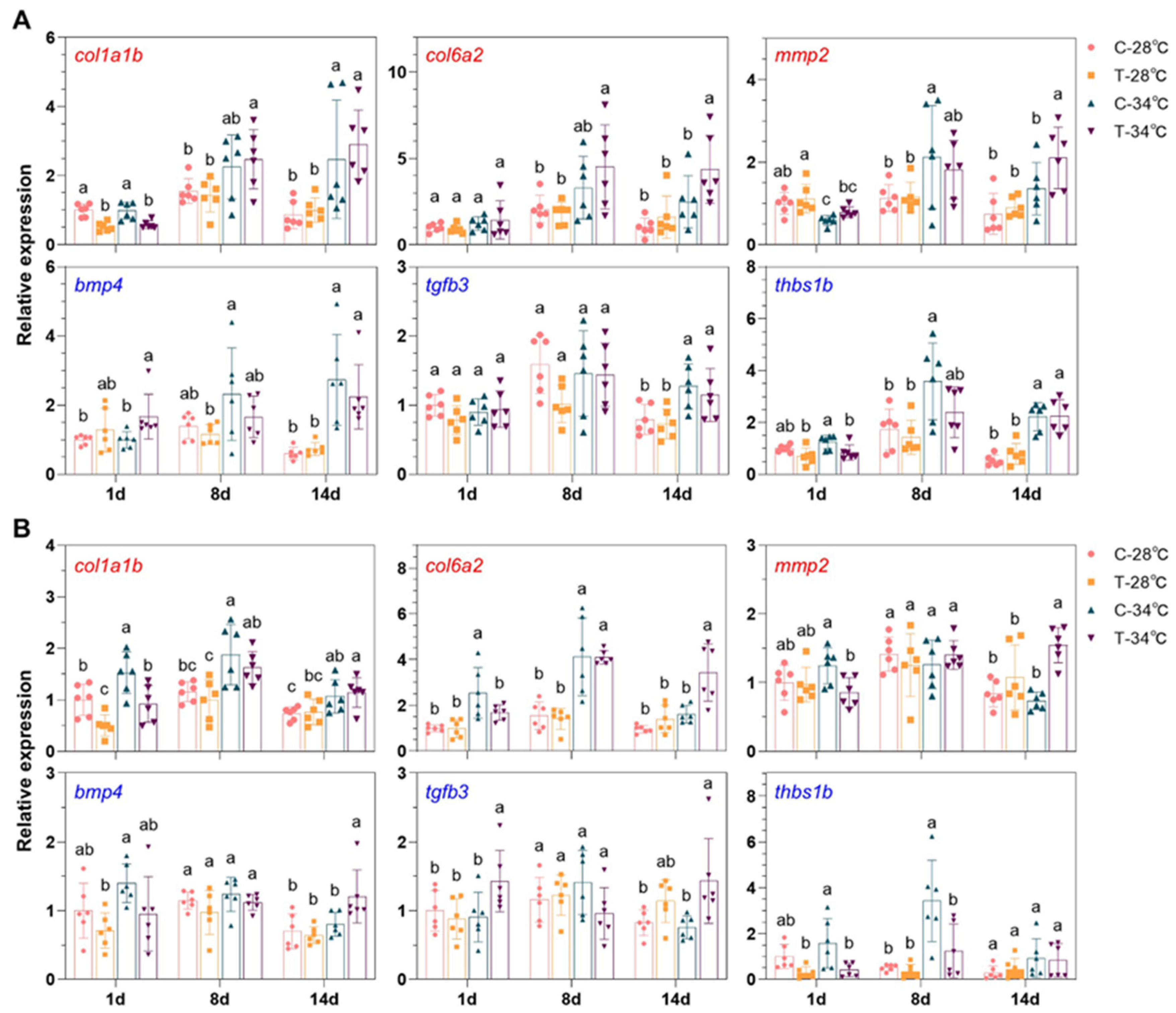
3.8. Gene Expressions Measured by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAT | Catalase |
| DEGs | Differentially expressed genes |
| DO | Dissolved oxygen |
| ECM | Extracellular matrix |
| FCR | Feed conversion ratio |
| GO | Gene ontology |
| HALN | High ammonia and low nitrite |
| H&E | Hematoxylin and eosin |
| ILCM | Interlamellar cell mass |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LAHN | Low ammonia and high nitrite |
| LALN | Low ammonia and low nitrite |
| LMB | Largemouth bass |
| MDA | Malondialdehyde |
| NH3 | Nonionic ammonia |
| NH4+ | Ionic ammonia |
| NO3-N | Nitrate |
| NO2-N | Nitrite |
| NXR | Nitrite oxidoreductase |
| PCA | Principal component analysis |
| PFA | Paraformaldehyde |
| SD | Standard deviation |
| SGR | Specific growth rate |
| SOD | Superoxide dismutase |
| SRA | Sequence read Archive |
| TAN | Total ammonia–nitrogen |
| TDT | Terminal deoxynucleotidy1 transferase |
| TN | Total nitrogen |
| TPM | Transcripts per million |
| WGCNA | Weighted gene co-expression network analysis |
| WGR | Weight gain rate |
References
- Islam, M.J.; Kunzmann, A.; Slater, M.J. Responses of aquaculture fish to climate change-induced extreme temperatures: A review. J. World Aquac. Soc. 2022, 53, 314–366. [Google Scholar] [CrossRef]
- Bagnyukova, T.V.; Lushchak, O.V.; Storey, K.B.; Lushchak, V.I. Oxidative stress and antioxidant defense responses by goldfish tissues to acute change of temperature from 3 to 23 °C. J. Therm. Biol. 2007, 32, 227–234. [Google Scholar] [CrossRef]
- Lee, S.; Ji, K.; Choi, K. Effects of water temperature on perchlorate toxicity to the thyroid and reproductive system of Oryzias latipes. Ecotoxicol. Environ. Saf. 2014, 108, 311–317. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, E.; Li, C.; Pan, C.; Zhao, X.; Wang, Y.; Ling, Q. Effects of heat stress on histopathology, antioxidant enzymes, and transcriptomic profiles in gills of pikeperch Sander lucioperca. Aquaculture 2021, 534, 736277. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Guo, Z.-X.; Luo, S.-W.; Wang, A.-L. Effects of high temperature on biochemical parameters, oxidative stress, DNA damage and apoptosis of pufferfish (Takifugu obscurus). Ecotoxicol. Environ. Saf. 2018, 150, 190–198. [Google Scholar] [CrossRef]
- Li, B.; Sun, S.; Zhu, J.; Yanli, S.; Wuxiao, Z.; Ge, X. Transcriptome profiling and histology changes in juvenile blunt snout bream (Megalobrama amblycephala) liver tissue in response to acute thermal stress. Genomics 2019, 111, 242–250. [Google Scholar] [CrossRef]
- Wang, W.; Gao, L.; Liu, W.; Tian, Z.; Wang, X.; Hu, H. Regulation of antioxidant defense in response to heat stress in Siberian sturgeon (Acipenser baerii). Aquaculture 2023, 572, 739551. [Google Scholar] [CrossRef]
- Besson, M.; Vandeputte, M.; van Arendonk, J.A.M.; Aubin, J.; de Boer, I.J.M.; Quillet, E.; Komen, H. Influence of water temperature on the economic value of growth rate in fish farming: The case of sea bass (Dicentrarchus labrax) cage farming in the Mediterranean. Aquaculture 2016, 462, 47–55. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Lai, J.; Liu, Y.; Song, M.; Li, F.; Gong, Q. Integrated biochemical, transcriptomic and metabolomic analyses provide insight into heat stress response in Yangtze sturgeon (Acipenser dabryanus). Ecotoxicol. Environ. Saf. 2023, 249, 114366. [Google Scholar] [CrossRef]
- Yan, D.; Long, X.; Zhang, X.; Dong, X.; Wang, Z.; Jiang, H.; An, M.; Chen, J.; Gan, L. Identification and characterization of long non-coding RNAs in intestinal immune regulation of largemouth bass, Micropterus salmoides, under acute heat stress. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 48, 101132. [Google Scholar] [CrossRef]
- Servili, A.; Canario, A.V.M.; Mouchel, O.; Muñoz-Cueto, J.A. Climate change impacts on fish reproduction are mediated at multiple levels of the brain-pituitary-gonad axis. Gen. Comp. Endocrinol. 2020, 291, 113439. [Google Scholar] [CrossRef] [PubMed]
- Xv, Z.; Chen, S.X.; Song, G.L.; Hu, H.; Lin, S.M.; Long, Y. Biochemical, histological and transcriptomic analyses for the immunological organs provide insights into heat stress-induced disease susceptibility in Largemouth Bass. Sci. Total Environ. 2024, 912, 168758. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Lin, J.; Zhou, Q.; Wang, H.; Han, Y. Effect of ammonia stress on transcriptome and endoplasmic reticulum stress pathway for common carp (Cyprinus carpio) hepatopancreas. Aquac. Rep. 2021, 20, 100694. [Google Scholar] [CrossRef]
- Liu, C.-H.; Chen, J.-C. Effect of ammonia on the immune response of white shrimpLitopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol. 2004, 16, 321–334. [Google Scholar] [CrossRef]
- Mangang, Y.A.; Pandey, P.K. Hemato-biochemical responses and histopathological alterations in the gill and kidney tissues of Osteobrama belangeri (Valenciennes, 1844) exposed to different sub-lethal unionized ammonia. Aquaculture 2021, 542, 736887. [Google Scholar] [CrossRef]
- Edwards, T.M.; Puglis, H.J.; Kent, D.B.; Durán, J.L.; Bradshaw, L.M.; Farag, A.M. Ammonia and aquatic ecosystems—A review of global sources, biogeochemical cycling, and effects on fish. Sci. Total Environ. 2024, 907, 167911. [Google Scholar] [CrossRef]
- Zhong, L.; Liu, S.; Zuo, F.Y.; Geng, Y.; Ouyang, P.; Chen, D.F.; Yang, S.Y.; Zheng, W.; Xiong, Y.L.; Cai, W.L.; et al. The IL17 signaling pathway: A potential signaling pathway mediating gill hyperplasia and inflammation under ammonia nitrogen stress was identified by multi-omics analysis. Sci. Total Environ. 2023, 867, 161581. [Google Scholar] [CrossRef]
- Parvathy, A.J.; Das, B.C.; Jifiriya, M.J.; Varghese, T.; Pillai, D.; Kumar, V.J.R. Ammonia induced toxico-physiological responses in fish and management interventions. Rev. Aquac. 2023, 15, 452–479. [Google Scholar] [CrossRef]
- Ching, B.; Chew, S.F.; Wong, W.P.; Ip, Y.K. Environmental ammonia exposure induces oxidative stress in gills and brain of Boleophthalmus boddarti (mudskipper). Aquat. Toxicol. 2009, 95, 203–212. [Google Scholar] [CrossRef]
- Zhang, M.; Li, M.; Wang, R.; Qian, Y. Effects of acute ammonia toxicity on oxidative stress, immune response and apoptosis of juvenile yellow catfish Pelteobagrus fulvidraco and the mitigation of exogenous taurine. Fish Shellfish Immunol. 2018, 79, 313–320. [Google Scholar] [CrossRef]
- Li, M.; Yu, N.; Qin, J.G.; Li, E.; Du, Z.; Chen, L. Effects of ammonia stress, dietary linseed oil and Edwardsiella ictaluri challenge on juvenile darkbarbel catfish Pelteobagrus vachelli. Fish Shellfish Immunol. 2014, 38, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Chen, C.; Liang, Y.; Wu, K.; Wen, X. Ammonia–nitrogen stress affects immune regulation via TNFα in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2024, 583, 740593. [Google Scholar] [CrossRef]
- Ip, Y.K.; Chew, S.F. Ammonia production, excretion, toxicity, and defense in fish: A review. Front. Physiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Wang, H.-J.; Xiao, X.-C.; Wang, H.-Z.; Li, Y.; Yu, Q.; Liang, X.-M.; Feng, W.-S.; Shao, J.-C.; Rybicki, M.; Jungmann, D.; et al. Effects of high ammonia concentrations on three cyprinid fish: Acute and whole-ecosystem chronic tests. Sci. Total Environ. 2017, 598, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Kroupová, H.K.; Valentová, O.; Svobodová, Z.; Sauer, P.; Máchová, J. Toxic effects of nitrite on freshwater organisms: A review. Rev. Aquac. 2018, 10, 525–542. [Google Scholar] [CrossRef]
- Gao, X.-Q.; Fei, F.; Huo, H.H.; Huang, B.; Meng, X.S.; Zhang, T.; Liu, B.-L. Effect of acute exposure to nitrite on physiological parameters, oxidative stress, and apoptosis in Takifugu rubripes. Ecotoxicol. Environ. Saf. 2020, 188, 109878. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Miao, L.-H.; Pan, W.-J.; Huang, X.; Dengu, J.M.; Zhang, W.-X.; Ge, X.-P.; Liu, B.; Ren, M.-C.; Zhou, Q.-L.; et al. Effect of nitrite exposure on the antioxidant enzymes and glutathione system in the liver of bighead carp, Aristichthys nobilis. Fish Shellfish Immunol. 2018, 76, 126–132. [Google Scholar] [CrossRef]
- Chand, R.K.; Sahoo, P.K. Effect of nitrite on the immune response of freshwater prawn Macrobrachium malcolmsonii and its susceptibility to Aeromonas hydrophila. Aquaculture 2006, 258, 150–156. [Google Scholar] [CrossRef]
- Sun, S.; Ge, X.; Xuan, F.; Zhu, J.; Yu, N. Nitrite-induced hepatotoxicity in Bluntsnout bream (Megalobrama amblycephala): The mechanistic insight from transcriptome to physiology analysis. Environ. Toxicol. Pharmacol. 2014, 37, 55–65. [Google Scholar] [CrossRef]
- Miao, L.H.; Lin, Y.; Pan, W.J.; Huang, X.; Ge, X.P.; Zhou, Q.L.; Liu, B.; Ren, M.C.; Zhang, W.X.; Liang, H.L.; et al. Comparative transcriptome analysis reveals the gene expression profiling in bighead carp (Aristichthys nobilis) in response to acute nitrite toxicity. Fish Shellfish Immunol. 2018, 79, 244–255. [Google Scholar] [CrossRef]
- Ciji, A.; Sahu, N.P.; Pal, A.K.; Akhtar, M.S. Physiological changes in Labeo rohita during nitrite exposure: Detoxification through dietary vitamin E. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2013, 158, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-Q.; Fei, F.; Huo, H.H.; Huang, B.; Meng, X.S.; Zhang, T.; Liu, W.B.; Liu, B.-L. Exposure to nitrite alters thyroid hormone levels and morphology in Takifugu rubripes. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 225, 108578. [Google Scholar] [CrossRef]
- Huong, D.T.T.; Gam, L.T.H.; Lek, S.; Ut, V.N.; Phuong, N.T. Effects of nitrite at different temperatures on physiological parameters and growth in clown knifefish (Chitala ornata, Gray 1831). Aquaculture 2020, 521, 735060. [Google Scholar] [CrossRef]
- Molayemraftar, T.; Peyghan, R.; Jalali, M.R.; Shahriari, A. Single and combined effects of ammonia and nitrite on common carp, Cyprinus carpio: Toxicity, hematological parameters, antioxidant defenses, acetylcholinesterase, and acid phosphatase activities. Aquaculture 2022, 548, 737676. [Google Scholar] [CrossRef]
- Li, Y.; Guo, H.; Ge, H.; Sha, H.; Wu, Y.; Zou, G.; Liang, H. A time-dependent interactive effect of nitrite and ammonia on inflammatory and immune response in the head kidney of silver carp (Hypophthalmichthys molitrix). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2025, 288, 110078. [Google Scholar] [CrossRef]
- Esam, F.; Khalafalla, M.M.; Gewaily, M.S.; Abdo, S.; Hassan, A.M.; Dawood, M.A.O. Acute ammonia exposure combined with heat stress impaired the histological features of gills and liver tissues and the expression responses of immune and antioxidative related genes in Nile tilapia. Ecotoxicol. Environ. Saf. 2022, 231, 113187. [Google Scholar] [CrossRef]
- Shen, C.; Cao, S.; Mohsen, M.; Li, X.S.; Wang, L.; Lu, K.L.; Zhang, C.X.; Song, K. Effects of chronic nitrite exposure on hematological parameters, oxidative stress and apoptosis in spotted seabass (Lateolabrax maculatus) reared at high temperature. Aquac. Rep. 2024, 35, 102022. [Google Scholar] [CrossRef]
- Egnew, N.; Renukdas, N.; Ramena, Y.; Yadav, A.K.; Kelly, A.M.; Lochmann, R.T.; Sinha, A.K. Physiological insights into largemouth bass (Micropterus salmoides) survival during long-term exposure to high environmental ammonia. Aquat. Toxicol. 2019, 207, 72–82. [Google Scholar] [CrossRef]
- Jia, R.; Han, C.; Lei, J.-L.; Liu, B.-L.; Huang, B.; Huo, H.-H.; Yin, S.-T. Effects of nitrite exposure on haematological parameters, oxidative stress and apoptosis in juvenile turbot (Scophthalmus maximus). Aquat. Toxicol. 2015, 169, 1–9. [Google Scholar] [CrossRef]
- Xu, Z.; Li, E.; Xu, C.; Gan, L.; Qin, J.G.; Chen, L. Response of AMP-activated protein kinase and energy metabolism to acute nitrite exposure in the Nile tilapia Oreochromis niloticus. Aquat. Toxicol. 2016, 177, 86–97. [Google Scholar] [CrossRef]
- Zou, J.H.; Hu, P.; Wang, M.Y.; Chen, Z.W.; Wang, H.; Guo, X.L.; Gao, J.; Wang, Q.C. Liver Injury and Metabolic Dysregulation in Largemouth Bass (Micropterus salmoides) after Ammonia Exposure. Metabolites 2023, 13, 274. [Google Scholar] [CrossRef] [PubMed]
- Randall, D.J.; Tsui, T.K.N. Ammonia toxicity in fish. Mar. Pollut. Bull. 2002, 45, 17–23. [Google Scholar] [CrossRef]
- Jia, R.; Liu, B.-L.; Han, C.; Huang, B.; Lei, J.-L. The physiological performance and immune response of juvenile turbot (Scophthalmus maximus) to nitrite exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2016, 181–182, 40–46. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, H.J.; Kim, K.W.; Hwang, I.K.; Kim, D.H.; Oh, C.W.; Lee, J.S.; Kang, J.C. Growth performance, oxidative stress, and non-specific immune responses in juvenile sablefish, Anoplopoma fimbria, by changes of water temperature and salinity. Fish. Physiol. Biochem. 2017, 43, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Du, J.X.; Xie, Y.J.; Li, M.; Zhu, T.; Lei, C.X.; Song, H.M.; Han, L.Q.; Li, S.J. Effects of chronic heat stress on growth performance, liver histology, digestive enzyme activities, and expressions of HSP genes in different populations of Largemouth bass (Micropterus salmoides). Aquac. Rep. 2024, 35, 101972. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Li, L.L.; Li, C.J.; Liu, E.U.; Zhu, H.; Ling, Q.F. Heat stress-induced endoplasmic reticulum stress promotes liver apoptosis in largemouth bass (Micropterus salmoides). Aquaculture 2022, 546, 737401. [Google Scholar] [CrossRef]
- Fantini, L.E.; Smith, M.A.; Jones, M.; Roy, L.A.; Lochmann, R.; Kelly, A.M. Growth parameters in northern largemouth bass Micropterus salmoides salmoides raised near their upper thermal tolerance for 28 days. Aquac. Rep. 2021, 21, 100845. [Google Scholar] [CrossRef]
- Chen, H.; Qiao, H.; Xv, Z.; Song, G.; Liu, S.; Luo, C.; Long, Y.; Lin, S. Assessing Heat Resistance and Selecting Heat-Resistant Individuals of Largemouth Bass (Micropterus salmoides) with Tiered Thermal Exposure. Animals 2025, 15, 128. [Google Scholar] [CrossRef]
- SEPA. Analytical Methods for Water and Wastewater Monitor, 4th ed.; Chinese Environmental Science Press: Beijing, China, 2002. (In Chinese) [Google Scholar]
- Liu, M.J.; Guo, H.Y.; Zhu, K.C.; Liu, B.S.; Liu, B.; Guo, L.; Zhang, N.; Yang, J.W.; Jiang, S.G.; Zhang, D.C. Effects of acute ammonia exposure and recovery on the antioxidant response and expression of genes in the Nrf2-Keap1 signaling pathway in the juvenile golden pompano (Trachinotus ovatus). Aquat. Toxicol. 2021, 240, 105969. [Google Scholar] [CrossRef]
- Liu, R.; Liu, R.Y.; Song, G.L.; Li, Q.; Cui, Z.B.; Long, Y. Mitochondria Dysfunction and Cell Apoptosis Limit Resistance of Nile Tilapia (Oreochromis niloticus) to Lethal Cold Stress. Animals 2022, 12, 2382. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. FASTQ: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Fang, H.; Harris, S.C.; Su, Z.; Chen, M.; Qian, F.; Shi, L.; Perkins, R.; Tong, W. ArrayTrack: An FDA and public genomic tool. Methods Mol. Biol. 2009, 563, 379–398. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Törönen, P.; Holm, L. PANNZER-A practical tool for protein function prediction. Protein Sci. 2022, 31, 118–128. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Bucking, C. A broader look at ammonia production, excretion, and transport in fish: A review of impacts of feeding and the environment. J. Comp. Physiol. B 2017, 187, 1–18. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.L.; Lehtovirta-Morley, L.E. Nitrification and beyond: Metabolic versatility of ammonia oxidising archaea. Isme J. 2023, 17, 1358–1368. [Google Scholar] [CrossRef]
- van Kessel, M.A.H.J.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.M.; Kartal, B.; Jetten, M.S.M.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef]
- Chicano, T.M.; Dietrich, L.; de Almeida, N.M.; Akram, M.; Hartmann, E.; Leidreiter, F.; Leopoldus, D.; Mueller, M.; Sánchez, R.; Nuijten, G.H.L.; et al. Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat. Microbiol. 2021, 6, 1129–1139. [Google Scholar] [CrossRef]
- Zhao, L.L.; Cui, C.; Liu, Q.; Sun, J.L.; He, K.; Adam, A.A.; Luo, J.; Li, Z.Q.; Wang, Y.; Yang, S. Combined exposure to hypoxia and ammonia aggravated biological effects on glucose metabolism, oxidative stress, inflammation and apoptosis in largemouth bass (Micropterus salmoides). Aquat. Toxicol. 2020, 224, 105514. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, S.L.; Huang, Y.; Hong, Y.H.; Xu, D.Y.; Jiang, C.W.; Huang, Z.Q. Unraveling the molecular mechanisms of nitrite-induced physiological disruptions in largemouth bass. Aquaculture 2024, 580, 740320. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, D.Y.; Chen, X.; Zhou, J.M.; Jiang, C.W.; Huang, Z.Q.; Qi, D.M. Deciphering the transcriptional regulatory networks of FOX genes in nitrite-induced spleen injury in largemouth bass. Aquat. Ecol. 2025, 59, 1–20. [Google Scholar] [CrossRef]
- Trenberth, K.E.; Dai, A.G.; van der Schrier, G.; Jones, P.D.; Barichivich, J.; Briffa, K.R.; Sheffield, J. Global warming and changes in drought. Nat. Clim. Change 2014, 4, 17–22. [Google Scholar] [CrossRef]
- Wang, W.; Yang, S.P.; Wang, C.G.; Shi, L.L.; Guo, H.; Chan, S.M. Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp. Fish Shellfish Immunol. 2017, 71, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Li, Q.Y.; Tu, J.P.; Chen, X.L.; Chen, X.H.; Liu, Q.Y.; Liu, H.; Zhou, X.Y.; Zhao, Y.Z.; Wang, H.L. Stress response and tolerance mechanisms of ammonia exposure based on transcriptomics and metabolomics in Litopenaeus vannamei. Ecotoxicol. Environ. Saf. 2019, 180, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.F.; Hoffmann, E.K.; Mills, J.W. The cytoskeleton and cell volume regulation. Comp. Biochem. Phys. A 2001, 130, 385–399. [Google Scholar] [CrossRef]
- Saraswathibhatla, A.; Indana, D.; Chaudhuri, O. Cell-extracellular matrix mechanotransduction in 3D. Nat. Rev. Mol. Cell Biol. 2023, 24, 495–516. [Google Scholar] [CrossRef]
- Pu, D.C.; Wang, Z.X.; Zheng, J.S.; Li, P.Y.; Wei, X.L.; Li, D.S.; Gao, L.H.; Zhou, L.; Wang, Y. Effects of Ammonia Stress on Liver Tissue Structure, Enzyme Activities, and Metabolome of Juvenile Largemouth Bass. Metabolites 2024, 14, 649. [Google Scholar] [CrossRef]
- Sinha, A.K.; Matey, V.; Giblen, T.; Blust, R.; De Boeck, G. Gill remodeling in three freshwater teleosts in response to high environmental ammonia. Aquat. Toxicol. 2014, 155, 166–180. [Google Scholar] [CrossRef]
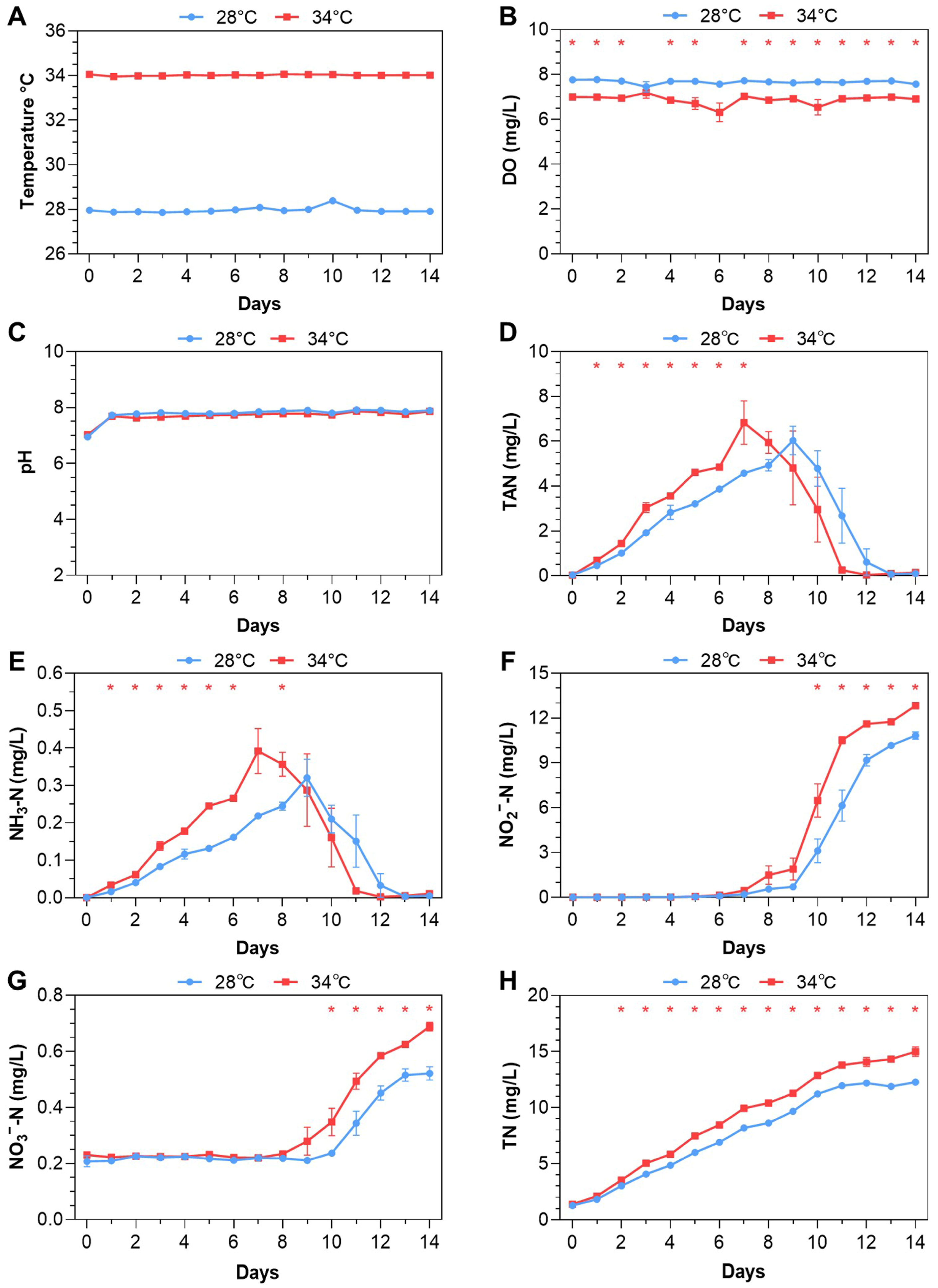
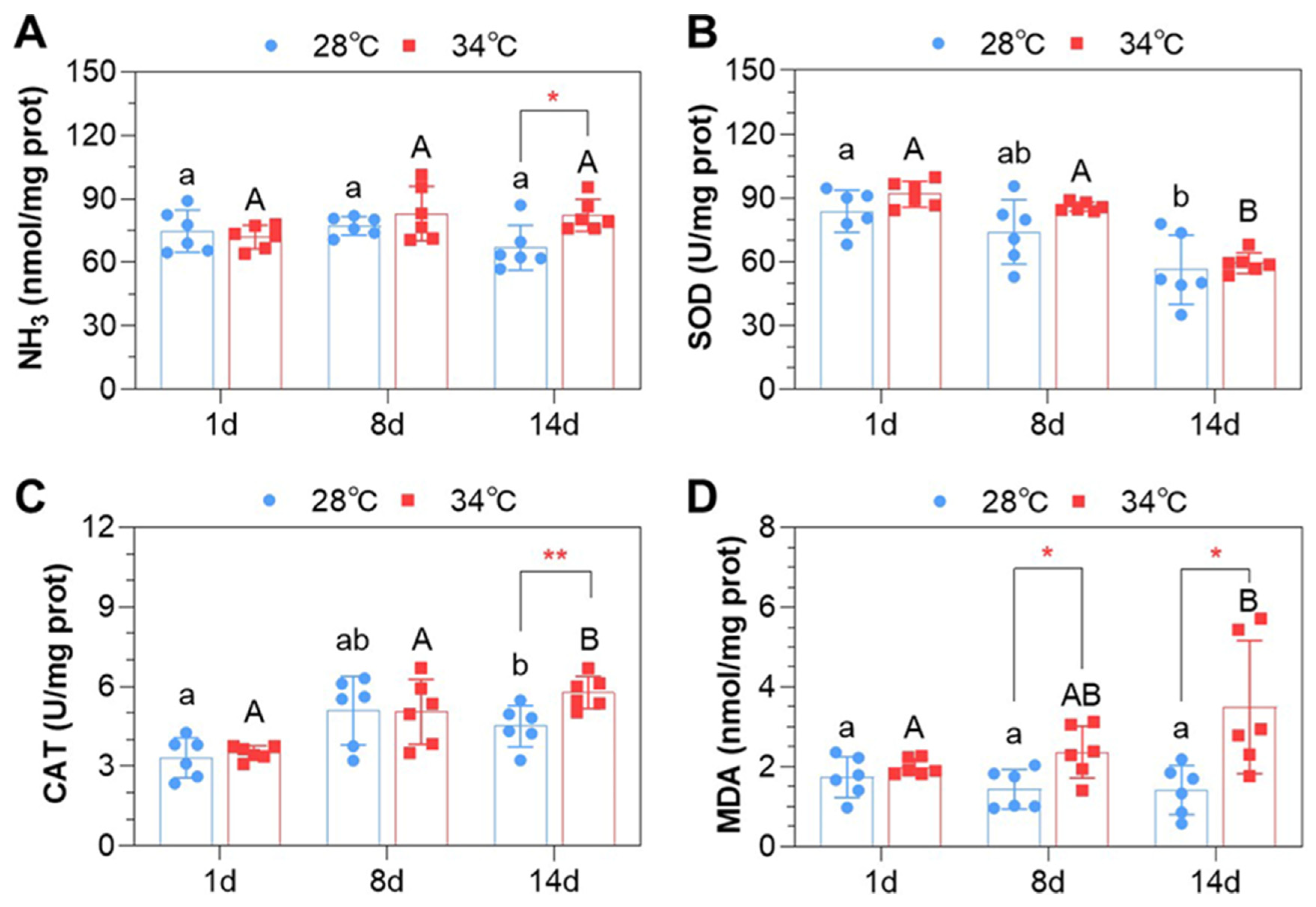

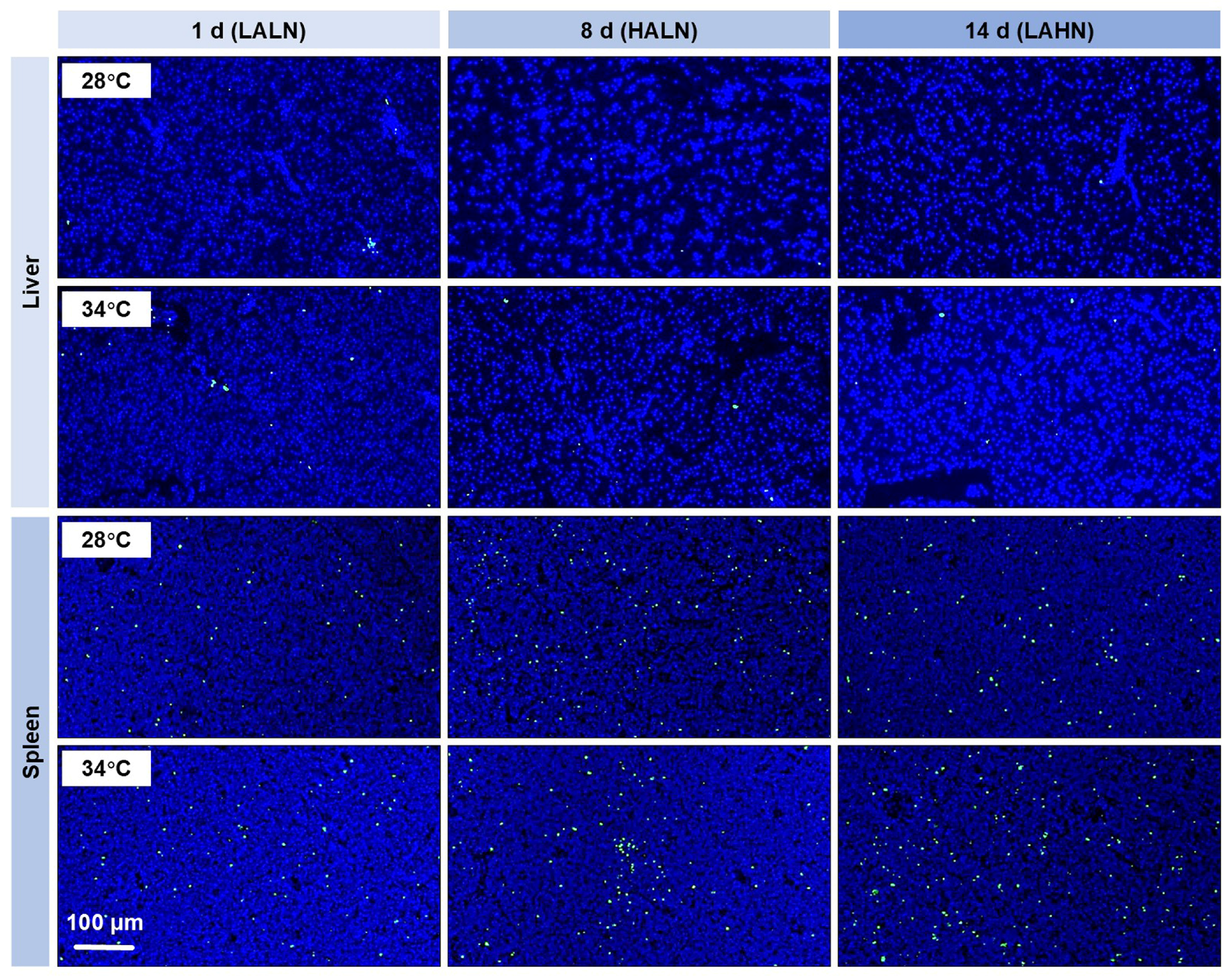

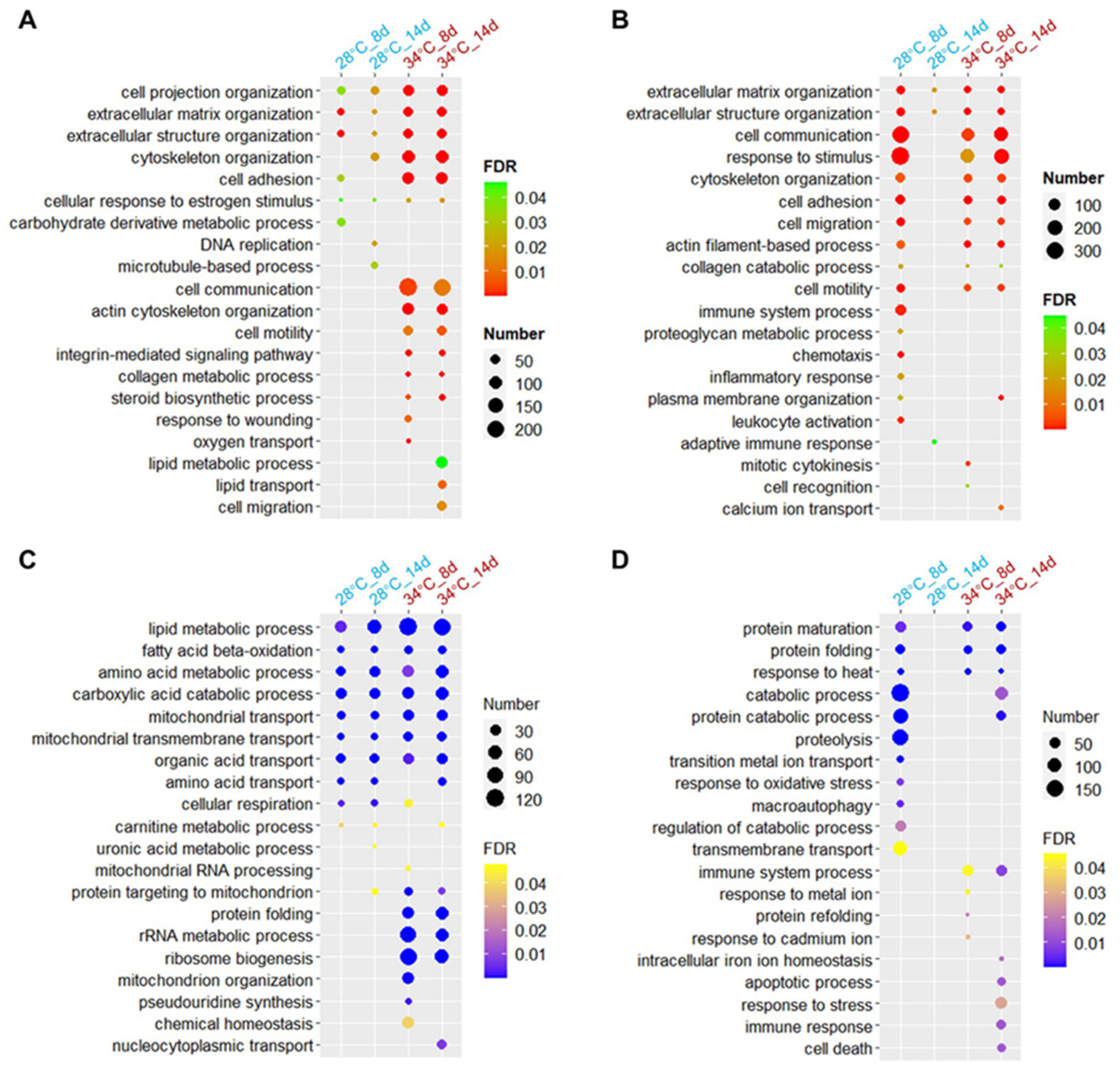
| Pollutant | Time (Days) | Experiment #1 | Experiment #2 | ||
|---|---|---|---|---|---|
| 28 °C | 34 °C | 28 °C | 34 °C | ||
| Ammonia (TAN, mg/L) | 1 | 0.45 ± 0.06 b | 0.68 ± 0.04 b,** | 0.53 ± 0.08 a | 0.16 ± 0.01 a,* |
| 8 | 4.93 ± 0.44 a | 5.95 ± 0.83 a | 0.02 ± 0.02 b | 0.03 ± 0.02 b | |
| 14 | 0.10 ± 0.01 b | 0.15 ± 0.09 b | 0.01 ± 0.02 b | 0.02 ± 0.02 b | |
| Unionized Ammonia (NH3-N, mg/L) | 1 | 0.02± 0.00 b | 0.03 ± 0.00 b,** | 0.02 ± 0.00 | 0.02 ± 0.01 |
| 8 | 0.24 ± 0.02 a | 0.36 ± 0.06 a,* | 0.00 ± 0.00 | 0.00 ± 0.00 | |
| 14 | 0.01 ± 0.00 b | 0.01 ± 0.01 b | 0.00 ± 0.00 | 0.00 ± 0.00 | |
| Nitrite (NO2−-N, mg/L) | 1 | 0.00 ± 0.00 b | 0.00 ± 0.00 c | 0.01 ± 0.00 | 0.01 ± 0.00 |
| 8 | 0.56 ± 0.35 b | 1.50 ± 1.08 b | 0.04 ± 0.00 | 0.00 ± 0.00 | |
| 14 | 10.83 ± 0.42 a | 12.83 ± 0.21 a,** | 0.00 ± 0.00 | 0.00 ± 0.00 | |
| Item | 28 °C | 34 °C |
|---|---|---|
| Survival (%) | 100.00 ± 0.00 | 94.87 ± 5.88 |
| Initial weight (g) | 632.00 ± 2.00 | 634.33 ± 1.46 |
| Average initial weight (g) | 21.07 ± 0.07 | 21.15 ± 0.05 |
| Final weight (g) | 523.33 ± 11.55 | 490.00 ± 10.00 * |
| Average final weight (g) | 23.79 ± 0.53 | 22.27 ± 0.46 * |
| Feeding amount (g) | 140.95 ± 2.14 | 141.69 ± 1.25 |
| Average weight gain rate (WGR, %) | 12.91 ± 2.19 | 5.33 ± 2.01 * |
| Average special growth rate (SGR, %) | 0.87 ± 0.14 | 0.37 ± 0.14 * |
| Feed conversion ratio (FCR) | 1.51 ± 0.11 | 2.98 ± 0.65 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Qiao, H.; Peng, L.; Meng, Y.; Song, G.; Luo, C.; Long, Y. Influence of High Temperature and Ammonia and Nitrite Accumulation on the Physiological, Structural, and Genetic Aspects of the Biology of Largemouth Bass (Micropterus salmoides). Antioxidants 2025, 14, 495. https://doi.org/10.3390/antiox14040495
Zhang Y, Qiao H, Peng L, Meng Y, Song G, Luo C, Long Y. Influence of High Temperature and Ammonia and Nitrite Accumulation on the Physiological, Structural, and Genetic Aspects of the Biology of Largemouth Bass (Micropterus salmoides). Antioxidants. 2025; 14(4):495. https://doi.org/10.3390/antiox14040495
Chicago/Turabian StyleZhang, Yuexing, Hui Qiao, Leyang Peng, Yujie Meng, Guili Song, Cheng Luo, and Yong Long. 2025. "Influence of High Temperature and Ammonia and Nitrite Accumulation on the Physiological, Structural, and Genetic Aspects of the Biology of Largemouth Bass (Micropterus salmoides)" Antioxidants 14, no. 4: 495. https://doi.org/10.3390/antiox14040495
APA StyleZhang, Y., Qiao, H., Peng, L., Meng, Y., Song, G., Luo, C., & Long, Y. (2025). Influence of High Temperature and Ammonia and Nitrite Accumulation on the Physiological, Structural, and Genetic Aspects of the Biology of Largemouth Bass (Micropterus salmoides). Antioxidants, 14(4), 495. https://doi.org/10.3390/antiox14040495






