Double Auxotrophy to Improve the Safety of a Live Anti-Pseudomonas aeruginosa Vaccine
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Manipulations
2.2. Growth, Viability and Phenotypic STABILITY
2.3. Animal Experiments
2.4. ELISA
2.5. Statistical Analysis
3. Results
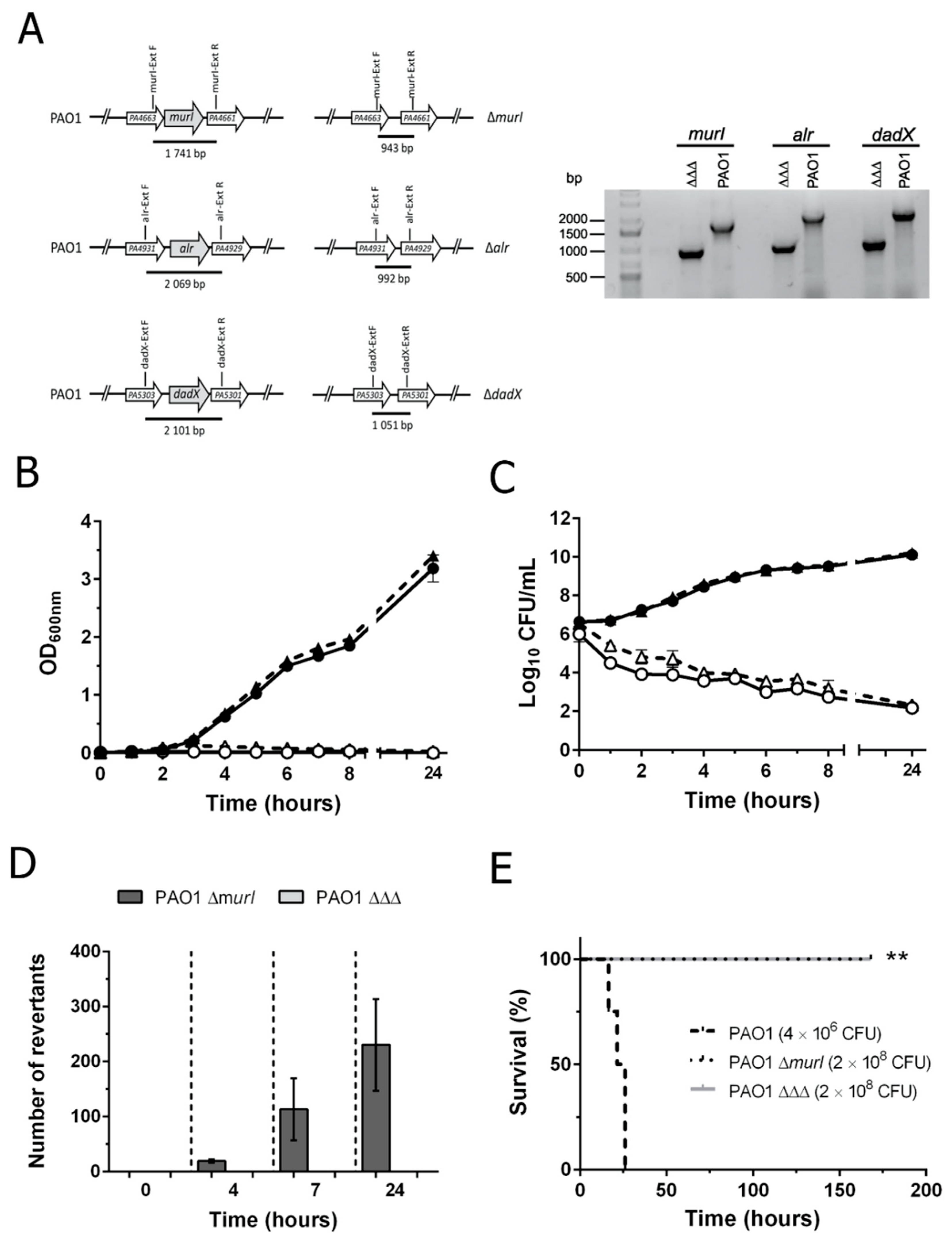
3.1. Construction and Characterization of the Double Auxotroph for D-Glutamate plus D-Alanine
3.2. P. aeruginosa PAO1 ΔΔΔ Has a More Stable and Safer Auxotrophic Phenotype Than PAO1 ΔmurI
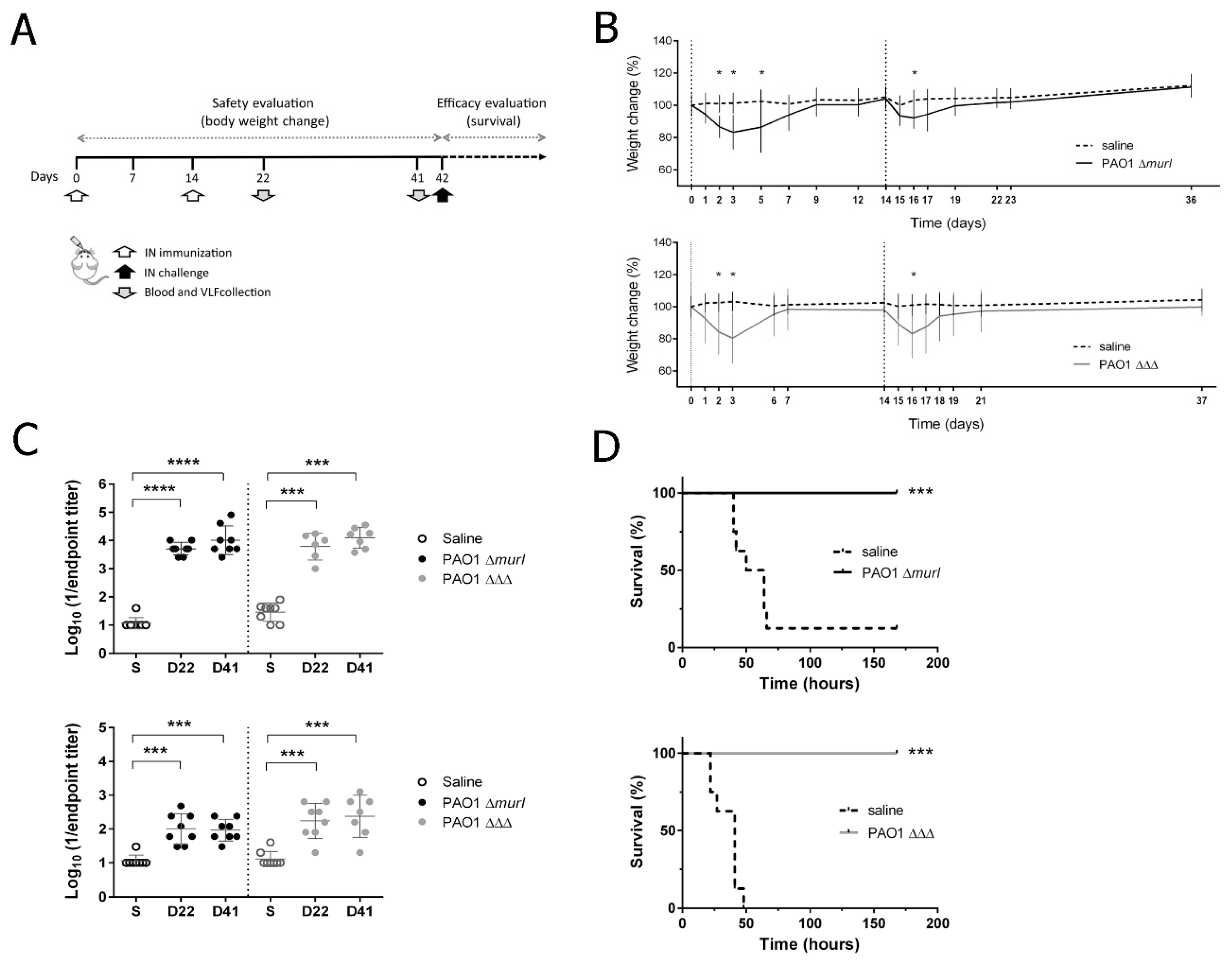
3.3. PAO1 ΔΔΔ Strain Is Attenuated in BALB/c Mice
3.4. PAO1 ΔΔΔ Strain Preserved Immunogenicity and Showed a Similar Level of Protective Efficacy to the Previous Vaccine Prototype, PAO1 ΔmurI, against Acute Lung Infection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Williams, B.J.; Dehnbostel, J.; Blackwell, T.S. Pseudomonas aeruginosa: Host defence in lung diseases. Respirology 2010, 15, 1037–1056. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Gregson, D.B.; Pitout, J.D.; Ross, T.; Laupland, K.B. Population-based study of the epidemiology and the risk factors for Pseudomonas aeruginosa bloodstream infection. Infection 2010, 38, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.M.; McLachlan, J.B.; Morici, L.A. Immunological considerations in the development of Pseudomonas aeruginosa vaccines. Hum. Vaccines Immunother. 2020, 16, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.; Seixas, A.M.M.; Marques, J.M.M.; Leitao, J.H. Immunization and Immunotherapy Approaches against Pseudomonas aeruginosa and Burkholderia cepacia Complex Infections. Vaccines 2021, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Sainz-Mejías, M.; Jurado-Martín, I.; McClean, S. Understanding Pseudomonas aeruginosa-Host Interactions: Ongoing Quest for an Efficacious Vaccine. Cells 2020, 9, 2617. [Google Scholar] [CrossRef] [PubMed]
- Belyakov, I.M.; Ahlers, J.D. What role does the route of immunization play in the generation of protective immunity against mucosal pathogens? J. Immunol. 2009, 183, 6883–6892. [Google Scholar] [CrossRef] [PubMed]
- Cabral, M.P.; Garcia, P.; Beceiro, A.; Rumbo, C.; Perez, A.; Moscoso, M.; Bou, G. Design of live attenuated bacterial vaccines based on D-glutamate auxotrophy. Nat. Commun. 2017, 8, 15480. [Google Scholar] [CrossRef] [PubMed]
- Cabral, M.P.; Correia, A.; Vilanova, M.; Gartner, F.; Moscoso, M.; Garcia, P.; Vallejo, J.A.; Perez, A.; Francisco-Tome, M.; Fuentes-Valverde, V.; et al. A live auxotrophic vaccine confers mucosal immunity and protection against lethal pneumonia caused by Pseudomonas aeruginosa. PLoS Pathog. 2020, 16, e1008311. [Google Scholar] [CrossRef] [PubMed]
- Strych, U.; Huang, H.C.; Krause, K.L.; Benedik, M.J. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 2000, 41, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: From bacterial pathogenesis to host response. J. Intensive Care 2014, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Muhl, D.; Filloux, A. Site-directed mutagenesis and gene deletion using reverse genetics. Methods Mol. Biol. 2014, 1149, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.G.; Laubier, A.; Bleves, S. Gene transfer: Conjugation. Methods Mol. Biol. 2014, 1149, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Frey, J. Biological safety concepts of genetically modified live bacterial vaccines. Vaccine 2007, 25, 5598–5605. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.M., Jr.; Popham, D.L.; Schmidt, A.L.; Neidle, E.L.; Stabb, E.V. Vibrio fischeri DarR Directs Responses to d-Aspartate and Represents a Group of Similar LysR-Type Transcriptional Regulators. J. Bacteriol. 2018, 200, e00773-17. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.W.; Chang, J.H.; Kim, K.J. Structural basis for an atypical active site of an L-aspartate/glutamate-specific racemase from Escherichia coli. FEBS Lett. 2015, 589, 3842–3847. [Google Scholar] [CrossRef] [PubMed]
- Palsson-McDermott, E.M.; O’Neill, L.A. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 2004, 113, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.; Rietschel, E.T. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 2001, 7, 167–202. [Google Scholar] [CrossRef] [PubMed]
- Cinel, I.; Dellinger, R.P. Advances in pathogenesis and management of sepsis. Curr. Opin. Infect. Dis. 2007, 20, 345–352. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Direction | Sequence (5′ to 3′) * | Comments |
|---|---|---|---|
| UP-PA4930(BamHI)-FW | Forward | CGCGGATCCGCACCGACAAGGCCGTGTTG | To amplify the upstream region of PA4930 (alr) |
| UP-PA4930(NotI)-RV | Reverse | CCCGCGGCCGCGGCATCGGGTCCTGCAAC | |
| DN-PA4930(NotI)-FW | Forward | CCCGCGGCCGCTTCAGGAGATACGCTCCG | To amplify the downstream region of PA4930 (alr) |
| DN-PA4930(ApaI)-RV | Reverse | TTTGGGCCCTGGCCCTGG | |
| UP-PA5302(SalI)-FW | Forward | CTTGTCGACCCCGATCGTCGGCGCC | To amplify the upstream region of PA5302 (dadX) |
| UP-PA5302(NotI)-RV | Reverse | CCCGCGGCCGCGGCGACGGGTCTCTCTTC | |
| DN-PA5302(NotI)-FW | Forward | CCCGCGGCCGCGAAAAACTTTCCGAATTC | To amplify the downstream region of PA5302 (dadX) |
| DN-PA5302(ApaI)-RV | Reverse | TTTGGGCCCGGCGGGGTC | |
| PA4662-EXTFW | Forward | GTATCGGCAAGGTGGAGT | To confirm PA4662 (murI) gene deletion |
| PA4662-EXTRV | Reverse | GAATGGCTTGATCGAGTC | |
| UpKn | Forward | CCCTGGATTTCACTGATGAG | Universal pKNG101 primers flanking the multiple cloning site |
| RpKn | Reverse | CATATCACAACGTGCGTGGA | |
| alr-EXT F | Forward | GATCATGATCGACTACCT | To confirm PA4930 (alr) gene deletion |
| alr-EXT R | Reverse | GATGGAGTTCGCCGAAAG | |
| dadX-EXT F | Forward | CGCAGATCAGTACCGAAG | To confirm PA5302 (dadX) gene deletion |
| dadX-EXT R | Reverse | CGTGGTTGAGCATTTCCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentes-Valverde, V.; García, P.; Moscoso, M.; Bou, G. Double Auxotrophy to Improve the Safety of a Live Anti-Pseudomonas aeruginosa Vaccine. Vaccines 2022, 10, 1622. https://doi.org/10.3390/vaccines10101622
Fuentes-Valverde V, García P, Moscoso M, Bou G. Double Auxotrophy to Improve the Safety of a Live Anti-Pseudomonas aeruginosa Vaccine. Vaccines. 2022; 10(10):1622. https://doi.org/10.3390/vaccines10101622
Chicago/Turabian StyleFuentes-Valverde, Víctor, Patricia García, Miriam Moscoso, and Germán Bou. 2022. "Double Auxotrophy to Improve the Safety of a Live Anti-Pseudomonas aeruginosa Vaccine" Vaccines 10, no. 10: 1622. https://doi.org/10.3390/vaccines10101622
APA StyleFuentes-Valverde, V., García, P., Moscoso, M., & Bou, G. (2022). Double Auxotrophy to Improve the Safety of a Live Anti-Pseudomonas aeruginosa Vaccine. Vaccines, 10(10), 1622. https://doi.org/10.3390/vaccines10101622





