The Advantage of Using Immunoinformatic Tools on Vaccine Design and Development for Coronavirus
Abstract
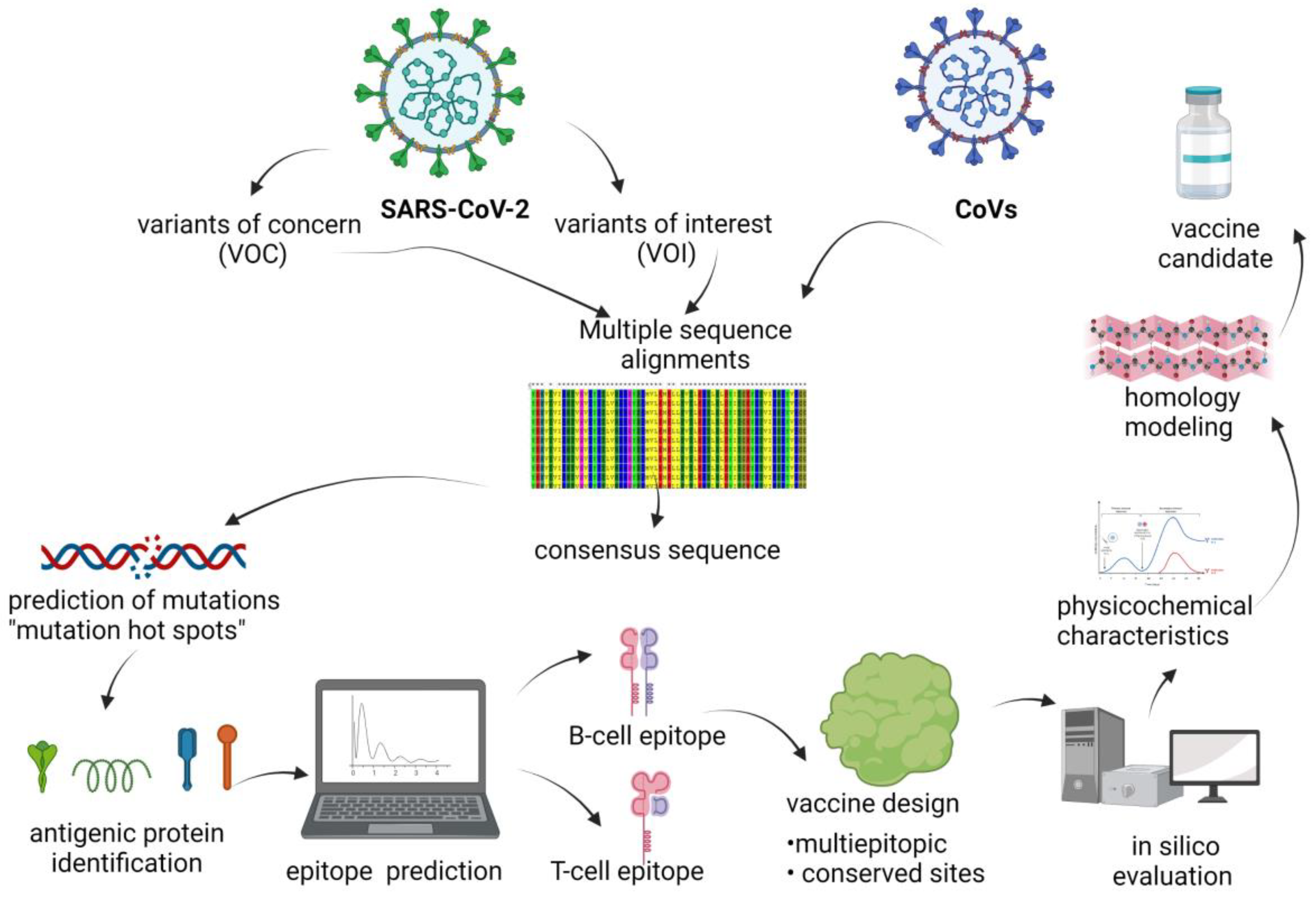
:1. Introduction
2. Epitope-Based Vaccines Using Bioinformatic Studies
2.1. Viral Vector-Based Vaccines
2.2. Nucleic Acid Vaccines
2.3. Dendrimer–Peptide Complexes
2.4. Gene Therapy Strategies
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef]
- Janik, E.; Niemcewicz, M.; Podogrocki, M.; Majsterek, I.; Bijak, M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens 2021, 10, 633. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 15 September 2022).
- Roy, U. Comparative structural analyses of selected spike protein-RBD mutations in SARS-CoV-2 lineages. Immunol. Res. 2022, 70, 143–151. [Google Scholar] [CrossRef]
- Waman, V.P.; Sen, N.; Varadi, M.; Daina, A.; Wodak, S.J.; Zoete, V.; Velankar, S.; Orengo, C. The impact of structural bioinformatics tools and resources on SARS-CoV-2 research and therapeutic strategies. Brief. Bioinform. 2021, 22, 742–768. [Google Scholar] [CrossRef]
- Higdon, M.M.; Wahl, B.; Jones, C.B.; Rosen, J.G.; Truelove, S.A.; Baidya, A.; Nande, A.A.; ShamaeiZadeh, P.A.; Walter, K.K.; Feikin, D.R.; et al. A Systematic Review of Coronavirus Disease 2019 Vaccine Efficacy and Effectiveness against Severe Acute Respiratory Syndrome Coronavirus 2 Infection and Disease. Open Forum Infect. Dis. 2022, 9, ofac138. [Google Scholar] [CrossRef]
- Liu, Y.; Rocklov, J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021, 28, taab124. [Google Scholar] [CrossRef]
- Sheikhi, F.; Yousefian, N.; Tehranipoor, P.; Kowsari, Z. Estimation of the basic reproduction number of Alpha and Delta variants of COVID-19 pandemic in Iran. PLoS ONE 2022, 17, e0265489. [Google Scholar] [CrossRef]
- Massard, M.; Eftimie, R.; Perasso, A.; Saussereau, B. A multi-strain epidemic model for COVID-19 with infected and asymptomatic cases: Application to French data. J. Theor. Biol. 2022, 545, 111117. [Google Scholar] [CrossRef]
- Du, Z.; Liu, C.; Wang, C.; Xu, L.; Xu, M.; Wang, L.; Bai, Y.; Xu, X.; Lau, E.H.Y.; Wu, P.; et al. Reproduction Numbers of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2022, 75, e293–e295. [Google Scholar] [CrossRef]
- Liu, Y.; Rocklov, J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J. Travel Med. 2022, 29, taac037. [Google Scholar] [CrossRef]
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses 2021, 13, 1192. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; Rits-Volloch, S.; et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022, 39, 110729. [Google Scholar] [CrossRef]
- Mohsin, M.; Mahmud, S. Omicron SARS-CoV-2 variant of concern: A review on its transmissibility, immune evasion, reinfection, and severity. Medicine 2022, 101, e29165. [Google Scholar] [CrossRef]
- Zimmer, C.; Corum, J.; Wee, S.; Kristoffersen, M. Coronavirus Vaccine Tracker. The New York Times, 31 August 2022. Available online: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html (accessed on 26 October 2022).
- Noor, R. A Review on the Effectivity of the Current COVID-19 Drugs and Vaccines: Are They Really Working against the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants? Curr. Clin. Microbiol. Rep. 2021, 8, 186–193. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Papakyriakou, A.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of SARS-CoV-2 Variants of Concern, Including Omicron, Highlights Their Common and Distinctive Amino Acid Substitution Patterns, Especially at the Spike ORF. Viruses 2022, 14, 707. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Markoulatos, P.; Van de Peer, Y.; Oliver, S.G.; Amoutzias, G.D. The Neighborhood of the Spike Gene Is a Hotspot for Modular Intertypic Homologous and Nonhomologous Recombination in Coronavirus Genomes. Mol. Biol. Evol. 2022, 39, msab292. [Google Scholar] [CrossRef] [PubMed]
- Pappas, N.; Roux, S.; Hölzer, M.; Lamkiewicz, K.; Mock, F.; Marz, M.; Dutilh, B.E. Virus Bioinformatics. Encycl. Virol. 2021, 1, 124–132. [Google Scholar] [CrossRef]
- Tay, J.H.; Porter, A.F.; Wirth, W.; Duchene, S. The Emergence of SARS-CoV-2 Variants of Concern Is Driven by Acceleration of the Substitution Rate. Mol. Biol. Evol. 2022, 39, msac013. [Google Scholar] [CrossRef] [PubMed]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Salama, M.A.; Hassanien, A.E.; Mostafa, A. The prediction of virus mutation using neural networks and rough set techniques. EURASIP J. Bioinform. Syst. Biol. 2016, 2016, 10. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, R.; Ghosh, M.; Sahoo, S.; Padhi, S.; Misra, N.; Raina, V.; Suar, M.; Son, Y.O. Next-Generation Bioinformatics Approaches and Resources for Coronavirus Vaccine Discovery and Development—A Perspective Review. Vaccines 2021, 9, 812. [Google Scholar] [CrossRef]
- Kaur, S.P.; Gupta, V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114. [Google Scholar] [CrossRef]
- Pushparajah, D.; Jimenez, S.; Wong, S.; Alattas, H.; Nafissi, N.; Slavcev, R.A. Advances in gene-based vaccine platforms to address the COVID-19 pandemic. Adv. Drug Deliv. Rev. 2021, 170, 113–141. [Google Scholar] [CrossRef]
- Hwang, W.; Lei, W.; Katritsis, N.M.; MacMahon, M.; Chapman, K.; Han, N. Current and prospective computational approaches and challenges for developing COVID-19 vaccines. Adv. Drug Deliv. Rev. 2021, 172, 249–274. [Google Scholar] [CrossRef]
- O’Neill, C.L.; Shrimali, P.C.; Clapacs, Z.P.; Files, M.A.; Rudra, J.S. Peptide-based supramolecular vaccine systems. Acta Biomater. 2021, 133, 153–167. [Google Scholar] [CrossRef]
- Jensen, K.K.; Rantos, V.; Jappe, E.C.; Olsen, T.H.; Jespersen, M.C.; Jurtz, V.; Jessen, L.E.; Lanzarotti, E.; Mahajan, S.; Peters, B.; et al. TCRpMHCmodels: Structural modelling of TCR-pMHC class I complexes. Sci. Rep. 2019, 9, 14530. [Google Scholar] [CrossRef] [Green Version]
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013, 3, 120139. [Google Scholar] [CrossRef] [Green Version]
- Martinez, L.; Malaina, I.; Salcines-Cuevas, D.; Teran-Navarro, H.; Zeoli, A.; Alonso, S.; De la Fuente, I.M.; Gonzalez-Lopez, E.; Ocejo-Vinyals, J.G.; Gozalo-Marguello, M.; et al. First computational design using lambda-superstrings and in vivo validation of SARS-CoV-2 vaccine. Sci. Rep. 2022, 12, 6410. [Google Scholar] [CrossRef]
- Martínez-Archundia, M.; Ramírez-Salinas, G.L.; García-Machorro, J.; Correa-Basurto, J. Searching Epitope-Based Vaccines Using Bioinformatics Studies. Methods Mol. Biol. 2022, 2412, 471–479. [Google Scholar] [CrossRef]
- Ramirez-Salinas, G.L.; Garcia-Machorro, J.; Rojas-Hernandez, S.; Campos-Rodriguez, R.; de Oca, A.C.; Gomez, M.M.; Luciano, R.; Zimic, M.; Correa-Basurto, J. Bioinformatics design and experimental validation of influenza A virus multi-epitopes that induce neutralizing antibodies. Arch. Virol. 2020, 165, 891–911. [Google Scholar] [CrossRef]
- Valiente, P.A.; Wen, H.; Nim, S.; Lee, J.; Kim, H.J.; Kim, J.; Perez-Riba, A.; Paudel, Y.P.; Hwang, I.; Kim, K.D.; et al. Computational Design of Potent D-Peptide Inhibitors of SARS-CoV-2. J. Med. Chem. 2021, 64, 14955–14967. [Google Scholar] [CrossRef]
- Rafi, M.O.; Al-Khafaji, K.; Sarker, M.T.; Taskin-Tok, T.; Rana, A.S.; Rahman, M.S. Design of a multi-epitope vaccine against SARS-CoV-2: Immunoinformatic and computational methods. RSC Adv. 2022, 12, 4288–4310. [Google Scholar] [CrossRef]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar] [CrossRef]
- Soria-Guerra, R.E.; Nieto-Gomez, R.; Govea-Alonso, D.O.; Rosales-Mendoza, S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J. Biomed. Inform. 2015, 53, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Loyola, P.K.; Campos-Rodriguez, R.; Bello, M.; Rojas-Hernandez, S.; Zimic, M.; Quiliano, M.; Briz, V.; Munoz-Fernandez, M.A.; Tolentino-Lopez, L.; Correa-Basurto, J. Theoretical analysis of the neuraminidase epitope of the Mexican A H1N1 influenza strain, and experimental studies on its interaction with rabbit and human hosts. Immunol. Res. 2013, 56, 44–60. [Google Scholar] [CrossRef]
- Vedamurthy, G.V.; Ahmad, H.; Onteru, S.K.; Saxena, V.K. In silico homology modelling and prediction of novel epitopic peptides from P24 protein of Haemonchus contortus. Gene 2019, 703, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Fonseca, R.A.; Bello, M.; de Los Munoz-Fernandez, M.A.; Luis Jimenez, J.; Rojas-Hernandez, S.; Fragoso-Vazquez, M.J.; Gutierrez-Sanchez, M.; Rodrigues, J.; Cayetano-Castro, N.; Borja-Urby, R.; et al. In silico search, chemical characterization and immunogenic evaluation of amino-terminated G4-PAMAM-HIV peptide complexes using three-dimensional models of the HIV-1 gp120 protein. Colloids Surf. B Biointerfaces 2019, 177, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, B.; Ullah, M.A.; Araf, Y.; Islam, N.N.; Zohora, U.S. Immunoinformatics-guided designing and in silico analysis of epitope-based polyvalent vaccines against multiple strains of human coronavirus (HCoV). Expert Rev. Vaccines 2021, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Nuismer, S.L.; Antia, R. Recombinant vector vaccine evolution. PLoS Comput. Biol. 2019, 15, e1006857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haan, P.; Van Diemen, F.R.; Toscano, M.G. Viral gene delivery vectors: The next generation medicines for immune-related diseases. Hum. Vaccines Immunother. 2021, 17, 14–21. [Google Scholar] [CrossRef]
- Raman, R.; Patel, K.J.; Ranjan, K. COVID-19: Unmasking Emerging SARS-CoV-2 Variants, Vaccines and Therapeutic Strategies. Biomolecules 2021, 11, 993. [Google Scholar] [CrossRef]
- National Institute of Health. 6 Questions (and Answers) about Viral Vector Vaccines; National Institute of Health: Bethesda, MD, USA, 2021. [Google Scholar]
- Seto, J.; Walsh, M.P.; Mahadevan, P.; Zhang, Q.; Seto, D. Applying Genomic and Bioinformatic Resources to Human Adenovirus Genomes for Use in Vaccine Development and for Applications in Vector Development for Gene Delivery. Viruses 2010, 2, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Tarasova, E.; Farafonov, V.; Khayat, R.; Okimoto, N.; Komatsu, T.S.; Taiji, M.; Nerukh, D. All-Atom Molecular Dynamics Simulations of Entire Virus Capsid Reveal the Role of Ion Distribution in Capsid’s Stability. J. Phys. Chem. Lett. 2017, 8, 779–784. [Google Scholar] [CrossRef] [Green Version]
- Reddy, T.; Sansom, M.S. Computational virology: From the inside out. Biochim. Biophys. Acta 2016, 1858, 1610–1618. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.P.; Tada, S.; Yumoto, A.; Kitao, A.; Ito, Y.; Uzawa, T.; Tsuda, K. Using molecular dynamics simulations to prioritize and understand AI-generated cell penetrating peptides. Sci. Rep. 2021, 11, 10630. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.P.; Argyros, O.; Harbottle, R.P. Sustained expression from DNA vectors. Adv. Genet. 2015, 89, 113–152. [Google Scholar] [CrossRef]
- Kramps, T.; Elbers, K. Introduction to RNA Vaccines. In RNA Vaccines; Methods in Molecular Biology Book Series; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1499, pp. 1–11. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.; Mendonca, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef]
- U.S. National Library of Medicine. ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 26 September 2022).
- Koyama, S.; Coban, C.; Aoshi, T.; Horii, T.; Akira, S.; Ishii, K.J. Innate immune control of nucleic acid-based vaccine immunogenicity. Expert Rev. Vaccines 2009, 8, 1099–1107. [Google Scholar] [CrossRef]
- Shahrear, S.; Islam, A. Immunoinformatics guided modeling of CCHF_GN728, an mRNA-based universal vaccine against Crimean-Congo hemorrhagic fever virus. Comput. Biol. Med. 2021, 140, 105098. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Kim, D.S.; Choe, C.A.; Nicol, J.J.; Wellington-Oguri, R.; Watkins, A.M.; Sperberg, R.A.P.; Huang, P.S.; Participants, E.; Das, R. Theoretical basis for stabilizing messenger RNA through secondary structure design. bioRxiv 2021, preprint. [Google Scholar] [CrossRef]
- Khan, T.; Abdullah, M.; Toor, T.F.; Almajhdi, F.N.; Suleman, M.; Iqbal, A.; Ali, L.; Khan, A.; Waheed, Y.; Wei, D.Q. Evaluation of the Whole Proteome of Achromobacter xylosoxidans to Identify Vaccine Targets for mRNA and Peptides-Based Vaccine Designing against the Emerging Respiratory and Lung Cancer-Causing Bacteria. Front. Med. 2021, 8, 825876. [Google Scholar] [CrossRef]
- Durojaye, O.A.; Sedzro, D.M.; Idris, M.O.; Yekeen, A.A.; Fadahunsi, A.A.; Alakanse, O.S. Identification of a Potential mRNA-based Vaccine Candidate against the SARS-CoV-2 Spike Glycoprotein: A Reverse Vaccinology Approach. ChemistrySelect 2022, 7, e202103903. [Google Scholar] [CrossRef]
- Bottger, R.; Hoffmann, R.; Knappe, D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS ONE 2017, 12, e0178943. [Google Scholar] [CrossRef]
- Ghosh, D.; Peng, X.; Leal, J.; Mohanty, R. Peptides as drug delivery vehicles across biological barriers. J. Pharm. Investig. 2018, 48, 89–111. [Google Scholar] [CrossRef]
- Flores-Mejia, R.; Fragoso-Vazquez, M.J.; Perez-Blas, L.G.; Parra-Barrera, A.; Hernandez-Castro, S.S.; Estrada-Perez, A.R.; Rodrigues, J.; Lara-Padilla, E.; Ortiz-Morales, A.; Correa-Basurto, J. Chemical characterization (LC-MS-ESI), cytotoxic activity and intracellular localization of PAMAM G4 in leukemia cells. Sci. Rep. 2021, 11, 8210. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.; Rodriguez-Fonseca, R.A.; Correa-Basurto, J. Complexation of peptide epitopes with G4-PAMAM dendrimer through ligand diffusion molecular dynamic simulations. J. Mol. Graph. Model 2020, 96, 107514. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Zhou, J.; Wang, Q.; Wang, Y.; Kang, C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics 2021, 11, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.D.; Soemardy, C.; et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226. [Google Scholar] [CrossRef]
- Ma, L.; Li, H.; Lan, J.; Hao, X.; Liu, H.; Wang, X.; Huang, Y. Comprehensive analyses of bioinformatics applications in the fight against COVID-19 pandemic. Comput. Biol. Chem. 2021, 95, 107599. [Google Scholar] [CrossRef]
- Cannataro, M.; Harrison, A. Bioinformatics helping to mitigate the impact of COVID-19—Editorial. Brief. Bioinform. 2021, 22, 613–615. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Patients. Med. Sci. Monit. 2022, 28, e935952. [Google Scholar] [CrossRef]
- Anser, M.K.; Sharif, M.; Khan, M.A.; Nassani, A.A.; Zaman, K.; Abro, M.M.Q.; Kabbani, A. Demographic, psychological, and environmental factors affecting student’s health during the COVID-19 pandemic: On the rocks. Environ. Sci. Pollut. Res. Int. 2021, 28, 31596–31606. [Google Scholar] [CrossRef]
- Debnath, M.; Banerjee, M.; Berk, M. Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J. 2020, 34, 8787–8795. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.e2379. [Google Scholar] [CrossRef]
- Mabrouk, M.T.; Huang, W.C.; Martinez-Sobrido, L.; Lovell, J.F. Advanced Materials for SARS-CoV-2 Vaccines. Adv. Mater. 2022, 34, e2107781. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Nikolaidis, M.; Tryfonopoulou, E.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses 2022, 14, 78. [Google Scholar] [CrossRef]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-CoV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2021, 11, 781429. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Q.; Liang, Z.; Li, T.; Liu, S.; Cui, Q.; Nie, J.; Wu, Q.; Qu, X.; Huang, W.; et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes Infect. 2022, 11, 1–5. [Google Scholar] [CrossRef]
- Herman, C.; Bradley, C.; Gordon, A.; Wang, C.; Cooke, M.; Kohrn, B.; Kennedy, S.; Lichtarge, O.; Ronca, S. RNA polymerase inaccuracy underlies SARS-CoV-2 variants and vaccine heterogeneity. Res. Sq. 2022, preprint. [Google Scholar] [CrossRef]
- Madhusoodanan, J. Animal Reservoirs—Where the Next SARS-CoV-2 Variant Could Arise. JAMA 2022, 328, 696–698. [Google Scholar] [CrossRef]
- Dhama, K.; Patel, S.K.; Sharun, K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Malik, Y.S.; Sah, R.; Rabaan, A.A.; Panwar, P.K.; et al. SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020, 37, 101830. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Brun, J.; Vasiljevic, S.; Gangadharan, B.; Hensen, M.; Anu, V.C.; Hill, M.L.; Kiappes, J.L.; Dwek, R.A.; Alonzi, D.S.; Struwe, W.B.; et al. Assessing Antigen Structural Integrity through Glycosylation Analysis of the SARS-CoV-2 Viral Spike. ACS Cent. Sci. 2021, 7, 586–593. [Google Scholar] [CrossRef]
- Mishra, M. End of COVID pandemic is ‘in sight’—WHO chief. Reuters, 14 September 2022. [Google Scholar]
- Barrera, A.; Salvador, E.; Gómez Pliego, R.; Espinosa Raya, J.; Correa Basurto, J.; García Machorro, J. Coronavirus de tipo 2 causante del síndrome respiratorio agudo severo, un virus que llegó para quedarse. Rev. Mex. Mastol. 2021, 1, 9–17. [Google Scholar] [CrossRef]
- Murray, C.J.L. COVID-19 will continue but the end of the pandemic is near. Lancet 2022, 399, 417–419. [Google Scholar] [CrossRef]
- Guo, F.; Yang, J.; Pan, J.; Liang, X.; Shen, X.; Irwin, D.M.; Chen, R.A.; Shen, Y. Origin and Evolution of H1N1/pdm2009: A Codon Usage Perspective. Front. Microbiol. 2020, 11, 1615. [Google Scholar] [CrossRef]
- Mohammad, S.; Bouchama, A.; Mohammad Alharbi, B.; Rashid, M.; Saleem Khatlani, T.; Gaber, N.S.; Malik, S.S. SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: Genomic Divergence and Functional Convergence. Pathogens 2020, 9, 677. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ji, J.; Chen, X.; Bi, Y.; Li, J.; Wang, Q.; Hu, T.; Song, H.; Zhao, R.; Chen, Y.; et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 2021, 184, 4380–4391.e4314. [Google Scholar] [CrossRef] [PubMed]
- Neches, R.Y.; Kyrpides, N.C.; Ouzounis, C.A. Atypical Divergence of SARS-CoV-2 Orf8 from Orf7a within the Coronavirus Lineage Suggests Potential Stealthy Viral Strategies in Immune Evasion. mBio 2021, 12, 12. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Cheng, Y.X.; Xu, L.; Li, J.Y.; Tao, C.Y.; Ji, C.Y.; Han, N.; Yang, R.; Wu, H.; Li, Y.; et al. Genomic evidence for divergent co-infections of co-circulating SARS-CoV-2 lineages. Comput. Struct. Biotechnol. J. 2022, 20, 4015–4024. [Google Scholar] [CrossRef]
- Dilucca, M.; Forcelloni, S.; Georgakilas, A.G.; Giansanti, A.; Pavlopoulou, A. Codon Usage and Phenotypic Divergences of SARS-CoV-2 Genes. Viruses 2020, 12, 498. [Google Scholar] [CrossRef]
- Yadav, P.D.; Potdar, V.A.; Choudhary, M.L.; Nyayanit, D.A.; Agrawal, M.; Jadhav, S.M.; Majumdar, T.D.; Shete-Aich, A.; Basu, A.; Abraham, P.; et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J. Med. Res. 2020, 151, 200–209. [Google Scholar] [CrossRef]
- He, J.; Huang, F.; Zhang, J.; Chen, Q.; Zheng, Z.; Zhou, Q.; Chen, D.; Li, J.; Chen, J. Vaccine design based on 16 epitopes of SARS-CoV-2 spike protein. J. Med. Virol. 2021, 93, 2115–2131. [Google Scholar] [CrossRef]
- Dolgin, E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022, 21, 324–326. [Google Scholar] [CrossRef]
- Li, M.; Zeng, J.; Li, R.; Wen, Z.; Cai, Y.; Wallin, J.; Shu, Y.; Du, X.; Sun, C. Rational Design of a Pan-Coronavirus Vaccine Based on Conserved CTL Epitopes. Viruses 2021, 13, 333. [Google Scholar] [CrossRef]
- Patel, R.S.; Agrawal, B. Heterologous immunity induced by 1(st) generation COVID-19 vaccines and its role in developing a pan-coronavirus vaccine. Front. Immunol. 2022, 13, 952229. [Google Scholar] [CrossRef]
- Mohammad, T.; Choudhury, A.; Habib, I.; Asrani, P.; Mathur, Y.; Umair, M.; Anjum, F.; Shafie, A.; Yadav, D.K.; Hassan, M.I. Genomic Variations in the Structural Proteins of SARS-CoV-2 and Their Deleterious Impact on Pathogenesis: A Comparative Genomics Approach. Front. Cell. Infect. Microbiol. 2021, 11, 765039. [Google Scholar] [CrossRef]
- Brand, I.; Gilberg, L.; Bruger, J.; Gari, M.; Wieser, A.; Eser, T.M.; Frese, J.; Ahmed, M.I.M.; Rubio-Acero, R.; Guggenbuehl Noller, J.M.; et al. Broad T Cell Targeting of Structural Proteins after SARS-CoV-2 Infection: High Throughput Assessment of T Cell Reactivity Using an Automated Interferon Gamma Release Assay. Front. Immunol. 2021, 12, 688436. [Google Scholar] [CrossRef]
- Lizbeth, R.G.; Jazmin, G.M.; Jose, C.B.; Marlet, M.A. Immunoinformatics study to search epitopes of spike glycoprotein from SARS-CoV-2 as potential vaccine. J. Biomol. Struct. Dyn. 2021, 39, 4878–4892. [Google Scholar] [CrossRef]
- Diez-Rivero, C.M.; Lafuente, E.M.; Reche, P.A. Computational analysis and modeling of cleavage by the immunoproteasome and the constitutive proteasome. BMC Bioinform. 2010, 11, 479. [Google Scholar] [CrossRef]
| Characteristic/ Variant of Concern | Initial Virus | Alpha | Beta | Gamma | Delta | Omicron |
|---|---|---|---|---|---|---|
| Reproduction number (R0) or transmissibility * | 2.7 [11] | 4.5 [12,13,14] | ≥2 [13,14] | 4 [13,14] | 8 [12,15] | ≥10 [15] |
| Ability to evade immune response ** | --- | --- | +++ [16,17] | ++ [16,17] | ++++ [16,17] | +++++ [18,19] |
| First reported | December 2019; China | 14 December 2020; United Kingdom | 18 December 2020; South Africa | 2 January 2021; Brazil | 24 March 2021; India | 24 November 2021; South Africa data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Machorro, J.; Ramírez-Salinas, G.L.; Martinez-Archundia, M.; Correa-Basurto, J. The Advantage of Using Immunoinformatic Tools on Vaccine Design and Development for Coronavirus. Vaccines 2022, 10, 1844. https://doi.org/10.3390/vaccines10111844
García-Machorro J, Ramírez-Salinas GL, Martinez-Archundia M, Correa-Basurto J. The Advantage of Using Immunoinformatic Tools on Vaccine Design and Development for Coronavirus. Vaccines. 2022; 10(11):1844. https://doi.org/10.3390/vaccines10111844
Chicago/Turabian StyleGarcía-Machorro, Jazmín, Gema Lizbeth Ramírez-Salinas, Marlet Martinez-Archundia, and José Correa-Basurto. 2022. "The Advantage of Using Immunoinformatic Tools on Vaccine Design and Development for Coronavirus" Vaccines 10, no. 11: 1844. https://doi.org/10.3390/vaccines10111844
APA StyleGarcía-Machorro, J., Ramírez-Salinas, G. L., Martinez-Archundia, M., & Correa-Basurto, J. (2022). The Advantage of Using Immunoinformatic Tools on Vaccine Design and Development for Coronavirus. Vaccines, 10(11), 1844. https://doi.org/10.3390/vaccines10111844




