Abstract
Sulfate transporters (SULTRs) are key players that regulate sulfur acquisition and distribution within plants, thereby influencing cellular redox hemostasis under pathogen attacks, such as Alternaria brassicicola (Ab). In this study, a total of 23 BolSULTR (Brassica oleracea SULTR) genes were identified from the Brassica genome. These BolSULTRs are distributed across nine chromosomes, with all collinear BolSULTR gene pairs undergoing purifying selections. Phylogenetic analysis reveals that the SULTR family is evolutionarily conserved among plant kingdoms. qRT-PCR analysis demonstrated that the expression of BolSULTRs varies across different plant organs and is modulated by hormonal signals. Furthermore, transcriptome analysis identified several BolSULTRs whose expression levels were depressed in Ab-challenged leaves in broccoli. Among them, the BolSULTR2;1 gene emerged as a key player in the plant’s response to Ab. Virus-induced gene silencing (VIGS) of BolSULTR2;1s resulted in elevated glutathione (GSH) levels and enhanced tolerance to Ab. Taken together, these findings underscore the role of BolSULTR2;1 in maintaining redox homeostasis and enhancing plant disease resistance, suggesting its potential as a target for genome editing to develop broccoli varieties with improved pathogen tolerance.
1. Introduction
Sulfur is one of the essential elements in all living organisms, and it plays a fundamental role in various biochemical processes in higher plants [1]. The leaves and roots of plants are essential tissues for the acquisition of external sulfur (S), playing a crucial role in the plant’s overall nutrient uptake and metabolism. Plants actively absorb hydrogen sulfide (H2S) through their leaves as a source of sulfur. The sulfur-deprived Brassica plants can be more easily restored to the level of plants grown in the presence of sulfate with H2S [2]. Plant roots take up sulfur from the soil mainly in the form of sulfates (SO42−), which is commonly mediated by several integral membrane proteins, namely sulfate transporters (SULTRs) [3,4]. Once imported into root cells, sulfate is translocated to plastids in leaves and also via SULTRs, where sulfate is converted to adenosine 5′-phosphosulfate (APS) [5]. Subsequently, APS undergoes two consecutive enzymatic reactions and is sequentially reduced to sulfite and sulfide by APS reductase (APR) and sulfite reductase (SiR), respectively. Finally, sulfide is incorporated into the carbon skeleton of O-acetylserine (OAS), and the resulting product cysteine is the first reduced sulfur donor involved in the biosynthesis of other sulfur biomolecules, such as methionine, sulfolipides, vitamins, glutathione (GSH), and glucosinolates [6,7,8,9].
It is reported that the typical SULTR proteins are generally H+/SO42− co-transporters with 12 transmembrane domains (TMDs), a Sulfate_transp domain in the N-terminal region, and an anti-sigma antagonist (STAS) domain in the C-terminal region [10,11]. Thus, the SULTR genes can be identified from the plant genome according to the sequences of conserved domains. For example, the model plant Arabidopsis thaliana contains 12 SULTR genes in its genomes [12,13]. Based on sequence similarity, the SULTR gene family can be divided into four groups. In Arabidopsis, Group I includes SULTR1;1, SULTR1;2, and SULTR1;3, known as high-affinity transporters distributed in the root surface cell layer of the roots [14,15]. However, Group II includes SULTR2;1 and SULTR2;2, which are low-affinity transporters responsible for sulfate transport from root to stem. It has been accepted that SULTRs in Group II mainly participate in long-distance transport, maintaining sulfur metabolite levels between the aerial parts [16]. Group III is the largest group with preferential expression in leaves, but their exact roles remain under investigation. It is proposed that SULTR3s facilitate SO42− uptake into chloroplasts and consequently influence downstream sulfate assimilation [17,18]. Interestingly, a yeast-based complementation experiment suggested that SULTR3;5 could directly interact with SULTR2;1, enhancing the transporter ability of SULTR2;1 under sulfur deficiency [19]. SULTR4;1 and SULTR4;2 belong to Group IV, and these SULTR4s are localized to the vacuolar membrane, where they actively export SO42− from the vacuole to the cytoplasm [20]. Thus, these functionally diversified SULTRs play central roles in the absorption and distribution of sulfate within different plant organs, cell types, and subcellular compartments.
Intensive studies have demonstrated the critical roles of SULTR genes in plant growth and development. In Arabidopsis, sulfur starvation strongly activates the expression of SULTR1;1, whereas under normal sulfate supply, SULTR1;2 is the major transporter [14,21]. Sulfate translocation from roots to shoots is largely mediated by microRNA395 (miR395) [22], which affects sulfate transport from root to stem and from old leaves to new leaves via direct suppression of SULTR2;1. The Arabidopsis sultr2;1 mutant exhibits an early flowering phenotype while keeping plant biomass unchanged [23], whereas several sultr3 mutants display stunted growth to varying degrees, resulting in a smaller thallus and shorter roots due to the reduced sulfate uptake to chloroplasts [17,24]. The SULTR4;1 and SULTR4;2 transporters are associated with sulfate transport in the vacuole, fulfilling the S requirements in young leaves and seeds [25,26]. Vacuoles isolated from callus of the sultr4;1 and sultr4;2 double knockout showed excess accumulation of sulfate [27]. However, our knowledge about their functions under abiotic and biotic stresses is still scarce. In rice, the miR395-OsSULTR2s module also regulates sulfate accumulation in leaves, resulting in broad-spectrum resistance to bacterial pathogens [28]. In the model legume Medicago truncatula, the majority of MtSULTR3 genes were strongly upregulated by drought or salinity [29], implying that they might be a potential target for abiotic stresses. Indeed, the loss of SULTR3;1 function led to lower ABA levels in Arabidopsis seedlings [30], further indicating that SULTRs could be the cross-talking point between sulfur metabolism and stress signaling pathways. Those findings suggest that SULTRs could help plants maintain sufficient sulfur supply, thereby protecting plants from environmental stresses as well as pathogen attacks.
Broccoli (Brassica oleracea L. var. italica) is a cruciferous vegetable that is widely cultivated worldwide owing to its nutritional value [31,32], and, in 2023, global production of broccoli (cauliflower included) reached approximately 25.9 million tons (http://faostat.fao.org/ (accessed on 15 October 2024)). Black spot disease, a necrotrophic pathogen caused by fungal Alternaria brassicicola (Ab), severely affects broccoli production. So far, fully Ab-resistant broccoli cultivars have not been reported yet, and, currently, all commercial varieties are highly susceptible to Ab [33]. Therefore, genetic manipulation is the most efficient method for the rapid improvement of broccoli resistance to Ab [34]. As sulfur is implicated in plant immunity, modulation of sulfur transport might benefit plant sulfur metabolism and consequently confer Ab resistance to broccoli.
Although the SULTR genes have been investigated in several plant species, their genomic distribution and biological functions in broccoli, to our knowledge, have been largely unexplored. Therefore, in this study, a genome-wide, comprehensive analysis of SULTR genes was performed to search for SULTRs associated with broccoli Ab resistance. In total, 23 SULTR genes were identified from broccoli and further confirmed through sequencing. The physical and chemical characteristics, genomic structures, chromosomal locations, evolutionary relationship, and expression profiles of the broccoli SULTR gene family were examined in detail. Furthermore, the roles of BolSULTR2;1a were analyzed through the virus-induced gene silencing (VIGS) approach, supporting its critical role in Ab resistance. Our study provides molecular information regarding the SULTR gene family in broccoli, providing a theoretical foundation for future genetic engineering to generate Ab-resistant vegetables.
2. Materials and Methods
2.1. Identification and Characterization of BolSULTRs
The newest Brassica oleracea cv. JZS V2.0 data were obtained from the Brassicaceae Database (BRAD) (http://brassicadb.cn (accessed on 10 December 2024)) [35]. The genome CDS sequence data were extracted and transformed into protein sequences using TBtools-II v2.154. Then, all Arabidopsis thaliana SULTR protein sequences were downloaded from the Arabidopsis Genome Database (TAIR) using SULTR as a keyword. These sequences were then used as seed sequences to search the B. oleracea protein database for the candidate BolSULTR using BLASTp with an e-value ≤ 1 × 10−5 [36]. In addition, the B. oleracea protein sequences of SULTR were blasted by using Hidden Markov Models (HMMs) for Sulfate_transp (PF00916) and STAS (PF01740) domains downloaded from the Pfam database [37]. Domain structures were verified using NCBI-CDD and the Pfams database [38,39], yielding 23 SULTR genes. Physicochemical properties were predicted using ExPASy (http://web.expasy.org/protparam (accessed on 10 November 2024)) [40], and subcellular localization was predicted using Plant-mPLoc [41].
2.2. Phylogenetic Analysis, Classification, and Evolutionary Analysis
Protein sequences of SULTR genes were retrieved from TAIR and BRAD. Multiple sequence alignment of broccoli and Arabidopsis SULTR proteins was performed using MUSCLE in MEGA X. Phylogenetic trees were constructed through the neighbor-joining method with 1000 bootstrap replicates [42]. Based on their kinship with Arabidopsis, the broccoli SULTR genes were designated as BolSULTR. Tree visualization and subfamily classification were performed using iTOL (https://itol.embl.de/login.cgi (accessed on 11 December 2024)). The neighbor-joining tree was constructed based on the alignment of SULTR amino acid sequences from Chlamydomonas reinhardtii, Physcomitrella patens, Oryza sativa, Solaunum tuberosum, Solanum lycoperiscum, Brassica oleracea L. var. italica, and Arabidopsis thaliana. The percent bootstrap support for 1000 replicates is shown on each branch.
2.3. Chromosome Location and Homologous Gene Analysis
The chromosome location of BolSULTR genes appeared on the BRAD. The density of each gene was analyzed in the entire chromosome region. We divided the number of genes by the region size and then multiplied it by a conversion factor of 1,000,000 to obtain the number of genes per million base pairs (Mb). Moreover, gene duplication events were examined using MCScanX with default parameters [43]. The events of gene duplication in broccoli were revealed through the Advanced Circus tool from TBtools. Meanwhile, the synonymous mutation rate (Ks), the nonsynonymous mutation rate (Ka), and the ratio of nonsynonymous to synonymous mutation rates (Ka/Ks) for duplicated BolSULTR genes were calculated using the Ka/Ks Calculator tool in TBtools. In addition, we downloaded the genome annotation files for Arabidopsis thaliana and rice from the Ensembl Plants database (https://plants.ensembl.org/index.html (accessed on 10 November 2024)) to create synteny maps and analyzed the distribution of homologous genes across these different species by using the TBtools.
2.4. Analysis of Conserved Motifs and Gene Structures of the BolSULTRs
We conducted a protein motif analysis of BolSULTR family members utilizing the MEME tool (http://meme-suite.org/tools/meme (accessed on 11 December 2024)) with default settings. Briefly, motif sites were expected to be distributed in sequences with zero or one occurrence per sequence, and a maximum of 12 patterns were expected. Other settings were as follows: the motif E-value threshold: no limit; sites per motif: 16–23; and motif width: 15–50. The motifs were annotated in the form of amino acid logo plots. The quantitative structures of exons and introns were determined using the genome annotation file (GFF file format). TBtools was utilized to integrate and visualize phylogenetic relationships, motif patterns, and gene structures.
2.5. Promoter Cis-Acting Element
We used TBtools to extract promoter sequences of upstream 2000 bp BolSULTR genes, and the cis-regulatory elements were identified using PlantCAR [44]. The redundant elements were removed, and, according to its potential functions, all elements were classified. TBtools was used to visualize element distribution (Figure S3).
2.6. Protein–Protein Interaction Prediction
The BolSULTR protein interactions based on data from Arabidopsis orthologs were analyzed using the online tool STRING (https://string-db.org/ (accessed on 11 November 2024)). The interacting proteins were functionally characterized through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses.
2.7. Plant Material and Treatment
Seeds of the susceptible cultivar ‘41-3’ of Brassica oleracea L. var. italica (broccoli) were germinated in a soil–perlite mixture (2:1) in a plant growth chamber with a photoperiod of 16 h of light and 8 h of darkness at a temperature of 25 °C ± 1 °C and a relative humidity of approximately 65%. The 8-week-old broccoli seedlings were used for Ab infection experiments. The A. brassicicola wild-type strain used in this study was routinely grown and maintained on potato dextrose agar at 24 °C.
2.8. Expression Patterns of BolSULTR Genes Based on RNA-Seq Data
The concentration of the A. brassicicola conidial suspension was 5 × 105 spores per milliliter with 0.05% (v/v) Tween 20 was added as a surfactant, which was applied using a spray bottle directly onto each leaf. The control group received the same volume of Tween 20 as the treatment groups. Additionally, the relative humidity of the environment was increased to 80%. We selected leaves at different time points (0 days, 1 day, 3 days, 5 days, and 7 days) after treatment to perform transcriptome sequencing (RNA-Seq); each time point included three samples, with each sample consisting of four biological replicates. Additionally, we searched seven RNA-seq datasets of Brassica from the GSE42891 (GEO database), specifically for the root, stem, leaf, flower, silique, bud, and callus. The BolSULTR genes were used as a template to screen for highly homologous genes in RNA-seq datasets, and then the expression pattern of the target gene was extracted based on the FPKM value. Subsequently, log2 transformation was applied to generate a heatmap using TBtool. RNA-seq dataset information is presented in Supplementary Tables S4 and S5.
2.9. Expression of SULTR Genes in Broccoli Treated with Different Phytohormones
We used different phytohormones to treat broccoli seedling leaves. The exogenous hormones were dissolved in a small amount of ethanol and then diluted to the desired concentration for each treatment with distilled water, including 100 mg/L of Ethephon (ETH, Solarbio, 16672-87-0, Beijing, China), 1 mM/L of MeJA (Macklin, 39924-52-2, Beijing, China), and 1 mM/L of SA (Biotopped, 69-72-7, Beijing, China). Working fluids were applied to the plants through spraying and distilled water was used as a control treatment while ensuring that the concentration of anhydrous ethanol in the spray solution was the same. Additionally, 0.1% (v/v) Tween 20 was added as a surfactant. We ensured that each plant leaf was sprayed thoroughly until it dripped completely. For each treatment, we selected three samples at different time points (0 h, 4 h, 24 h), and for each sample we used 3 biological replicates. The samples were verified through qRT-PCR. These samples were then rapidly frozen in liquid nitrogen and stored at −80 °C for further experiments.
2.10. RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from young broccoli leaves using the FastPure Plant Total RNA Isolation Kit (Vazyme, Nanjing, China), with modifications [45]. RNA was then treated with DNase I to remove genomic DNA in one step, and 3 μg of total RNA was used for first-strand cDNA synthesis using a cDNA synthesis kit (Transgen, Beijing, China). The resulting cDNA sequences served as templates for quantitative reverse transcription real-time PCR (qRT-PCR) using a Roche LightCycler 480 II and the LightCycler 480 SYBR Green I Master Mix (Toyobo, Shanghai, China), following the manufacturer’s instructions. Fold changes were calculated from 2−ΔΔCt values, with each assay replicated at least three times using three biological replicates (independent RNA preparations). The Actin-2 was employed as a reference gene. The relevant primer sequences are presented in Supplemental Table S3.
2.11. Virus-Induced Gene Silencing (VIGS) of BolSULTR2;1
We used the tobacco rattle virus (TRV) vector system, consisting of pTRV1 and pTRV2 vectors, to achieve virus-induced BolSULTR2;1 silencing (VIGS). The 315 bp sequence from CDS was subcloned into the viral vector pTRV2. The Agrobacterium tumefaciens strain GV3101 harboring the recombinant plasmids pTRV2:BolSULTR2;1 and pTRV1 was used for in vitro agroinoculation through leaf injection. The Brassica oleracea 15-cis-phytoene desaturase (PDS) was used as a positive control, and the negative control was the pTRV2 empty vector (TRV:00). The Agrobacterium tumefaciens strain GV3101 carrying the constructs was cultured overnight at 28 °C and centrifuged to remove culture medium containing antibiotics when OD600 reached 1.5–2.0. The concentrated bacterial solutions were fully suspended with 1 mL of 10 mM magnesium chloride solution containing 200 μM of acetylsyringone and then diluted into a working solution (OD600 = 0.5). Then, a needleless syringe was used to infiltrate the abaxial surface of the broccoli cotyledons. Each treatment was inoculated with fifty broccoli seedlings. All of the plants were grown in the same growth environment at 25/23 °C (day/night), with a 16 h light/8 h dark cycle.
When observing a bleached leaf phenotype in seedlings infiltrated with pTRV:PDS, we performed a qRT-PCR analysis on the selected seedlings (pTRV:BolSULTR2;1). Significant downregulation of BolSULTR2;1 in the bleached seedlings compared to negative controls indicated successful silencing. At that moment, we collected a sufficient amount of leaf tissue for further analysis. Subsequently, the silenced seedlings and control plants with comparable growth conditions were selected for inoculation with the Ab suspension liquid (5 × 105 conidia/mL). Determination of the stress-related physiological index contents of the leaves was performed on the fifth day after inoculation, and ImageJ software (Ver. 1.54k) was used to calculate the area of the lesions. The VIGS experiments were repeated at least three times, and each treatment was applied to more than 15 plants. The primers used for vector construction are listed in Supplemental Table S3.
2.12. Measurements of ROS-Related Physiological and Biochemical Parameters
Quantifications of reduced glutathione (GSH), superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX) were conducted using commercial kits (BC1175 for GSH, BC0175 for SOD, BC0205 for CAT, and BC0225 for APX) purchased from Solarbio Life Sciences Co., Ltd. (Beijing, China) following the manufacturer’s instructions. The spectrophotometry was performed with an Agilent BioTek microplate reader (Santa Clara, CA, USA). Each assay was performed with three biological replicates. The detection of reactive oxygen species (ROS) was conducted using Coolaber DAB (3,3′-diaminobenzidine) detection kit (Beijing, China). The leaf staining procedure was conducted as previously described [46].
3. Results
3.1. Identification of SULTR Genes in Brassica Oleracea Broccoli
In this study, 23 unique BolSULTR members were obtained from broccoli and named according to their homology with Arabidopsis counterparts (Figure S1 and Table S1). Information on the gene name, loci number chromosomal location, and physicochemical characteristics of putative proteins of individual BolSULTR genes is listed in Table 1. Of these, the molecular weight of putative BolSULTR proteins ranged from 43.33 kDa (BolSULTR3;1c) to 108.00 kDa (BolSULTR4;2). Furthermore, the range of isoelectric points extends from a slightly acidic pI of 5.96 (BolSULTR3;1b) to a more basic pI of 9.63 (BolSULTR3;4b). Notably, except for BolSULTR3;1b protein, which was distinguished by its hydrophilic character, the other members, which exhibit positive GRAVY values, showed a generally hydrophobic nature. Consistent with transporter roles, 19 BolSULTR proteins were predicted to be localized on the cell membrane, while all BolSULTR4 members might be localized to the chloroplasts or vacuoles. Surprisingly, BolSULTR3;1b could be targeted to the nucleus, probably reflecting its distinct functions other than as a transporter. Together, the results reinforced the assumption that these BolSULTRs might functionally diverge and participate in different biochemical processes under disparate environments.

Table 1.
Physicochemical properties of SULTR protein in broccoli.
3.2. Phylogenetic Analysis of SULTR Gene Family
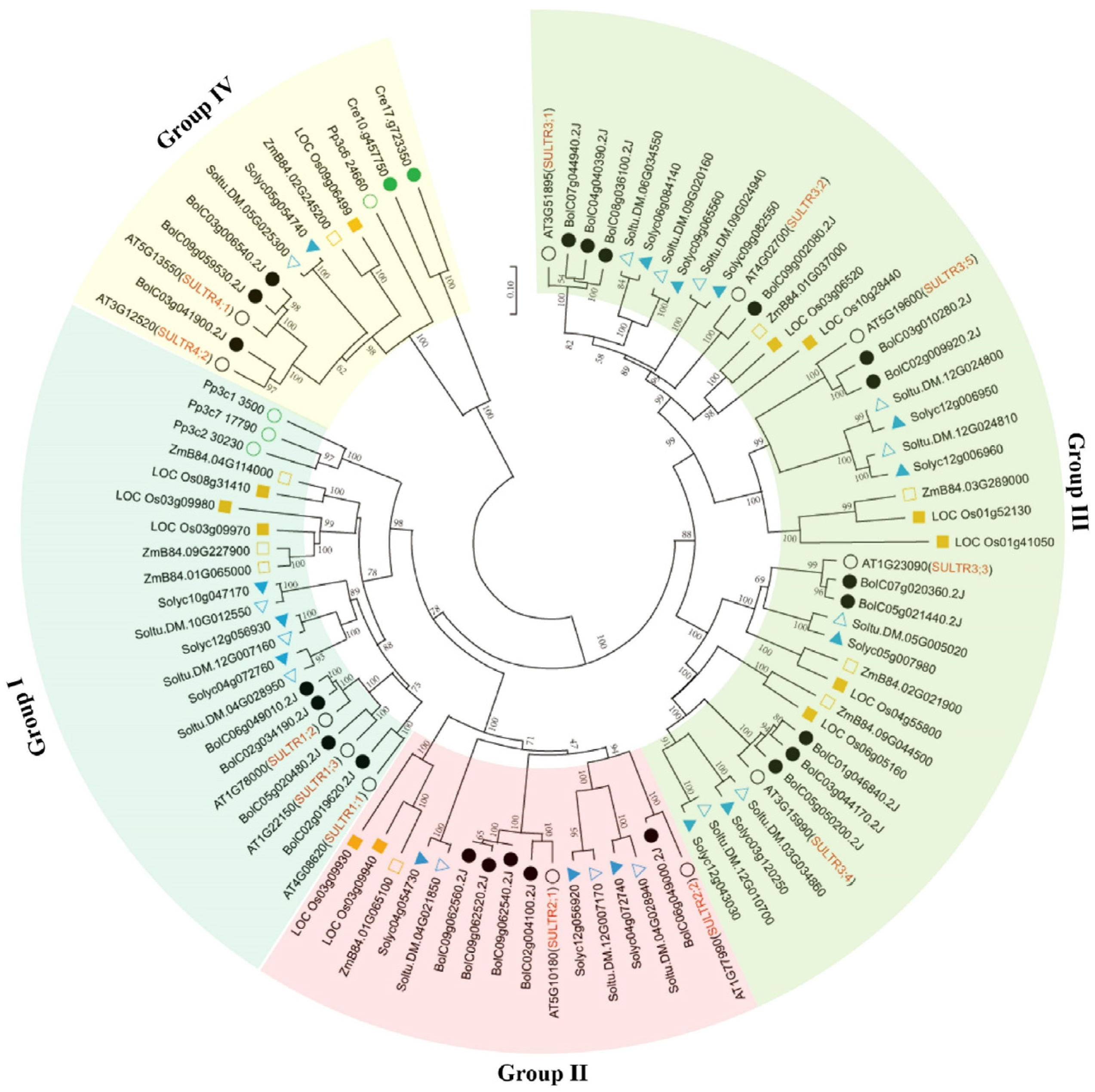
To investigate the phylogenetic relationship between the members of the BolSULTR family, a neighbor-joining tree was constructed based on the multiple alignment of SULTR protein sequences from green algae (Chlamydomonas reinhardtii), bryophyte (Physcomitrium patens), green algae (Chlamydomonas reinhardtii), rice (Oryza Stativa), tomato (Solanum tuberosum), potato (Solanum lycopersicum), Arabidopsis (Arabidopsis thaliana), and broccoli. Among these subfamilies, subclade III stands out with 34 members, which makes it the largest group; subclade I includes 26 members, followed by subclade IV (16 SULTRs) and subclade II (11 SULTRs). Only two and four SULTR proteins were identified from the genome of green algae and bryophytes, respectively (Figure 1). The reduced number of SULTRs in lower photosynthetic organisms may reflect evolutionary adaptations to their specific environmental conditions and physiological requirements. Conversely, in higher plants, such as Arabidopsis, tomato, and rice, dozens of SULTRs were found in their genomes, which indicates the expansion of the SULTR gene family in higher plants.
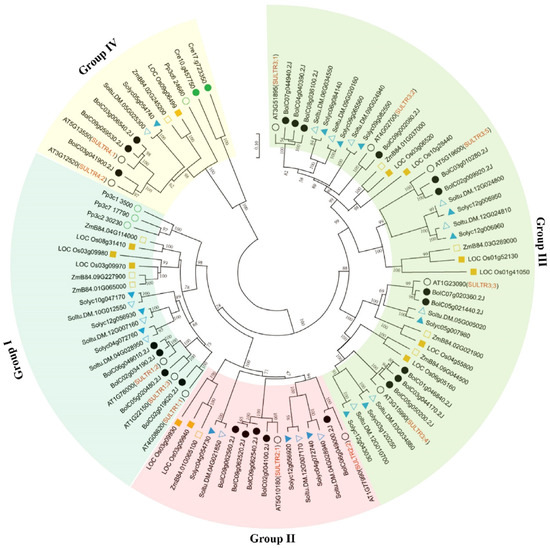
Figure 1.
Phylogenetic analysis of SULTR proteins in algae, liverwort, moss, and selected flowering plants. The neighbor-joining tree was constructed based on the alignment of SULTR amino acid sequences from Chlamydomonas reinhardtii (green circle), Physcomitrella patens (empty green circle), Solanum tuberosum (blue triangle), Solanum lycopersicum (empty blue triangle), Oryza sativa (empty yellow square), Zea mays (yellow square), Brassica oleracea L. var. Italica (black circle), and Arabidopsis thaliana (empty black circle). The position of the root was determined from an outgroup consisting of two SULTR-like sequences from yeast (Saccharomyces cerevisiae). The percent bootstrap support for 1000 replicates is shown on each branch.
3.3. Conserved Domains of BolSULTRs
To better understand the structural similarity of these SULTRs, the Hidden Markov Models were used to identify the TMD, STAS, and other conserved domains of BolSULTR, and the predicted domains were visualized in ChiPlot (Figure S2). Almost all BolSULTR proteins possess the Sulfate_transp domain (PF00916) and the STAS domain (PF01740), indicating that the BolSULTRs are highly conserved in broccoli. However, SULTR2;2 only has the intact TMD region, and its STAS domain was abolished. Thus, it is plausible that the transporter ability of SULTR2;2 could be impaired due to the sequence truncations. Other domains were identified in some BolSULTRs, which might confer novel functions. For example, in addition to TMD and STAS, BolSULTR4;2 also contains the Chitin-binding and glycoside hydrolase family 19 domains in its C-terminal. Similarly, LSm7 RNA-binding and high mobility group (HMG)-box motifs were found in BolSULTR3;1a and BolSULTR3;1b, respectively (Table S1).
3.4. Analysis of Conserved Motifs and Gene Structure of BolSULTRs
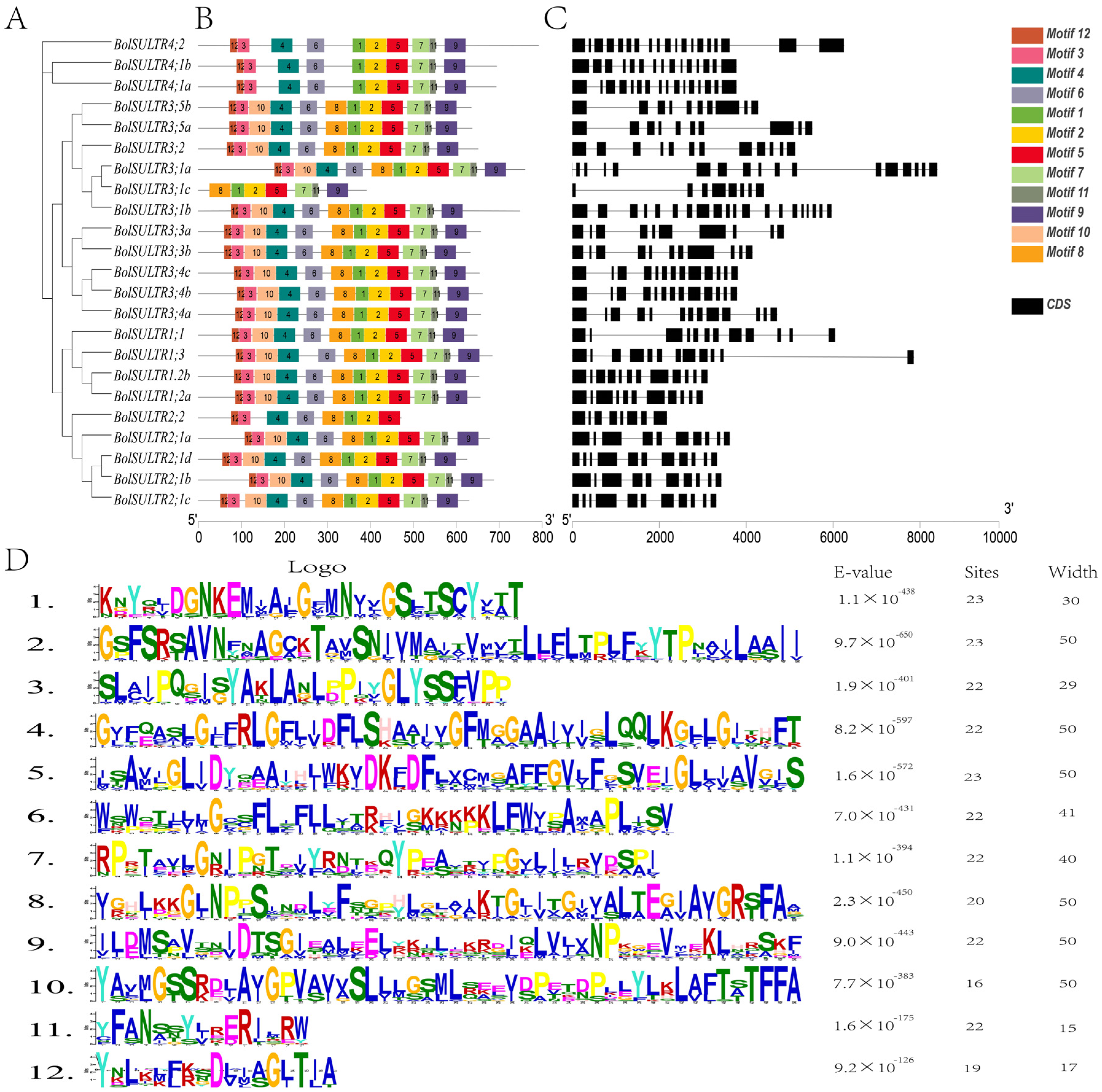
A total of 12 different motifs were detected among the BolSULTR proteins, with lengths ranging from 15 to 50 amino acids. The number of motifs varied from 7 to 12 among 23 BolSULTR proteins. The motifs 1, 2, and 5, containing the Sulfate_transp motif, or motifs 7, 9, and 11, containing the STAS signature motif, were characteristic in all BolSULTR proteins. These findings are consistent with the domain analysis of the SULTR family (Figure 2B,D). Additionally, motif 8 was exclusively absent in Group IV, and several motifs were deleted in BolSULTR2;2, BolSULTR3;1c, implying the different functions of these BolSULTRs. Additionally, most BolSULTRs within the same subfamily generally exhibit similar genetic structures in terms of the number and length of introns and exons (Figure 2C). BolSULTR2;1 members might arise from recent duplication events, as evidenced by the extremely high similarity in gene structure and sequences. In Group III, the number of exons ranged from 8 to 21, exhibiting a complex pattern. The SULTR members in Group IV had 15 to 19 exons, and BolSULTR4;2 possessed 2 longer exons at the 3′ end due to the additional domains. Overall, the conserved motifs and complex gene structure might provide more chances of heredity and variation for BolSULTR genes.
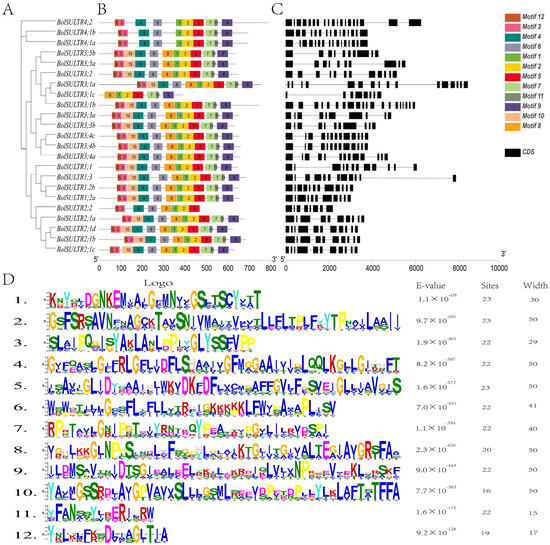
Figure 2.
Conserved motifs and gene structural analyses of the BolSULTRs. (A) Creating a phylogenetic tree involved arranging 23 BolSULTR conserved motifs. (B) The two conserved domains are divided into 12 motifs. The specific motifs or sequence patterns are retained across different BolSULTR proteins due to their functional importance. (C) General gene structure of BolSULTRs. Black lines and black boxes indicate introns, exons, and UTR, respectively. (D) The functional active site or binding site corresponds to the motif’s detailed sequence.
3.5. The Chromosome Localization, Gene Duplication, Ka/Ks, and Synteny Analysis
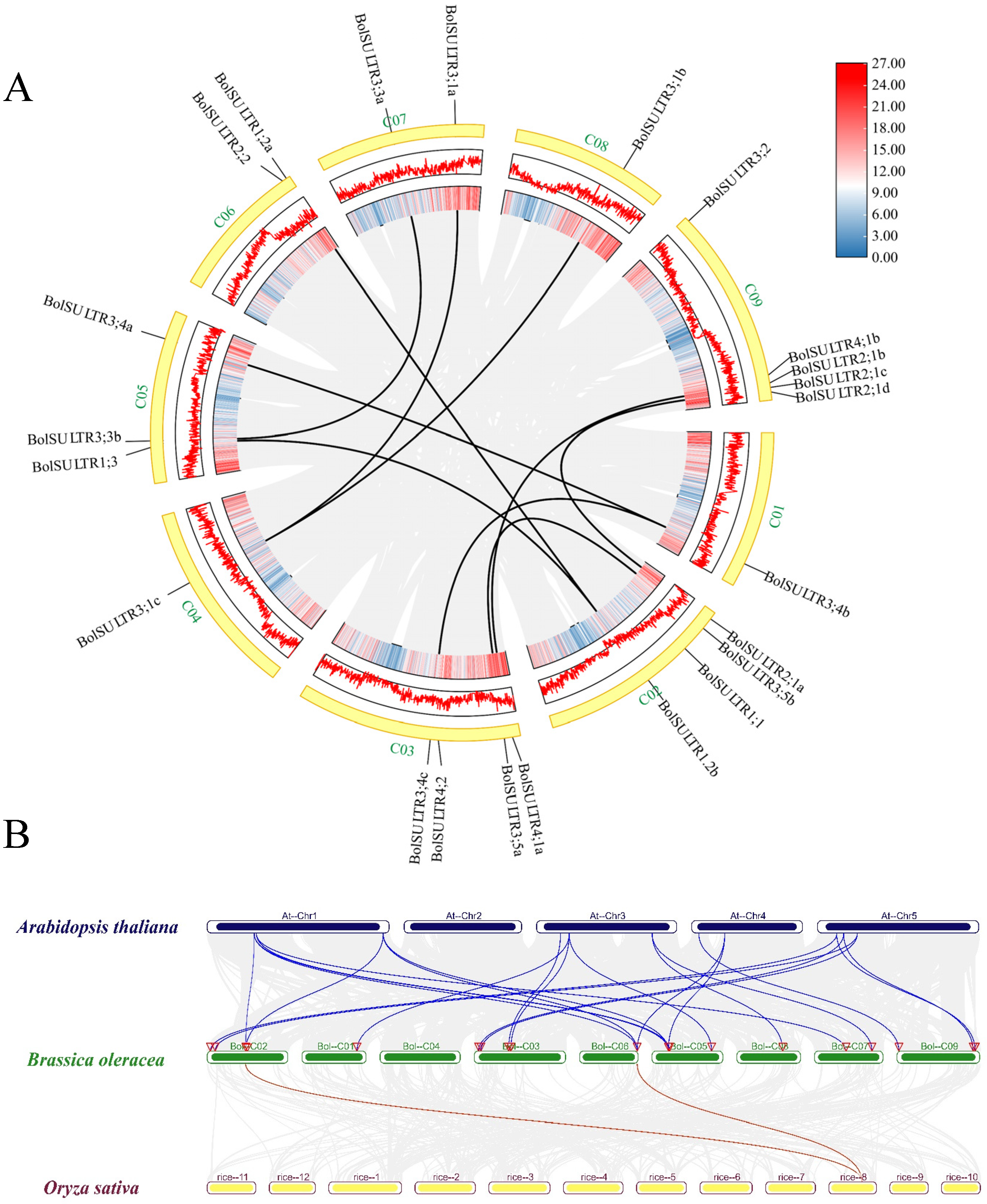
The results showed that 23 BolSULTR genes were unevenly distributed in nine chromosomes (Figure 3A). The majority of BolSULTR genes were mainly located on chromosomes C02, C03, and C09. Notably, only one BolSULTR3 gene was found on chromosomes C01, C04, and C08, while most BolSULTR2s were clustered on chromosome C09. In addition, the collinearity relationships showed BolSULTR1;1, BolSULTR2;2, BolSULTR3;2, and BolSULTR4;2 without associated collinear genes and a high degree of functional conservation among cruciferous species (Figure 3B). This may be due to biased gene loss in redundant gene members lacking collinear relationships [47]. Furthermore, consistent with previous observations, all collinear BolSULTR gene pairs had a Ka/Ks ratio of less than one, indicating that these genes underwent purifying selection. Interestingly, the genes BolSULTR2;1a and BolSULTR2;1b were involved in segmental duplication, while BolSULTR2;1b, BolSULTR2;1c, and the BolSULTR2;1d resulted from tandem duplication (Figure 3A and Table S2).
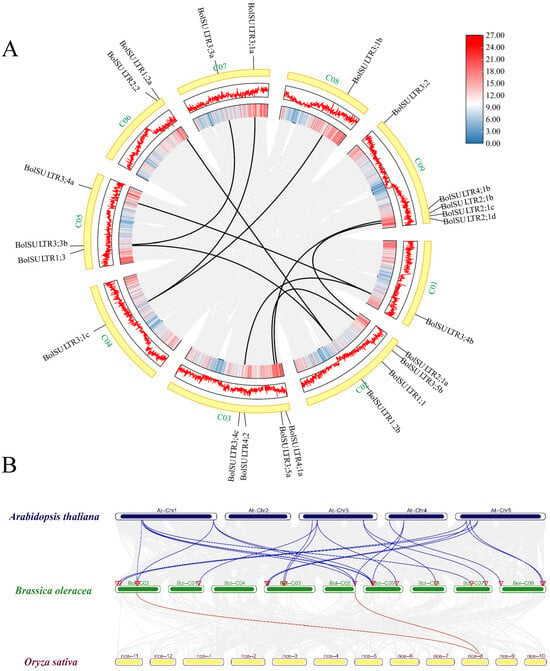
Figure 3.
Synteny analysis of BolSULTRs in the Brassica oleracea genome and between different species. (A) Chromosomal locations and synteny analysis of BolSULTRs in broccoli. Gray lines represent all synteny blocks in the B. oleracea genome, while black lines indicate segmental duplication genes between BolSULTRs. Chromosome gene density is represented by a hot map (inner circle) and column map (middle circle), with the outer circle showing the length of chromosomes. (B) Cognate relationship of BolSULTRs in broccoli, Arabidopsis, and rice; colored lines emphasize the pairs of syntenic BolSULTRs.
3.6. Analysis of the Promoter Region of the BolSULTRs
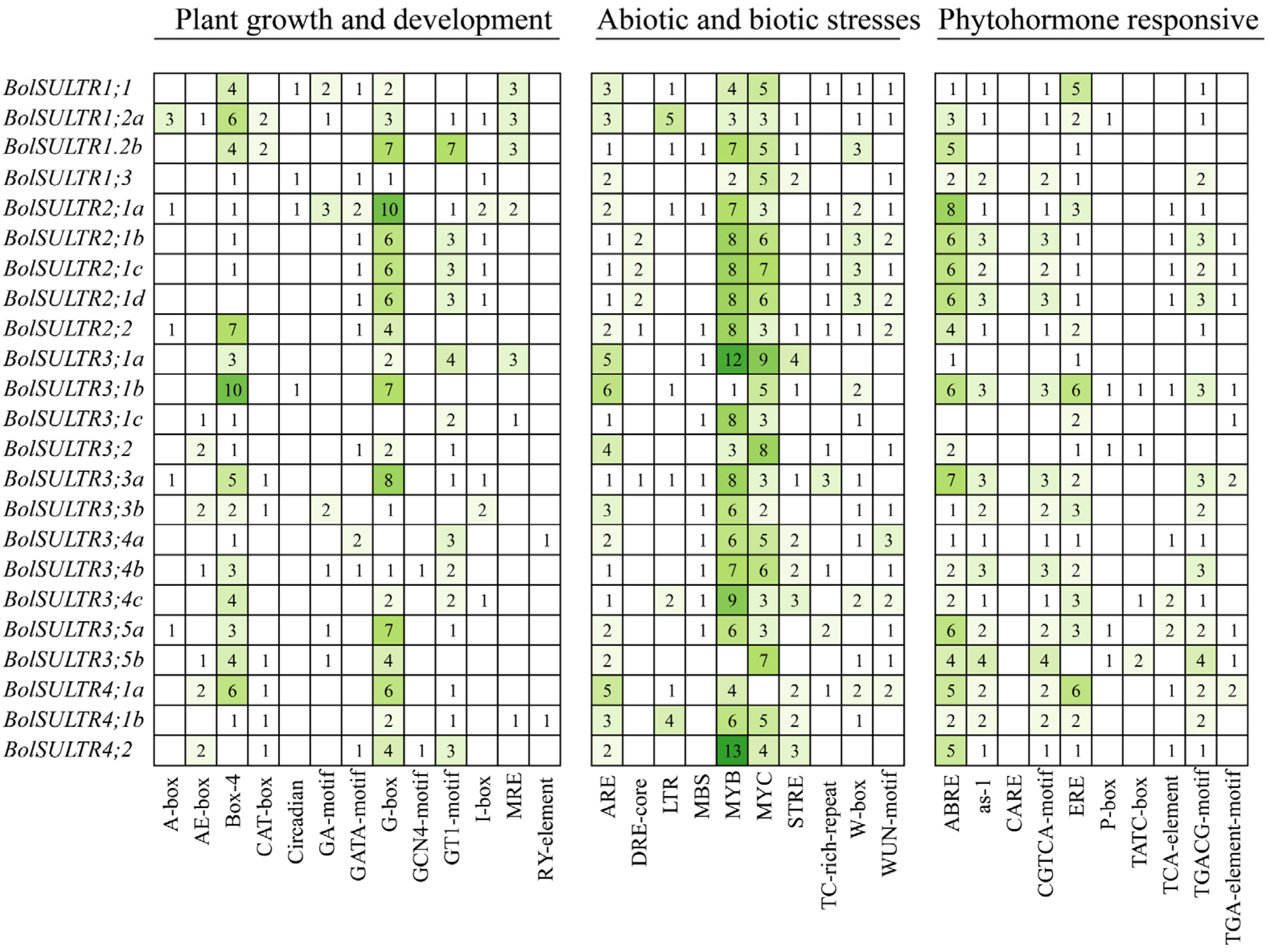
Through PlantCare analytics, we can identify the potential regulatory regime through the cis elements in the 2 kb upstream of all identified BolSULTR genes. It was clear of cis-acting element distribution (Figure S3), and we drew on their functional annotations to categorize three types: plant growth and development, abiotic and biotic stresses, and phytohormone responses (Figure 4). The results of type I showed that BolSULTR genes were associated with plant growth and development. This is because there were abundant optical signal sensing and transduction elements distributed in the promoter region, like Box-4 (ATTAAT), G-box (TACGTG), and GT1-motif, which can regulate gene expression related to photomorphogenesis. Important cis-acting elements for the cell cycle and metal ion response were found, such as CAT-box and MRE. These elements were also related to responses to environmental changes.
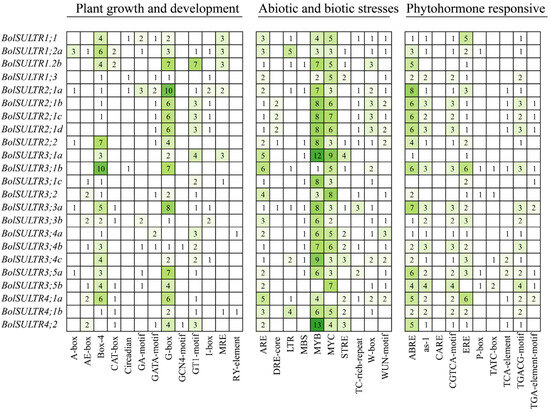
Figure 4.
Analysis of cis-acting elements of 23 BolSULTR gene promoters. Cis-acting elements of the 23 BolSULTR gene promoters are grouped, including phytohormone-responsive, biotic/abiotic stress, and plant growth and development. The different colors and number of boxes indicate different promoter elements in the BolSULTR genes.
The abiotic and biotic stress response elements were divided into the second category, which is associated with stress responses, particularly those related to defense against pathogens and other environmental stresses. Among them, the elements responding to abiotic stress were widely distributed, including MYB (CCAAT), MYC (CACATG), ARE (anaerobic response element), MBS (drought inducibility), and LTR (low-temperature-responsive) binding sites. The second was the functional element in response to biological stress, including TC-rich-repeat, W-box (WRKY-box), WUN-motif, and STRE (Stress-Responsive Element). These elements are involved in the activation of BolSULTR genes that are part of the plant’s response to various biotic stresses. The phytohormone response elements were grouped into the third category, which mainly included ABRE (ABA-responsive element), CGTCA-motif (MeJA-responsiveness), TGACG-motif (cis-acting regulatory element involved in MeJA-responsiveness), as-1element, and TCA-element (salicylic acid responsiveness). Additionally, ERE (Ethylene Response Element), TGA-element (auxin-responsive element), and P-box (gibberellin-responsive element) were identified in those BolSULTR promoters. These motifs are crucial for the regulation of genes involved in secondary metabolite production, defense compound biosynthesis, and other protective responses. Together, cis-element analyses indicate that BolSULTRs might be regulated by light, hormones, stress responses, and developmental cues.
3.7. BolSULTR Interaction Prediction
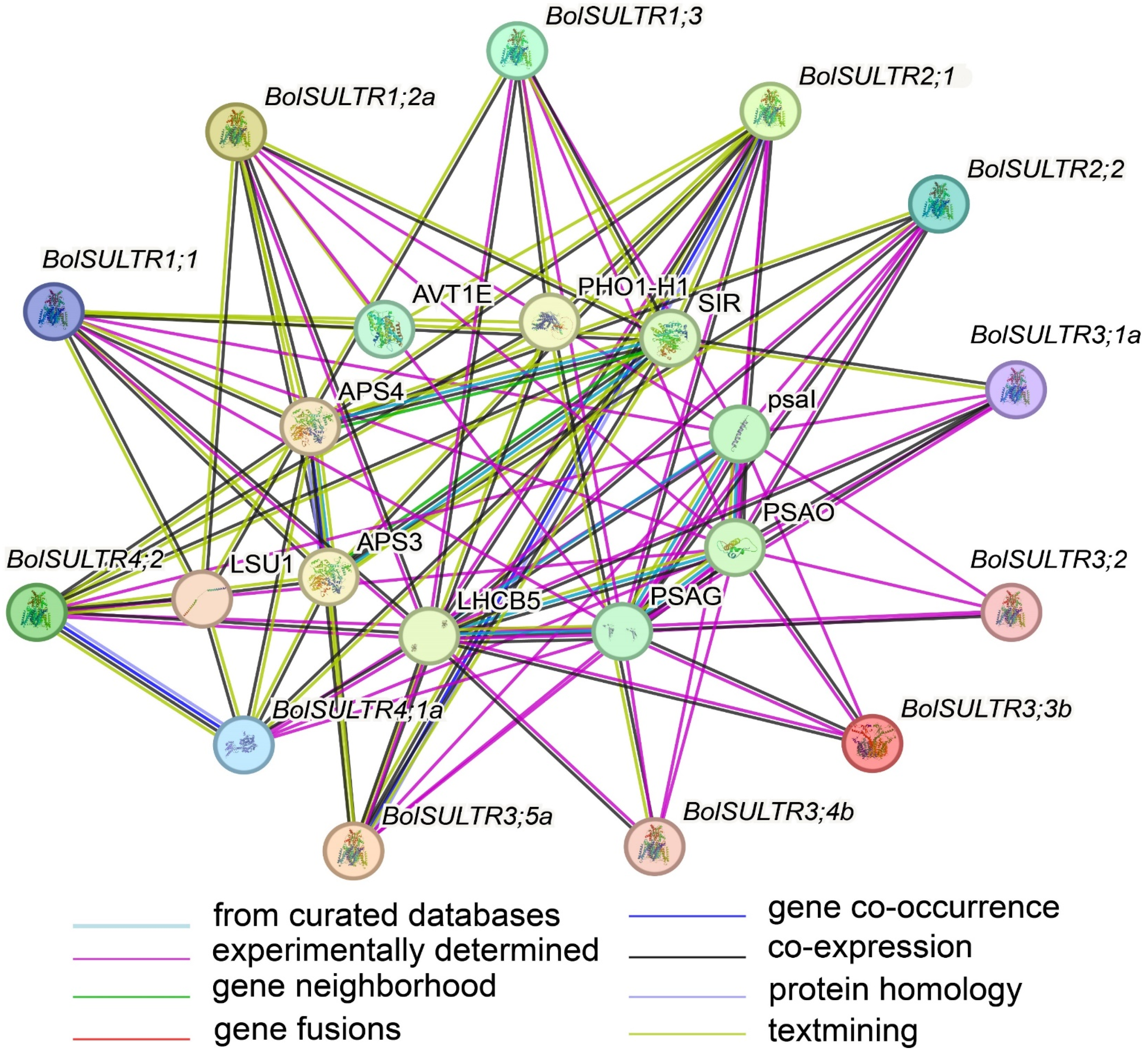
As interacting objects, amino acid transporter AVT1E (the amino acid/polyamine transporter 2 family), LHCB5 (Chlorophyll a-b binding protein), PSAO (Photosystem I subunit O), and PSAG (Photosystem I reaction center subunit V) were predicted to be the potential partners of BolSULTRs (Figure 5 and Table S6), which suggests that sulfur assimilation is closely associated with the antioxidant defense mechanisms of photosynthesis [48]. As expected, the BolSULTR proteins could interact with some proteins related to sulfur metabolism, including APS4 (ATP sulfurylase 4) and SIR (assimilatory sulfite reductase). This shows that BolSULTR proteins could form complexes with other proteins involved in the sulfur assimilation pathway to enhance efficiency and regulation.
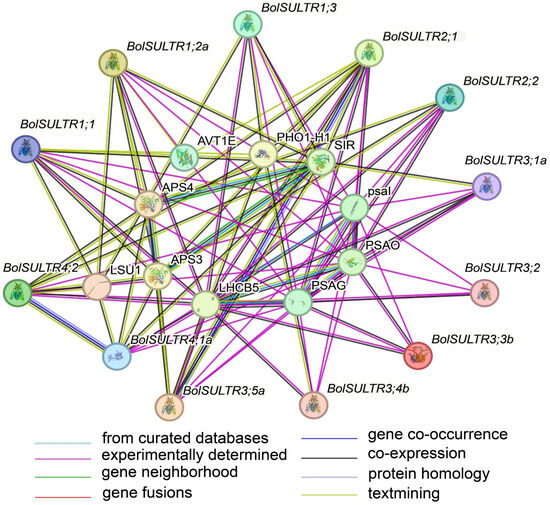
Figure 5.
Interactions between BolSULTR proteins and other proteins. Line color indicates the type of interaction evidence; edges represent protein–protein associations.
3.8. Expression Patterns of the BolSULTR Genes
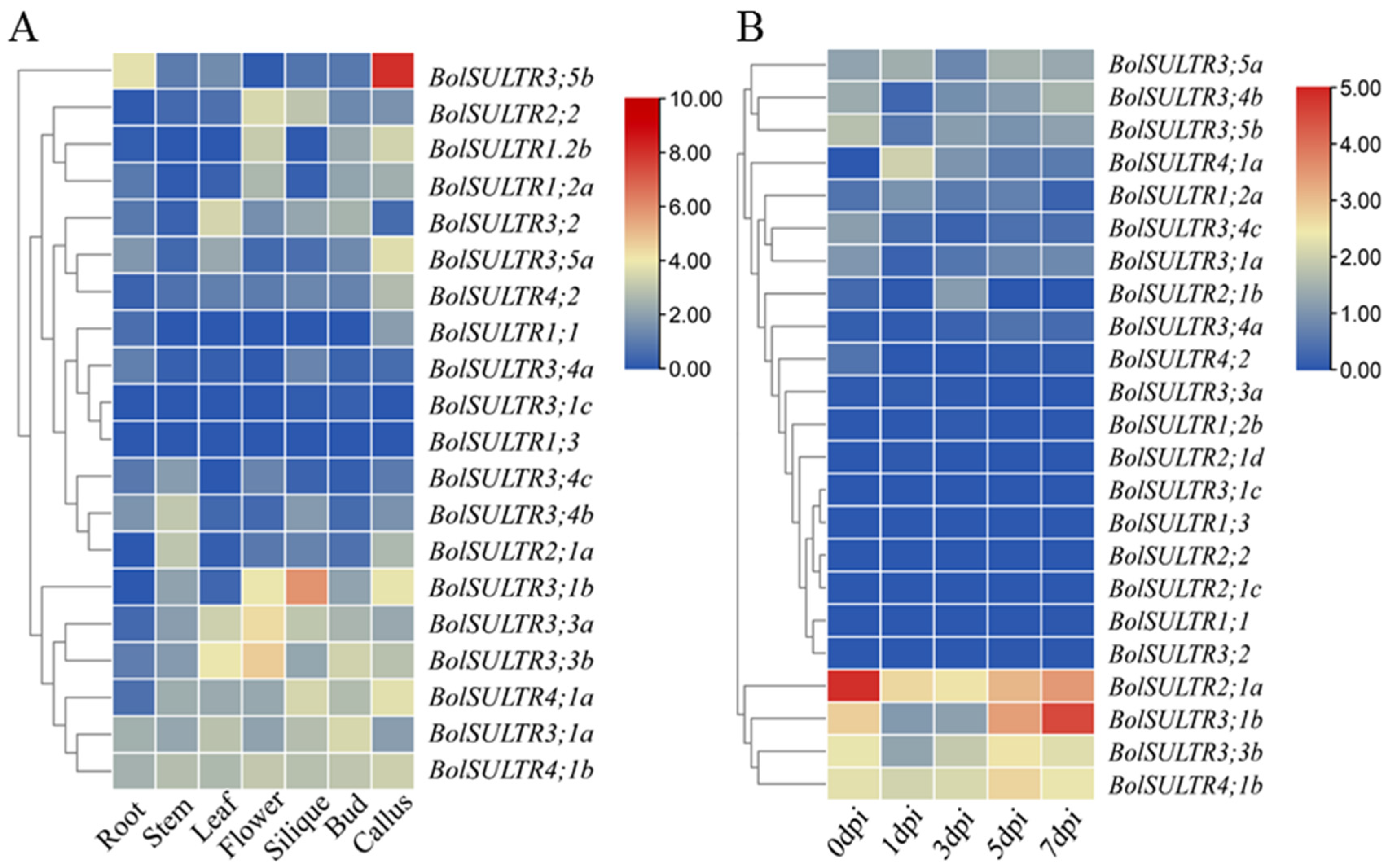
The expression profiles of BolSULTR were obtained from transcriptome analysis. Expression patterns of BolSULTR genes in different organs and under the Ab are shown in Figure 6A,B. The seven RNA-seq datasets of Brassica told us that BolSULTR3;1a and BolSULTR4;1b were ubiquitously expressed throughout the different organs, whereas other genes exhibited organ-preferential patterns. Specifically, BolSULTR2;1a was highly expressed in the stem. BolSULTR3;1b, BolSULTR2;2, and BolSULTR3;3b were highly expressed in flowers and the silique, and BolSULTR3;5b was predominantly expressed in the callus and roots. In contrast, BolSULTR3;1c, BolSULTR1;3, BolSULTR1;1, and BolSULTR3;4a displayed relatively lower expression levels across all organs. In summary, the expression of BolSULTR genes could be detected in diverse tissues and organs. The transcriptome datasets showed that initial gene expression levels (BolSULTR2;1a, BolSULTR3;1a/b, and BolSULTR4;1b and BolSULTR3;3b, BolSULTR3;4b, and BolSULTR3;5b) exhibited much higher expression levels than the other BolSULTRs. After the broccoli plants were challenged with Ab, their gene expression levels were robust biology stress responses and remained low until a correction occurred later in the course of the disease, particularly for BolSULTR2;1a. But, the homologous genes of BolSULTR2;1b/c/d did not respond. This suggests that BolSULTR2;1a may be important in early infections. Furthermore, the expression of BolSULT3;1b reached its maximum at 7 dpi, implying that it might be late-responsive genes during Ab infections.
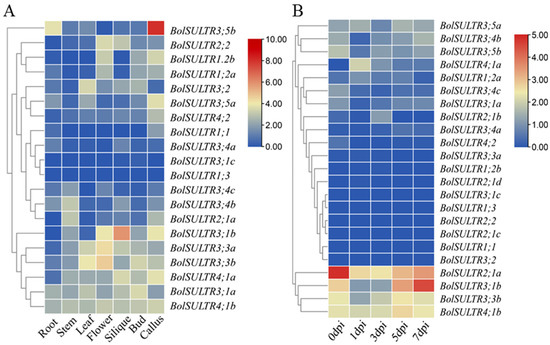
Figure 6.
Expression patterns of the BolSULTR genes. (A) Different tissues’ and organs’ expression of BolSULTR genes; RNA-seq datasets belong to the GSE42891 (GEO database). (B) Expression of the BolSULTR genes under Ab challenges at different points in time (0, 1, 3, 5, and 7 days). The transcriptome data were averaged through log2 transformation and TBtools software to generate a heatmap. The FPKM values are from three independent biological replicates.
3.9. Expression of BolSULTR Genes Under the Phytohormone Treatments
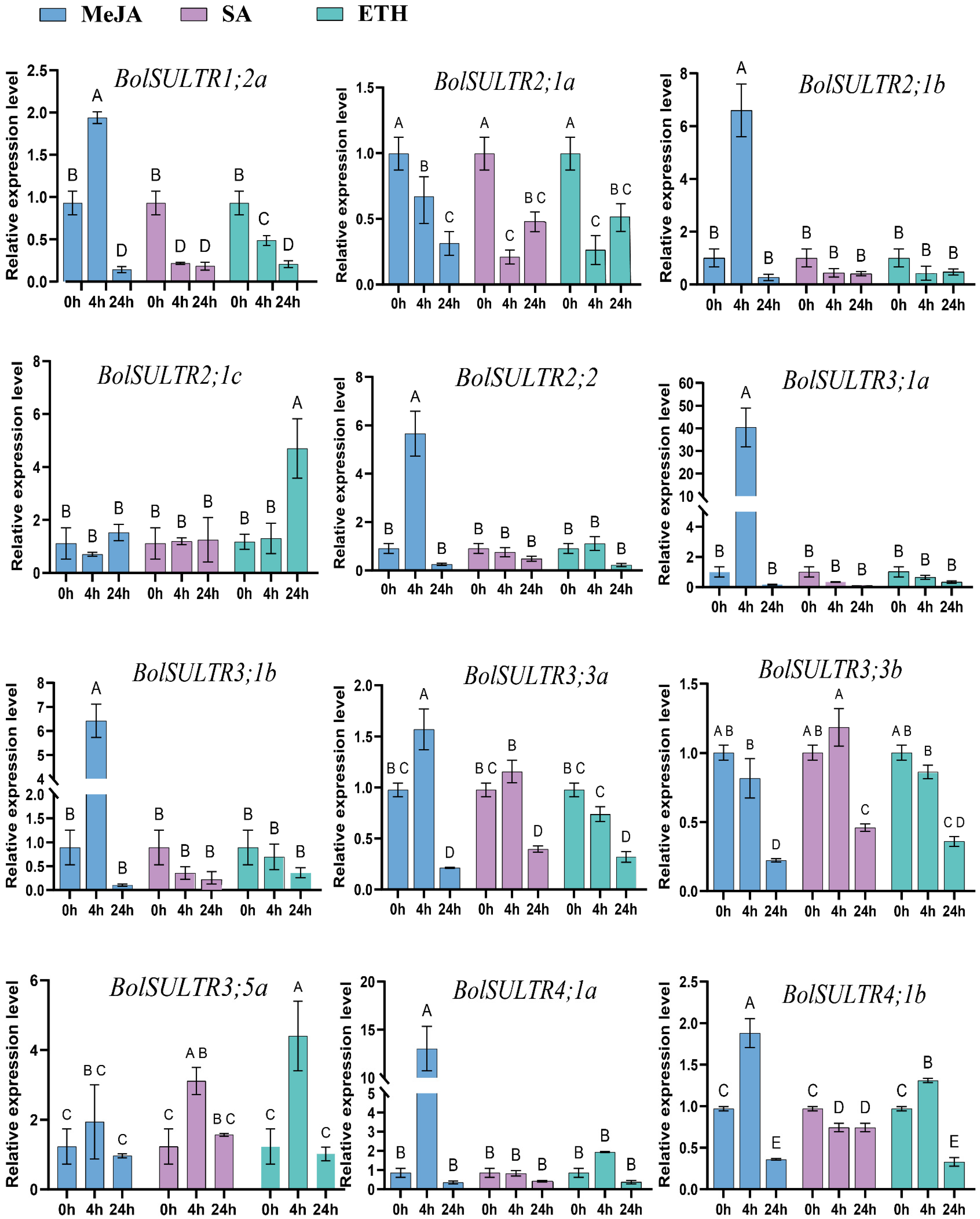
To further investigate the potential roles of the BolSULTR gene family in response to the effects of different hormones on BolSULTR expression in broccoli, we selected and examined the relative expression levels of twelve BolSULTR genes after various hormone treatments for 0 h, 4 h, and 12 h using qRT-PCR (Figure 7). The results indicate that the expression of most BolSULTR genes was upregulated robustly under exogenous MeJA treatment conditions, suggesting that MeJA could induce plant resistance to saprophytic pathogens [49,50,51]. Interestingly, the BolSULTR2;1a gene was downregulated in response to various hormones.
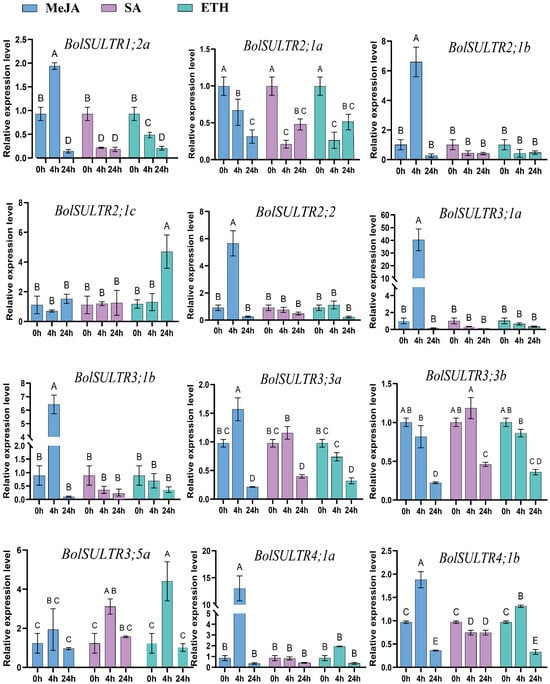
Figure 7.
The expression levels of BolSULTRs in different hormones using qRT-PCR analysis. In total, 100 mg/L of Ethephon, 1 mM/L of MeJA, and 1 mM/L of SA were sprayed onto seedling leaves. The leaves of mature broccoli were selected at different time points (0 h, 4 h, 24 h) and three from independent biological replicates. Capital letters indicate significant differences (p < 0.01). Error bars represent the standard deviation.
3.10. Silencing of BolSULTR2;1 Resulted in Reduced Leaf Spot Stress
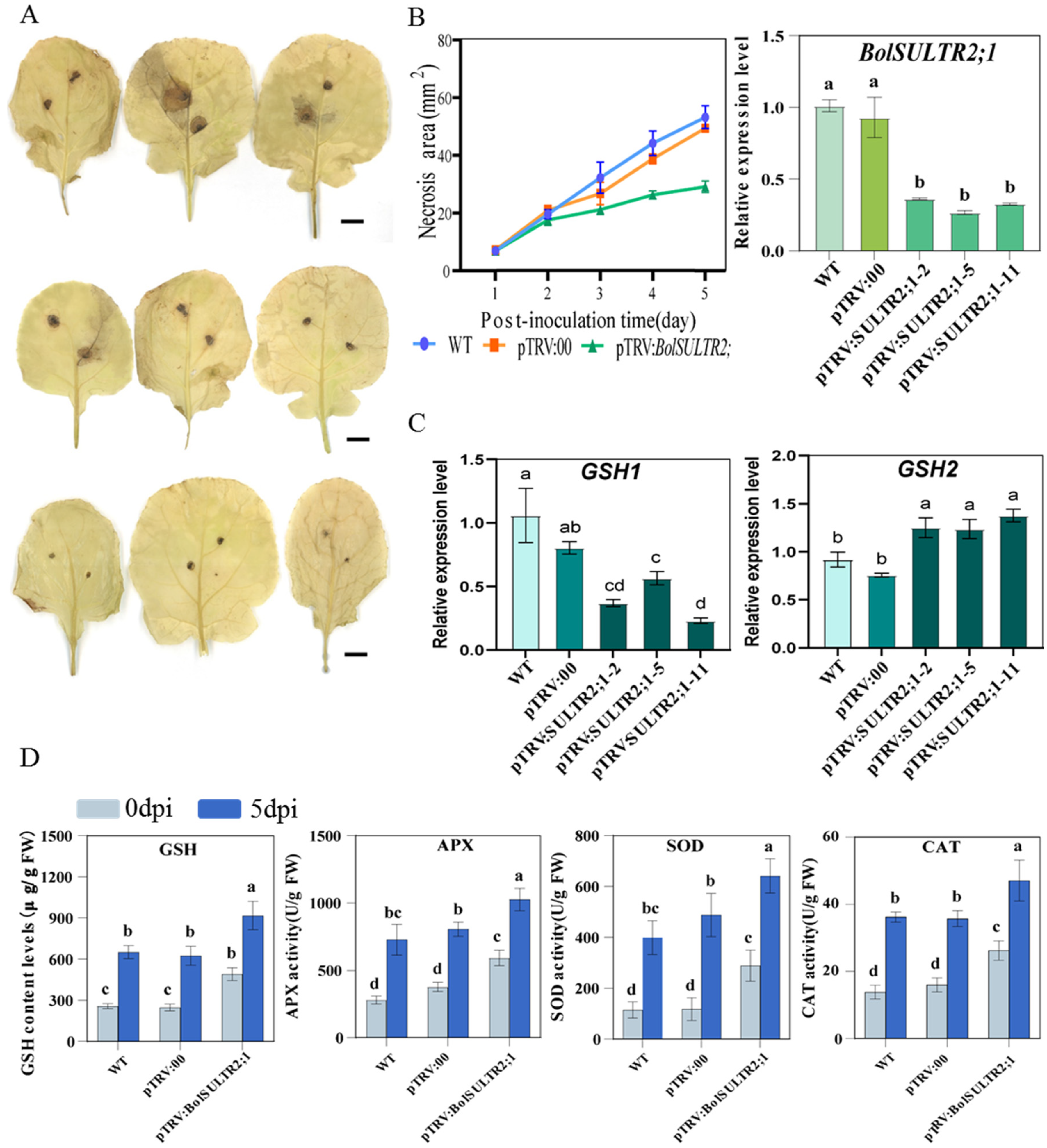
To further verify the functions of the BolSULTR2;1a gene in broccoli immunity, we obtained 25 independent silent plants of pTRV:BolSULTR2;1 via the VIGS approach. The qRT-PCR analysis suggested that the genes’ expression of BolSULTR2;1 was specifically downregulated in silent plants compared with the control group, and glutathione synthetases2 (GSH2) gene expression was upregulated (Figure 8C and Figure S5B). Then, these silent plants were subjected to phenotypic analysis and physiological parameter measurements. After 5 days of A. brassicicola inoculation, the silent plants exhibited local necrosis, and the areas of the necrotic spots were significantly smaller than those of the control plants. DAB staining revealed less extensive lesions of reactive oxygen species (ROS) in the silent plant leaves (Figure 8A). As glutathione (GSH) is the major antioxidant molecule in plants, we measured the GSH (glutathione) content in the leaves’ tissue surrounding the lesion. Consistent with the observed phenotypes, the GSH levels in silent plants were much higher than those of the control. And, with the development of the disease, the activity of various antioxidant enzymes in the silenced plants increased to a higher level than that of the control group (Figure 8D). Thus, these results illustrated that the repression of BolSULTR2;1 could lead to enhanced tolerance under Ab infections.
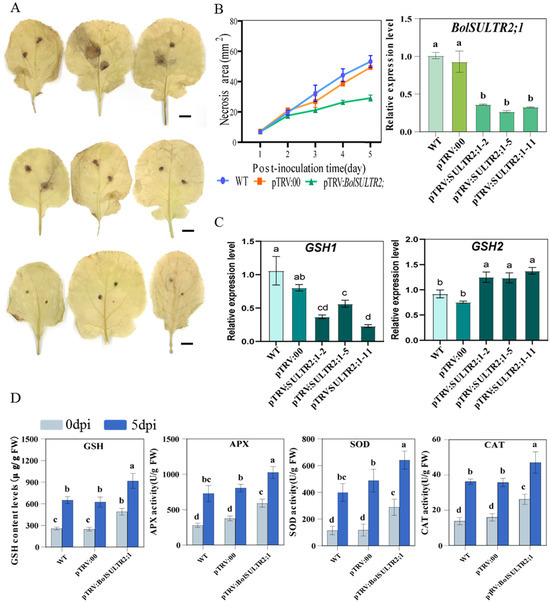
Figure 8.
Physiological parameter measurements of BolSULTR2;1-silencing and control plants. (A) DAB reactive oxygen species staining in leaves. Bars = 2 cm. (B) Analysis of infection site area; BolSULTR2;1 was specifically downregulated in silent plants. (C) Analysis of expression level of glutathione synthetases gene. (D) Physiological parameters of GSH, APX, SOD, and CAT with comparison between 0 days and 5 days under Ab stress in BolSULTR2;1-silencing plants. The mean values were derived from three independent biological replicates. Letters above each column indicate significant differences (p < 0.05 and p < 0.01). Error bars represent the standard deviation.
4. Discussion
4.1. The Paradox Between Genome Complexity and SULTR Copy Number in Broccoli
Plants use sulfate (SO42−) as a primary sulfur source to produce a wide range of secondary organic metabolites that have imperative roles in plant development, metabolism, and stress responses [52]. Once sulfate is acquired from the rhizosphere, it is assimilated to biosynthesize cysteine and other sulfur-containing metabolites in the leaf chloroplasts. Thus, higher plants need to translocate the sulfate from underground to aerial organs, which is accomplished by SULTR transporters [4,53,54]. The sulfate movements between organs, cells, and organelles are strictly regulated by internal and external signals. In the last two decades, the SULTR gene family has been studied much more intensively in model plant Arabidopsis and, to varying degrees, in other plant species, such as rice, maize, and cabbage [12,55,56,57]. In this study, a total of 23 BolSULTR genes were identified where 2, 4, 15, 15, 9, and 12 SULTRs were found in the genomes of green algae, moss, tomato, potato, maize, and rice, respectively. The results suggested that SULTRs might be encoded by a moderate-sized gene family.
Regardless of the variable size of their genome, the number of SULTRs is comparable in the species investigated. Among them, Zea mays B84 has the largest genome of 2.1 gigabases (Gb), while only nine ZmSULTRs were reported in the maize genome. However, the Arabidopsis with the smallest genome of 130 megabases (Mb) contains 12 SULTRs. As a close relative of Arabidopsis, broccoli belongs to the Brassicaceae family. Due to several rounds of whole genome duplication (WGD) events [47,58,59], the broccoli genome (613 Mb) is nearly five times larger than its Arabidopsis counterpart. Interestingly, the members of BolSULTR only slightly doubled compared to their Arabidopsis counterparts because the phylogenetic analysis did identify duplication events within the BolSULTR gene family. It seems that the increased complexity of the genome has little effect on the expansion of the SULTR gene family in these plants. Accordingly, the observations might provide a plausible hypothesis for the paradox between the increased genome complexity and the relatively stable copy number of SULTRs in diverse species. In this scenario, although several WGD events during Brassicaceae evolution technically increase the copy number of SULTRs in the genome, the duplicated SULTRs are eventually lost through natural selection that favors the deletion of redundant copies unless the duplicated SULTR can acquire new functions [47,60]. Indeed, Ka/Ks ratio analysis indicated that the majority of BolSULTRs have undergone strongly purifying selection in their evolutionary history. However, we are unable to deny other possibilities. For instance, other plant ion transporters, instead of SULTRs, might also have the ability to absorb sulfate from the environment, which might eliminate the need for more SULTRs in these higher plants with more complex genomes. Moreover, Group III represents the largest subfamily of SULTRs according to our analysis, which is in good agreement with previous findings. Although the functional identities of BolSULTR members in Groups II and III have not been determined in broccoli, the increase in BolSULTR copies might contribute to the sulfur assimilations and the biosynthesis of glucosinolates and their derivatives because broccoli can accumulate glucosinolates, a group of sulfur-rich secondary metabolites abundantly found in broccoli.
4.2. Group II and III SULTRs Might Represent Flowering-Plant-Specific Transporters Associated with Long-Distance Movement of Sulfate
The SULTRs are widely distributed across the kingdom Plantae. In previous studies, two and four SULTRs were reported in Chlamydomonas reinhardtii and moss Physcomitrium patens, respectively. According to the phylogenetic trees, two green algae SULTRs were present only in Group IV, implying that SULTR duplication predated the divergence of unicellular and multicellular life, suggesting that they may represent the ancestral precursors of SULTR found in land plants. Likewise, four bryophyte SULTRs were found in Groups I and IV, suggesting that the SULTR family diverged into at least two groups in the common ancestor of bryophytes. It is noteworthy that all SULTR members in Groups II and III belong to flowering plants, which leads to the suggestion that they might be associated with the long-distance transport of sulfate in plants. In support of this concept, Arabidopsis SULTR2;1 is specifically expressed in the pericycle and xylem parenchyma cells, controlling the amount of sulfate to be transported from the rhizosphere to the aboveground organs. Similarly, Arabidopsis SULTR3;5 was found to be preferably expressed in root vasculature, and the sultr3;5 mutant displayed a severe decrease in root to shoot sulfate movement [19]. Hence, it is not surprising that Group II and II SULTRs are absent in the non-vascular plants moss and algae. Therefore, we hypothesize that the expansion of two groups of SULTRs might have evolved in the vascular lineage as the multicellular land plants need long-distance sulfate transport from root to shoot.
4.3. BolSULTR2 Might Be Involved in Plant Defense Against Alternaria brassicicola (Ab), Presumably Through the Regulation of Glutathione Biosynthesis
It has been widely recognized that a sufficient sulfur supply is a prerequisite for plant responses to biotic stresses [29,61]. Because SULTRs are the major contributors to sulfate distributions within plants, they should theoretically impact plant immunity. In our study, the expression levels of several BolSULTR genes, including BolSULTR2;1a/b, BolSULTR3;1a/b, BolSULTR3;3b, and BolSULTR3;4b/c, were decreased at early stages, although some of them gradually returned to initial levels. Our results support the notion that some BolSULTRs might be repressed during the early stage of Ab attack, presumably altering the sulfate movements that favor plant immunity. Accordingly, promoter analysis of these BolSULTRs revealed the existence of the cis elements related to the MeJA, SA, and ethylene signaling pathways that are implicated in biotic stresses. Consistent with our findings, the expression levels of OsSultr2;1/2;2 and OsSultr3;1 were significantly reduced in the rice challenged by the fugal Magnaporthe grisea [28]. Similarly, the accumulation of StuSULTR2 transcripts was reduced significantly when potato leaves were inoculated by the oomycetes Phytophthora infestans [62]. Thus, these findings indicate that plants could control sulfate transport through the regulation of SULTR genes, particularly at the early stage of pathogen infections. Similarly, we also observed that transient silencing of BolSULTR2;1a resulted in enhanced tolerance to Ab in broccoli, implying that BolSULTR2;1a might be a negative regulator of Ab resistance. However, stable transgenic lines are still needed to further determine how sulfur metabolism and plant resistance are regulated by BolSULTR2;1a. In the future, we will generate the bolsultr2;1 knockout line through a gene editing method and investigate the distribution of sulfate in bolsultr2;1 lines.
Glutathione (GSH) is an important sulfur compound and a key cellular redox-buffering tripeptide with irreplaceable roles in plant defense mechanisms. It has been well-established that glutathione is oxidized by reactive oxygen species (ROS) as the antioxidant barrier that ameliorates oxidative stress imposed on sensitive cellular components [63,64,65]. GSH acts as a non-enzymatic antioxidant that directly removes reactive oxygen species (ROS) and works in concert with enzymatic antioxidant systems, such as the ASA–GSH cycle. For example, ascorbate peroxidase (APX) relies on GSH to regenerate ascorbic acid (ASA), and elevated levels of GSH may enhance APX activity, facilitating the co-removal of hydrogen peroxide (H2O2) [66]. In our study, the GSH content of the BolSULTR2;1-silencing in the leaves was much higher in comparison to the wild-type and TRV:00 lines. As expected, elevated levels of GSH can enhance antioxidant enzymes’ activity to work together to remove ROS, thereby protecting cells from oxidative stress, which provides an explanation for the BolSULTR2;1-silencing broccoli’s increased resistance to Ab.
The SULTR2.1 gene is involved in long-distance sulfate transport between leaves in Arabidopsis, and it is likely that BolSULTR2;1s might have similar capabilities in controlling sulfate movement. Therefore, our results perhaps show that upon Ab attack, broccoli leaves reduce the expression of BolSULTR2;1s, which transitorily reduces sulfate movement to non-infected counterparts. It is thus reasonable to assume that the accumulation of intracellular sulfate is conducive to elevated GSH biosynthesis during early infection, thus conferring increased tolerance to oxidative injuries imposed by Ab attack.
5. Conclusions
Sulfur is an essential element in plant growth resistance, and plant sulfur transporters play an essential role in the acquisition and distribution of sulfur. In this study, we identified 23 BolSULTR genes from the Brassica oleracea genome, classified and named them, studied the evolutional relationship, gene structure, and cis-regulatory elements of BolSULTRs in detail, and predicted the interaction proteins of the BolSULTR families. We characterized the expression patterns of BolSULTRs in different tissues and under different biological stresses. The VIGS experiment confirmed that silencing of BolSULTR2;1 resulted in enhanced tolerance to Ab by regulating GSH biosynthesis. These findings provide new insights into broccoli sulfate transporters in plant disease tolerance. Furthermore, functional studies are needed to reveal the molecular mechanism of BolSULTRs in sulfate transport and Ab infections.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antiox14040496/s1, Figure S1: The 23 BolSULTR members were named according to their homology with their Arabidopsis counterparts; Figure S2: The conserved regions of Sulfate_transp (PF00916) or STAS (PF01740) were visualized on the ChiPlot; Figure S3: Distribution map of promoter action elements for 2 kb upstream BolSULTR genes; Figure S4: Highly homologous gene sequences of BolSULTR2;1a; Figure S5: Schematic diagram of contagious disease; Table S1: The 23 BolSULTR members were identified and named according to their homology with their Arabidopsis counterparts; Table S2: The BolSULTR gene pairs had a Ka/Ks ratio and duplication type; Table S3: The relevant primer sequences; Table S4: RNA-seq datasets of Brassica from the GSE42891 (GEO database); Table S5: RNA-seq data of BolSULTR genes from leaves at five time points under Alternaria brassicicola stress in broccoli susceptible cultivar ‘41-3’; Table S6: Browse interactions in tabular form (lists only one-way edges: A-B).
Author Contributions
Conceptualization, N.L. (Ning Liu); funding acquisition, H.Z. and N.L. (Ning Liu); investigation, G.W. and N.L. (Ning Li); resources, Y.D.; supervision, H.Z. and N.L. (Ning Liu); writing—original draft, G.W.; writing—review and editing, N.L. (Ning Liu). All authors have read and agreed to the published version of the manuscript.
Funding
This project was financially supported by the “Collaborative Innovation Program (XTCX202302)”, the “Innovation and Development Program (KYCX202303)”, the “Reform & Development Program (KYCX202406)”, the “Cultivation of Green and High-quality Vegetable Varieties (030360007)” program of the Beijing Vegetable Research Center, and the “Young Talent Supporting Program (YCXTD00002-10)” of the Beijing Academy of Agriculture and Forestry Sciences.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author.
Acknowledgments
We thank to Decai Yu (Yunnan Agricultural University) for fruitful discussions on this manuscript.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Zenda, T.; Liu, S.; Dong, A.; Duan, H. Revisiting Sulphur—The Once Neglected Nutrient: It’s Roles in Plant Growth, Metabolism, Stress Tolerance and Crop Production. Agriculture 2021, 11, 626. [Google Scholar] [CrossRef]
- Ausma, T.; De Kok, L.J. Atmospheric H2S: Impact on Plant Functioning. Front. Plant Sci. 2019, 10, 743. [Google Scholar] [CrossRef]
- Buchner, P.; Stuiver, C.E.E.; Westerman, S.; Wirtz, M.; Hell, R.d.; Hawkesford, M.J.; De Kok, L.J. Regulation of Sulfate Uptake and Expression of Sulfate Transporter Genes in Brassica oleracea as Affected by Atmospheric H2S and Pedospheric Sulfate Nutrition. Plant Physiol. 2004, 136, 3396–3408. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.-H. Evolutionary Relationships and Functional Diversity of Plant Sulfate Transporters. Front. Plant Sci. 2012, 2, 119. [Google Scholar] [CrossRef]
- Mugford, S.G.; Yoshimoto, N.; Reichelt, M.; Wirtz, M.; Hill, L.; Mugford, S.T.; Nakazato, Y.; Noji, M.; Takahashi, H.; Kramell, R.; et al. Disruption of Adenosine-5′-Phosphosulfate Kinase in Arabidopsis Reduces Levels of Sulfated Secondary Metabolites. Plant Cell 2009, 21, 910–927. [Google Scholar] [CrossRef] [PubMed]
- Kopriva, S.; Calderwood, A.; Weckopp, S.C.; Koprivova, A. Plant Sulfur and Big Data. Plant Sci. 2015, 241, 1–10. [Google Scholar] [CrossRef]
- Saito, K. Sulfur Assimilatory Metabolism. The Long and Smelling Road. Plant Physiol. 2004, 136, 2443–2450. [Google Scholar] [CrossRef]
- Kopriva, S. Control of Sulphate Assimilation and Glutathione Synthesis: Interaction With N and C Metabolism. J. Exp. Bot. 2004, 55, 1831–1842. [Google Scholar] [CrossRef]
- Kopriva, S. Regulation of Sulfate Sssimilation in Arabidopsis and Beyond. Ann. Bot. 2006, 97, 479–495. [Google Scholar] [CrossRef]
- Shibagaki, N.; Grossman, A.R. Binding of Cysteine Synthase to the STAS Domain of Sulfate Transporter and Its Regulatory Consequences. J. Biol. Chem. 2010, 285, 25094–25102. [Google Scholar] [CrossRef]
- Wang, L.; Chen, K.; Zhou, M. Structure and Function of an Arabidopsis thaliana Sulfate Transporter. Nat. Commun. 2021, 12, 4455. [Google Scholar] [CrossRef] [PubMed]
- Hawkesford, M.J. Transporter Gene Families in Plants: The Sulphate Transporter Gene Family—Redundancy or Specialization? Physiol Plant. 2003, 117, 155–163. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, N.; Takahashi, H.; Smith, F.W.; Yamaya, T.; Saito, K. Two Distinct High-Affinity Sulfate Transporters with Different Inducibilities Mediate Uptake of Sulfate in Arabidopsis Roots. Plant J. 2002, 29, 465–473. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Inoue, E.; Saito, K.; Yamaya, T.; Takahashi, H. Phloem-Localizing Sulfate Transporter, Sultr1;3, Mediates Re-distribution of Sulfur From Source to Sink Organs in Arabidopsis. Plant Physiol. 2003, 131, 1511–1517. [Google Scholar] [CrossRef]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The Roles of Three Functional Sulphate Transporters Involved in Uptake and Translocation of Sulphate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, P.-X.; Miao, Z.-Q.; Qi, G.-F.; Wang, Z.; Yuan, Y.; Ahmad, N.; Cao, M.-J.; Hell, R.; Wirtz, M.; et al. SULTR3s Function in Chloroplast Sulfate Uptake and Affect ABA Biosynthesis and the Stress Response. Plant Physiol. 2019, 180, 593–604. [Google Scholar] [CrossRef]
- Cao, M.J.; Wang, Z.; Wirtz, M.; Hell, R.; Oliver, D.J.; Xiang, C.B. SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 2013, 73, 607–616. [Google Scholar] [CrossRef]
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot Transport of Sulfate in Arabidopsis. Evidence for the Role of SULTR3;5 as a Component of Low-Affinity Sulfate Transport System in the Root Vasculature. Plant Physiol. 2004, 136, 4198–4204. [Google Scholar] [CrossRef]
- Kataoka, T.; Watanabe-Takahashi, A.; Hayashi, N.; Ohnishi, M.; Mimura, T.; Buchner, P.; Hawkesford, M.J.; Yamaya, T.; Takahashi, H. Vacuolar Sulfate Transporters Are Essential Determinants Controlling Internal Distribution of Sulfate in Arabidopsis. Plant Cell 2004, 16, 2693–2704. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Inoue, E.; Watanabe-Takahashi, A.; Yamaya, T.; Takahashi, H. Transcriptome Profiling of Sulfur-Responsive Genes in Arabidopsis Reveals Global Effects of Sulfur Nutrition on Multiple Metabolic Pathways. Plant Physiol. 2003, 132, 597–605. [Google Scholar] [CrossRef]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 Mediates Regulation of Sulfate Accumulation and Allocation in Arabidopsis thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Soudthedlath, K.; Nakamura, T.; Ushiwatari, T.; Fukazawa, J.; Osakabe, K.; Osakabe, Y.; Maruyama-Nakashita, A. SULTR2;1 Adjusts the Bolting Timing by Transporting Sulfate from Rosette Leaves to the Primary Stem. Plant Cell Physiol. 2024, 65, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H. Regulation of Sulfate Transport and Assimilation in Plants. Int. Rev. Cell Mol. Biol. 2010, 281, 129–159. [Google Scholar] [CrossRef] [PubMed]
- Bachelet, F.; Sanchez, M.; Aimé, D.; Naudé, F.; Rossin, N.; Ourry, A.; Deulvot, C.; Le Signor, C.; Vernoud, V.; Neiers, F.; et al. The Vacuolar Sulfate Transporter PsSULTR4 is a Key Determinant of Seed Yield and Protein Composition in Pea. Plant J. 2024, 119, 2919–2936. [Google Scholar] [CrossRef]
- Dubousset, L.; Abdallah, M.; Desfeux, A.S.; Etienne, P.; Meuriot, F.; Hawkesford, M.J.; Gombert, J.; Ségura, R.; Bataillé, M.P.; Rezé, S.; et al. Remobilization of Leaf S Compounds and Senescence in Response to Restricted Sulphate Supply During the Vegetative Stage of Oilseed Rape are Affected by Mineral N Availability. J. Exp. Bot. 2009, 60, 3239–3253. [Google Scholar] [CrossRef]
- Liu, N.; Hu, M.; Liang, H.; Tong, J.; Xie, L.; Wang, B.; Ji, Y.; Han, B.; He, H.; Liu, M.; et al. Physiological, Transcriptomic, and Metabolic Analyses Reveal that Mild Salinity Improves the Growth, Nutrition, and Flavor properties of Hydroponic Chinese Chive (Allium tuberosum Rottler ex Spr). Front. Nutr. 2022, 9, 1000271. [Google Scholar] [CrossRef]
- Yang, Z.; Hui, S.; Lv, Y.; Zhang, M.; Chen, D.; Tian, J.; Zhang, H.; Liu, H.; Cao, J.; Xie, W.; et al. miR395-Regulated Sulfate Metabolism Exploits Pathogen Sensitivity to Sulfate to Boost Immunity in Rice. Mol. Plant 2022, 15, 671–688. [Google Scholar] [CrossRef]
- Gallardo, K.; Courty, P.E.; Le Signor, C.; Wipf, D.; Vernoud, V. Sulfate Transporters in the Plant’s Response to Drought and Salinity: Regulation and Possible Functions. Front. Plant Sci. 2014, 5, 580. [Google Scholar] [CrossRef]
- Cao, M.J.; Wang, Z.; Zhao, Q.; Mao, J.L.; Speiser, A.; Wirtz, M.; Hell, R.; Zhu, J.K.; Xiang, C.B. Sulfate Availability Affects ABA Levels and Germination Response to ABA and Salt Stress in Arabidopsis thaliana. Plant J. 2014, 77, 604–615. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Noor, N.M. Cruciferous Vegetables: Dietary Phytochemicals for Cancer Prevention. Asian Pac. J. Cancer Prev. 2013, 14, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Quispe, C.; Butnariu, M.; Sarac, I.; Marmouzi, I.; Kamle, M.; Tripathi, V.; Kumar, P.; Bouyahya, A.; Capanoglu, E.; et al. Phytotherapy and Food Applications from Brassica Genus. Phytother. Res. 2021, 35, 3590–3609. [Google Scholar] [CrossRef]
- Zhang, Q.; Branca, F.; Li, N.; Liu, N.; Ding, Y. Identification of Black Spot Resistance in Broccoli (Brassica oleracea L. var. italica) Germplasm Resources. Appl. Sci. 2024, 14, 2883. [Google Scholar] [CrossRef]
- Kumar, P.; Srivastava, D.K. Biotechnological Advancement in Genetic Improvement of Broccoli (Brassica oleracea L. var. italica), an Important Vegetable Crop. Biotechnol. Lett. 2016, 38, 1049–1063. [Google Scholar] [CrossRef]
- Cai, X.; Wu, J.; Liang, J.; Lin, R.; Zhang, K.; Cheng, F.; Wang, X. Improved Brassica oleracea JZS Assembly Reveals Significant Changing of LTR-RT Dynamics in Different Morphotypes. Theor. Appl. Genet. 2020, 133, 3187–3199. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Newbigin, E.; Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xie, L.; Hu, M.; Tong, J.; Wang, B.; Ji, Y.; Chen, J.; Liang, H.; Liu, W.; Liu, M.; et al. Effects of Blanching Cultivation on the Chemical Composition and Nutritional Quality of Chinese Chive. Food Chem. 2025, 464, 141824. [Google Scholar] [CrossRef]
- Lai, M.; Cheng, Z.; Xiao, L.; Klosterman, S.J.; Wang, Y. The bZip Transcription Factor VdMRTF1 is a Negative Regulator of Melanin Biosynthesis and Virulence in Verticillium dahliae. Microbiol. Spectr. 2022, 10, e02581-21. [Google Scholar] [CrossRef]
- Schnable, J.C.; Wang, X.; Pires, J.C.; Freeling, M. Escape from Preferential Retention Following Repeated Whole Genome Duplications in Plants. Front. Plant Sci. 2012, 3, 94. [Google Scholar] [CrossRef]
- Rahman, M.M.; Ghosh, P.K.; Akter, M.; Al Noor, M.M.; Rahman, M.A.; Keya, S.S.; Roni, M.S.; Biswas, A.; Bulle, M. Green Vanguards: Harnessing the Power of Plant Antioxidants, Signal Catalysts, and Genetic Engineering to Combat Reactive Oxygen Species under Multiple Abiotic Stresses. Plant Stress 2024, 13, 100547. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Hada, A.; Biswas, S.; Mishra, S.; Prusty, M.R.; Das, S.P.; Ray, S.; Kumar, A.; Sarker, U. Jasmonic Acid (JA) in Plant Immune Response: Unravelling Complex Molecular Mechanisms and Networking of Defence Signalling Against Pathogens. J. Plant Growth Regul. 2024, 44, 89–114. [Google Scholar] [CrossRef]
- Uji, Y.; Kashihara, K.; Kiyama, H.; Mochizuki, S.; Akimitsu, K.; Gomi, K. Jasmonic Acid-induced VQ-motif-Containing Protein OsVQ13 Influences the OsWRKY45 Signaling Pathway and Grain Size by Associating with OsMPK6 in Rice. Int. J. Mol. Sci. 2019, 20, 2917. [Google Scholar] [CrossRef]
- Wang, Z.; Tan, X.; Zhang, Z.; Gu, S.; Li, G.; Shi, H. Defense to Sclerotinia Sclerotiorum in Oilseed Rape is Associated with the Sequential Activations of Salicylic Acid Signaling and Jasmonic Acid Signaling. Plant Sci. 2012, 184, 75–82. [Google Scholar] [CrossRef]
- Capaldi, F.R.; Gratão, P.L.; Reis, A.R.; Lima, L.W.; Azevedo, R.A. Sulfur Metabolism and Stress Defense Responses in Plants. Trop. Plant Biol. 2015, 8, 60–73. [Google Scholar] [CrossRef]
- Leustek, T.; Saito, K. Sulfate Transport and Assimilation in Plants. Plant Physiol. 1999, 120, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Kopriva, S.; Takahashi, H. Sulfate Transport Systems in Plants: Functional Diversity and Molecular Mechanisms Underlying Regulatory Coordination. J. Exp. Bot. 2019, 70, 4075–4087. [Google Scholar] [CrossRef]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Trivedi, P.K. Differential Expression and Alternative Splicing of Rice Sulphate Transporter Family Members Regulate Sulphur Status during Plant Growth, Development and Stress Conditions. Funct. Integr. Genom. 2011, 11, 259–273. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, M.; Xia, Z. The SULTR Gene Family in Maize (Zea mays L.): Gene Cloning and Expression Analyses under Sulfate Starvation and Abiotic Stress. J. Plant Physiol. 2018, 220, 24–33. [Google Scholar] [CrossRef]
- Yang, X.; Liao, X.; Yu, L.; Rao, S.; Chen, Q.; Zhu, Z.; Cong, X.; Zhang, W.; Ye, J.; Cheng, S.; et al. Combined Metabolome and Transcriptome Analysis Reveal the Mechanism of Selenate Influence on the Growth and Quality of Cabbage (Brassica oleracea var. capitata L.). Food Res. Int. 2022, 156, 111135. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P.C.J. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef]
- Liang, Z.; Schnable, J.C. Functional Divergence between Subgenomes and Gene Pairs after Whole Genome Duplications. Mol. Plant 2018, 11, 388–397. [Google Scholar] [CrossRef]
- De Smet, R.; Adams, K.L.; Vandepoele, K.; Van Montagu, M.C.E.; Maere, S.; Van de Peer, Y. Convergent Gene Loss Following Gene and Genome Duplications Creates Single-Copy Families in Flowering Plants. Proc. Natl. Acad. Sci. USA 2013, 110, 2898–2903. [Google Scholar] [CrossRef]
- Kruse, C.; Haas, F.H.; Jost, R.; Reiser, B.; Reichelt, M.; Wirtz, M.; Gershenzon, J.; Schnug, E.; Hell, R. Improved Sulfur Nutrition Provides the Basis for Enhanced Production of Sulfur-Containing Defense Compounds in Arabidopsis thaliana upon Inoculation with Alternaria brassicicola. J. Plant Physiol. 2012, 169, 740–743. [Google Scholar] [CrossRef]
- Vatansever, R.; Koc, I.; Ozyigit, I.I.; Sen, U.; Uras, M.E.; Anjum, N.A.; Pereira, E.; Filiz, E. Genome-wide Identification and Expression Analysis of Sulfate Transporter (SULTR) Genes in Potato (Solanum tuberosum L.). Planta 2016, 244, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; Bai, W.; Liang, Y.; Yang, R.; Zhao, M.; Muhammd, S.M.; Zhang, D.; Li, X. ROS Scavenging Enzyme-encoding Genes Play Important Roles in the Desert Moss Syntrichia Caninervis Response to Extreme Cold and Desiccation Stresses. Int. J. Biol. Macromol. 2024, 254, 127778. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ohkama-Ohtsu, N.; Marsolais, F. Degradation of Glutathione and Glutathione Conjugates in Plants. J. Exp. Bot. 2023, 74, 3313–3327. [Google Scholar] [CrossRef] [PubMed]
- Singh, D. Juggling with Reactive Oxygen Species and Antioxidant Defense System—A Coping Mechanism under Salt Stress. Plant Stress 2022, 5, 100093. [Google Scholar] [CrossRef]
- Li, S. Novel Insight into Functions of Ascorbate Peroxidase in Higher Plants: More than a Simple Antioxidant Enzyme. Redox Biol. 2023, 64, 102789. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).