Influenza A(H7N9) Pandemic Preparedness: Assessment of the Breadth of Heterologous Antibody Responses to Emerging Viruses from Multiple Pre-Pandemic Vaccines and Population Immunity
Abstract
:1. Introduction
2. Materials and Methods
2.1. A(H7N9) Clinical Trial Vaccine Sera
2.2. Seasonal Influenza Vaccine Sera and Population Immunity Sera
2.3. Influenza Viruses and Sequence Analysis
2.4. Hemagglutination Inhibition (HI) Assays
2.5. Microneutralization (MN) Assays
2.6. Enzyme-Linked Lectin Assay (ELLA)
2.7. Neuraminidase ELISA
2.8. Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) Natural Killer (NK) Cell Activation Assay
2.9. Statistical Analysis
3. Results
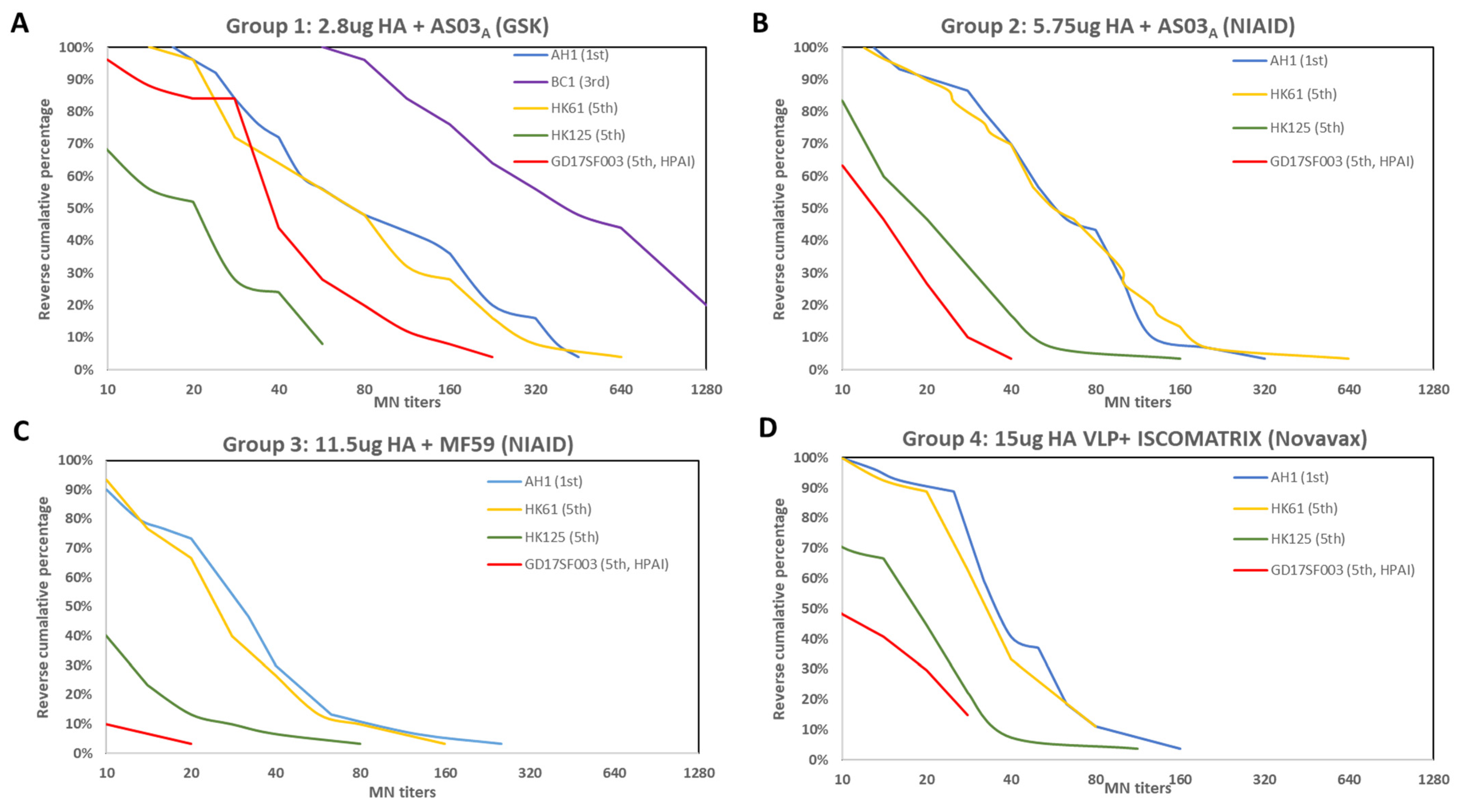
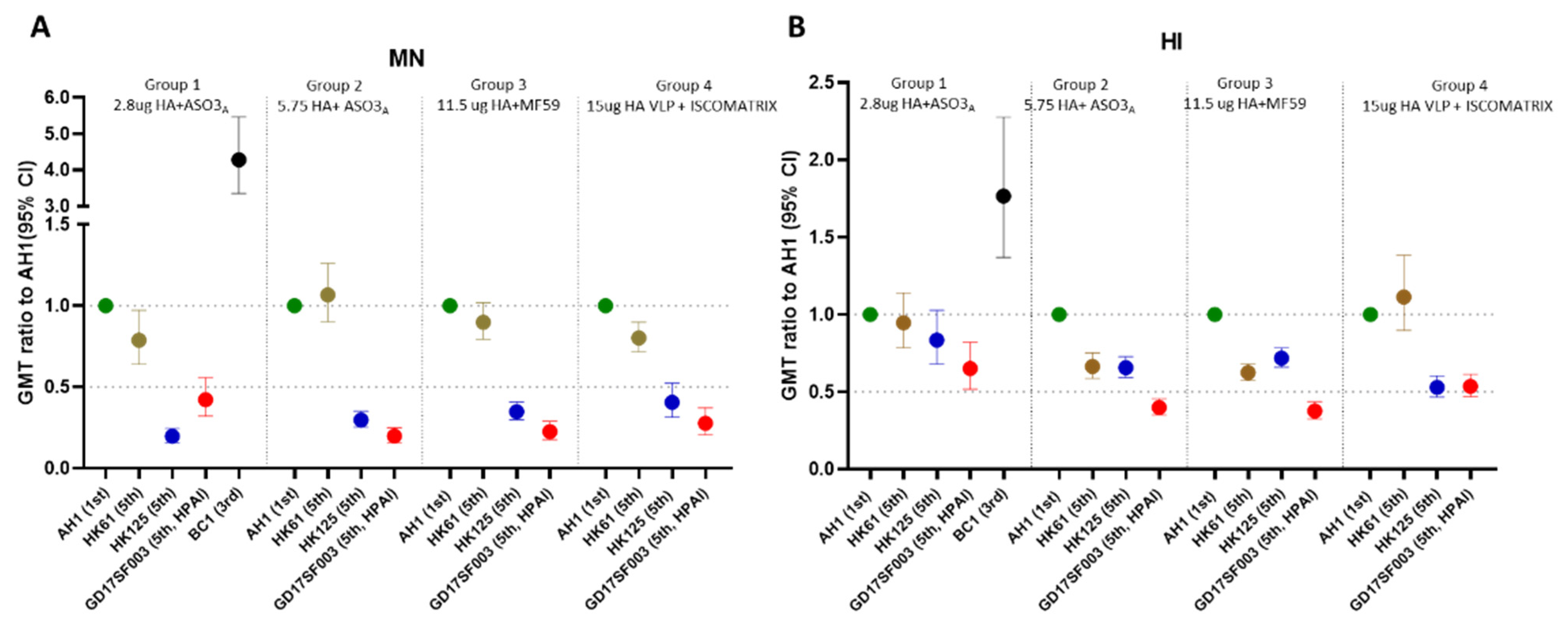
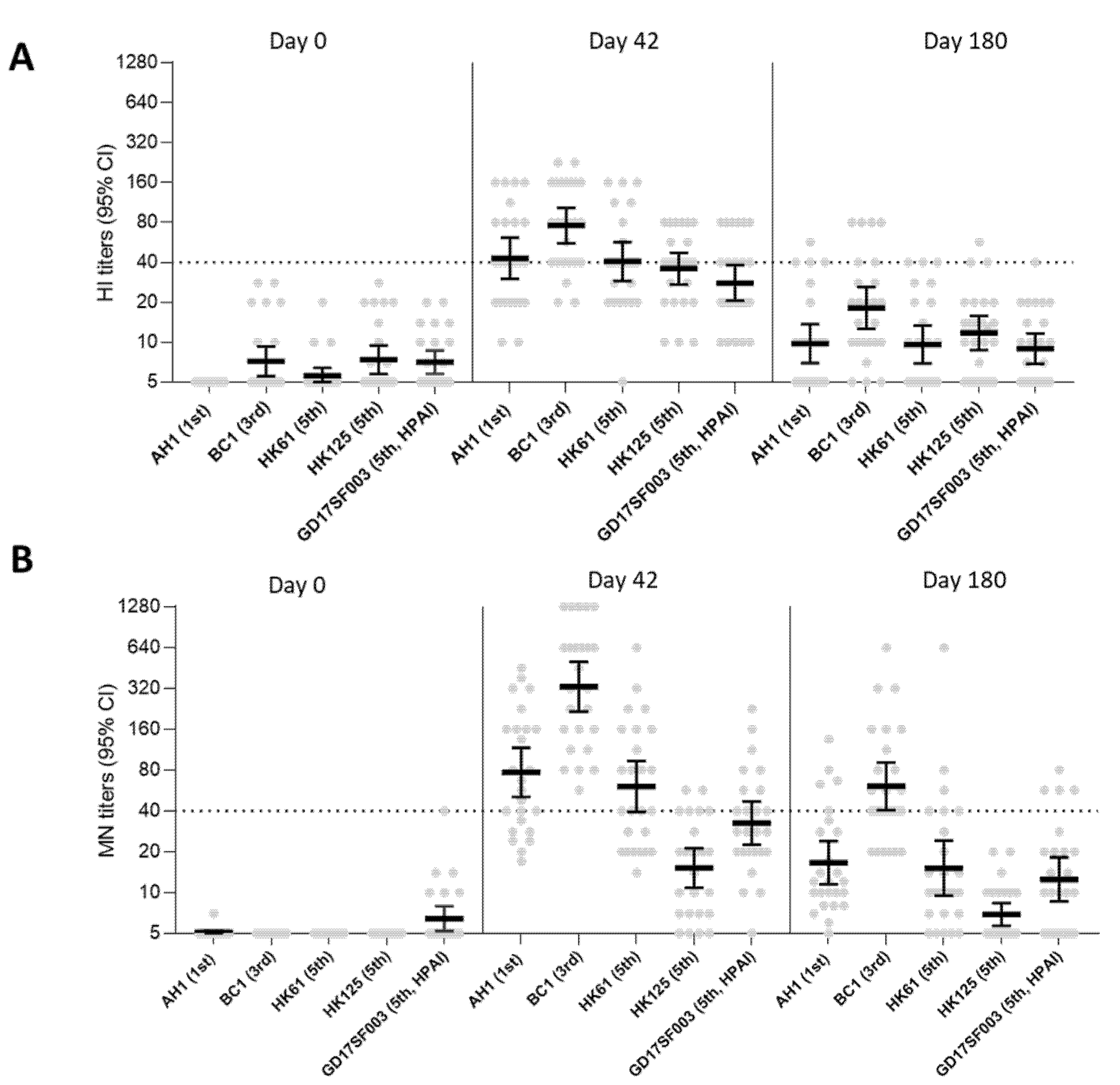
3.1. Breadth of the HI and Neutralizing Antibody Responses Elicited by the 1st Wave AS03A and MF59 Adjuvanted Inactivated Vaccines and ISCOMATRIX Adjuvanted Recombinant VLP Vaccine
3.2. No Pre-Existing Neutralizing Antibodies to HA of A (H7N9) Viruses in the US Population
3.3. Seasonal Influenza Vaccination Elicited Heterologous Cross-Reactive NAI Antibodies, but Not HI, MN and ADCC Antibody Responses to A(H7N9)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quan, C.; Wang, Q.; Zhang, J.; Zhao, M.; Dai, Q.; Huang, T.; Zhang, Z.; Mao, S.; Nie, Y.; Liu, J.; et al. Avian Influenza A Viruses among Occupationally Exposed Populations, China, 2014–2016. Emerg. Infect. Dis. 2019, 25, 2215–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skowronski, D.M.; Chambers, C.; Gustafson, R.; Purych, D.B.; Tang, P.; Bastien, N.; Krajden, M.; Li, Y. Avian Influenza A(H7N9) Virus Infection in 2 Travelers Returning from China to Canada, January 2015. Emerg. Infect. Dis. 2016, 22, 71–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, C.; Shi, W.; Yang, Y.; Yang, Y.; Liu, X.; Xu, W.; Li, H.; Li, J.; Wang, Q.; Tong, Z.; et al. New Threats from H7N9 Influenza Virus: Spread and Evolution of High- and Low-Pathogenicity Variants with High Genomic Diversity in Wave Five. J. Virol. 2018, 92, e00301-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Angriculture Organization (FAO). H7N9 Situation Update. Available online: https://www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html (accessed on 3 May 2022).
- World Health Organization (WHO). Influenza at the human-Animal Interface. Available online: https://cdn.who.int/media/docs/default-source/influenza/human-animal-interface-risk-assessments/influenza_summary_ira_ha_interface_jan_2022.pdf?sfvrsn=1893dd9f_5&download=true (accessed on 3 May 2022).
- CDC. Summary of Influenza Risk Assessment Tool (IRAT) Results. Available online: https://www.cdc.gov/flu/pandemic-resources/monitoring/irat-virus-summaries.htm?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fflu%2Fpandemic-resources%2Ftools%2Firat-virus-summaries.htm#H7N9hongkong (accessed on 13 May 2022).
- Yang, S.; Chen, Y.; Cui, D.; Yao, H.; Lou, J.; Huo, Z.; Xie, G.; Yu, F.; Zheng, S.; Yang, Y.; et al. Avian-origin influenza A(H7N9) infection in influenza A(H7N9)-affected areas of China: A serological study. J. Infect. Dis. 2014, 209, 265–269. [Google Scholar] [CrossRef] [Green Version]
- WHO. Antigenic and Genetic Characteristics of Zoonotic Influenza Viruses and Development of Candidate Vaccine Viruses for Pandemic Preparedness. Available online: https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-northern-hemisphere-recommendation-2017-2018/201703_zoonotic_vaccinevirusupdate.pdf?sfvrsn=4c4920df_14 (accessed on 13 May 2022).
- Jackson, L.A.; Campbell, J.D.; Frey, S.E.; Edwards, K.M.; Keitel, W.A.; Kotloff, K.L.; Berry, A.A.; Graham, I.; Atmar, R.L.; Creech, C.B.; et al. Effect of Varying Doses of a Monovalent H7N9 Influenza Vaccine With and Without AS03 and MF59 Adjuvants on Immune Response: A Randomized Clinical Trial. JAMA 2015, 314, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, M.J.; Bernstein, D.I.; Winokur, P.; Rupp, R.; Anderson, E.; Rouphael, N.; Dickey, M.; Stapleton, J.T.; Edupuganti, S.; Spearman, P.; et al. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: A randomized clinical trial. JAMA 2014, 312, 1409–1419. [Google Scholar] [CrossRef] [Green Version]
- Oshansky, C.M.; King, J.; Lu, D.; Zhou, J.; Pavetto, C.; Horwith, G.; Biscardi, K.; Nguyen, B.; Treanor, J.J.; Chen, L.M.; et al. Adjuvanted recombinant hemagglutinin H7 vaccine to highly pathogenic influenza A(H7N9) elicits high and sustained antibody responses in healthy adults. NPJ Vaccines 2021, 6, 41. [Google Scholar] [CrossRef]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, Ö.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef]
- Fries, L.F.; Smith, G.E.; Glenn, G.M. A recombinant viruslike particle influenza A (H7N9) vaccine. N. Engl. J. Med. 2013, 369, 2564–2566. [Google Scholar] [CrossRef]
- Madan, A.; Segall, N.; Ferguson, M.; Frenette, L.; Kroll, R.; Friel, D.; Soni, J.; Li, P.; Innis, B.L.; Schuind, A. Immunogenicity and Safety of an AS03-Adjuvanted H7N9 Pandemic Influenza Vaccine in a Randomized Trial in Healthy Adults. J. Infect. Dis. 2016, 214, 1717–1727. [Google Scholar] [CrossRef]
- Smith, G.E.; Flyer, D.C.; Raghunandan, R.; Liu, Y.; Wei, Z.; Wu, Y.; Kpamegan, E.; Courbron, D.; Fries, L.F., 3rd; Glenn, G.M. Development of influenza H7N9 virus like particle (VLP) vaccine: Homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine 2013, 31, 4305–4313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Network, W.G.I.S. Manual for the Labratory Diagnosis and Virological Surveillence of Influenza. 2011. Available online: http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf (accessed on 13 May 2022).
- Levine, M.Z.; Holiday, C.; Liu, F.; Jefferson, S.; Gillis, E.; Bellamy, A.R.; Tumpey, T.; Katz, J.M. Cross-Reactive Antibody Responses to Novel H5Nx Influenza Viruses Following Homologous and Heterologous Prime-Boost Vaccination with a Prepandemic Stockpiled A(H5N1) Vaccine in Humans. J. Infect. Dis. 2017, 216, S555–S559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couzens, L.; Gao, J.; Westgeest, K.; Sandbulte, M.; Lugovtsev, V.; Fouchier, R.; Eichelberger, M. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J. Virol. Methods 2014, 210C, 7–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, W.; Liu, F.; Wilson, J.R.; Holiday, C.; Li, Z.N.; Bai, Y.; Tzeng, W.P.; Stevens, J.; York, I.A.; Levine, M.Z. Antibody-Dependent Cell-Mediated Cytotoxicity to Hemagglutinin of Influenza A Viruses After Influenza Vaccination in Humans. Open Forum Infect. Dis. 2016, 3, ofw102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, W.; Levine, M.Z. Stockpiled Avian Influenza A(H7N9) Vaccines Induce Robust, Nonneutralizing Functional Antibodies Against Antigenically Drifted Fifth-Wave A(H7N9) Viruses. J. Infect. Dis. 2019, 220, 1276–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, T.K.; Cauchemez, S.; Perera, R.A.P.M.; Freeman, G.; Fang, V.J.; Ip, D.K.M.; Leung, G.M.; Peiris, J.S.M.; Cowling, B.J. Association between antibody titers and protection against influenza virus infection within households. J. Infect. Dis. 2014, 210, 684–692. [Google Scholar] [CrossRef] [Green Version]
- Levine, M.Z.; Holiday, C.; Jefferson, S.; Gross, F.L.; Liu, F.; Li, S.; Friel, D.; Boutet, P.; Innis, B.L.; Mallett, C.P.; et al. Heterologous prime-boost with A(H5N1) pandemic influenza vaccines induces broader cross-clade antibody responses than homologous prime-boost. NPJ Vaccines 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.G.; Stenehjem, E.; Grannis, S.; Ball, S.W.; Naleway, A.L.; Ong, T.C.; DeSilva, M.B.; Natarajan, K.; Bozio, C.H.; Lewis, N.; et al. Effectiveness of Covid-19 Vaccines in Ambulatory and Inpatient Care Settings. N. Engl. J. Med. 2021, 385, 1355–1371. [Google Scholar] [CrossRef]
- Reber, A.; Katz, J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev. Vaccines 2013, 12, 519–536. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. 1972, 70, 767–777. [Google Scholar] [CrossRef]
- Benoit, A.; Beran, J.; Devaster, J.M.; Esen, M.; Launay, O.; Leroux-Roels, G.; McElhaney, J.E.; Oostvogels, L.; van Essen, G.A.; Gaglani, M.; et al. Hemagglutination Inhibition Antibody Titers as a Correlate of Protection Against Seasonal A/H3N2 Influenza Disease. Open Forum Infect. Dis. 2015, 2, ofv067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, A.; Le Mai, Q.; Le Thanh, T.; Wolbers, M.; Le Khanh Hang, N.; Thai, P.Q.; Thi Thu Yen, N.; Le Minh Hoa, N.; Bryant, J.E.; Duong, T.N.; et al. Hemagglutination inhibiting antibodies and protection against seasonal and pandemic influenza infection. J. Infect. 2015, 70, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajendran, M.; Krammer, F.; McMahon, M. The Human Antibody Response to the Influenza Virus Neuraminidase Following Infection or Vaccination. Vaccines 2021, 9, 846. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Petrie, J.G.; Cross, R.T.; Johnson, E.; Liu, M.; Zhong, W.; Levine, M.; Katz, J.M.; Ohmit, S.E. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. J. Infect. Dis. 2015, 212, 1191–1199. [Google Scholar] [CrossRef] [PubMed]

| # | Vaccine Groups | Clinical Trials NCT# | Clinical Trial Sponsor | Vaccine (Antigens) | Actual Vaccine Dose (μg HA per Dose) | Target Vaccine Dose (μg HA/Dose) | Adjuvant | N | Age (Median Range) in yrs | A(H7N9) Viruses | HI Titers | MN Titers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain Name | Epidemic Wave/Lineage | Pathogenicity | Pre-Vaccination | 21–28 Days Post-2nd Dose | Pre-Vaccination | 21–28 Days Post-2nd Dose | ||||||||||||||
| GMT (95% CI) | GMT 95% CI | % Seroconversion | % SPR | GMT (95% CI) | GMT 95% CI | % Seroconversion | % SPR | |||||||||||||
| 1 | 2.8 μg HA + ASO3A | NCT01999842 | GSK | Inactivated A/Shanghai/2/2013 | 2.8 | 3.75 | ASO3A | 25 | ≥18 | A/AH/1/2013 | 1st | LPAI | 5 | 43 (30–61) | 64 | 64 | 5 (5–5) | 77 (51–117) | 72 | 72 |
| A/BC/1/2015 | 3rd | LPAI | 7 (6–9) | 76 (56–103) | 80 | 88 | 5 | 329 (216–502) | 100 | 100 | ||||||||||
| A/HK/61/2016 | 5th/PRD | LPAI | 6 (5–6) | 41 (29–57) | 64 | 64 | 5 | 61 (39–94) | 64 | 64 | ||||||||||
| A/HK/125/2016 | 5th/YRD | LPAI | 7 (6–9) | 36 (27–47) | 52 | 64 | 5 | 15 (11–21) | 24 | 24 | ||||||||||
| A/GD/17SF003/2016 | 5th/YRD | HPAI | 7 (6–9) | 28 (20–38) | 36 | 44 | 5 (5–8) | 32 (23–47) | 36 | 44 | ||||||||||
| 2 | 5.75 μg HA + ASO3A | NCT01942265 | NIAID | inactivated (A/Shanghai/2/2013) | 5.75 | 3.75 | ASO3A | 30 | 40 (19–59) | A/AH/1/2013 | 1st | LPAI | 5 (5–6) | 38 (29–49) | 53 | 53 | 5 (–) | 53 (40–71) | 70 | 70 |
| A/HK/61/2016 | 5th/PRD | LPAI | 5 | 25 (19–33) | 40 | 40 | 5 | 57 (41–79) | 70 | 70 | ||||||||||
| A/HK/125/2016 | 5th/YRD | LPAI | 5 | 25 (19–32) | 37 | 37 | 5 (5–5) | 16 (12–21) | 17 | 17 | ||||||||||
| A/GD/17SF003/2016 | 5th/YRD | HPAI | 5 | 15 (12–20) | 17 | 17 | 5 (5–6) | 11 (8–13) | 3 | 3 | ||||||||||
| 3 | 11.5 μg HA + MF59 | NCT01938742 | NIAID | inactivated (A/Shanghai/2/2013) | 11.5 | 7.5 | MF59 | 30 | 30 (21–58) | A/AH/1/2013 | 1st | LPAI | 5 | 25 (19–32) | 23 | 23 | 5 | 25 (18–34) | 30 | 30 |
| A/HK/61/2016 | 5th/PRD | LPAI | 5 | 15 (12–20) | 10 | 10 | 5 | 22 (16–30) | 27 | 27 | ||||||||||
| A/HK/125/2016 | 5th/YRD | LPAI | 5 | 18 (13–23) | 17 | 17 | 5 | 9 (7–11) | 7 | 7 | ||||||||||
| A/GD/17SF003/2016 | 5th/YRD | HPAI | 5 | 9 (7–12) | 3 | 3 | 5 (5–5) | 6 (5–6) | 0 | 0 | ||||||||||
| 4 | 15 μg HA + ISCOMATRIX | NCT01897701 | Novavax | recombinant VLP (A/Anui/1/2013) | 15 | NA | ISCO-MATRIX | 27 | 33 (18–49) | A/AH/1/2013 | 1st | LPAI | 5 | 40 (33–50) | 67 | 67 | 5 | 35 (28–44) | 41 | 41 |
| A/HK/61/2016 | 5th/PRD | LPAI | 5 | 45 (38–53) | 81 | 81 | 5 | 28 (23–35) | 33 | 33 | ||||||||||
| A/HK/125/2016 | 5th/YRD | LPAI | 5 | 21 (17–27) | 26 | 26 | 5 | 14 (11–19) | 7 | 7 | ||||||||||
| A/GD/17SF003/2016 | 5th/YRD | HPAI | 5 | 22 (17–28) | 22 | 22 | 5 (5–6) | 10 (7–13) | 0 | 0 | ||||||||||
| Age Groups (Age Range in Years) | Median Age (Years) | No of Subjects | HI | MN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A(H7N9) (AH1) | A(H3N2) (Perth16) | A(H7N9) (AH1) | A(H3N2) (Perth16) | |||||||
| GMT (95% CI) | SPR (%) | GMT (95% CI) | SPR (%) | GMT (95% CI) | SPR (%) | GMT (95% CI) | SPR (%) | |||
| 6–11 | 9 | 100 | 5 (-) | 0 | 34 (25–45) | 55 | 5 (-) | 0 | 99 (65–150) | 73 |
| 12–19 | 15 | 100 | 5 (-) | 0 | 19 (11–34) | 32 | 5 (-) | 0 | 59 (32–108) | 59 |
| 20–29 | 24 | 100 | 5 (-) | 0 | 9 (7–12) | 13 | 5 (-) | 0 | 50 (33–77) | 54 |
| 30–39 | 34 | 100 | 5 (-) | 0 | 7 (6–8) | 5 | 5 (-) | 0 | 21 (15–30) | 34 |
| 40–49 | 44 | 100 | 5 (-) | 0 | 8 (6–10) | 8 | 5 (-) | 0 | 22 (15–32) | 29 |
| 50–59 | 54 | 100 | 5 (-) | 0 | 12 (7–20) | 19 | 5 (-) | 0 | 33 (17–66) | 46 |
| 60–69 | 62 | 100 | 5 (-) | 0 | 11 (8–14) | 19 | 5 (-) | 0 | 32 (18–56) | 42 |
| 70–79 | 73.5 | 100 | 5 (-) | 0 | 14 (10–18) | 27 | 5 (-) | 0 | 49 (32–72) | 61 |
| ≥80 | 83 | 100 | 5(5–6) | 1 | 11 (9–15) | 23 | 5 (-) | 0 | 41 (30–57) | 55 |
| Age Groups | 6–35 Mos (Pediatrics) | 18–49 Yrs (Adult) | ≥ 65 Yrs (Elderly) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | 30 | 30 | 30 | |||||||||
| Viruses | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | NT # | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | ||
| HI | GMT | Pre * (95% CI) | 6 (5–6) | 5 (-) | 5 (-) | NT | 16 (9–29) | 5 (-) | 5 (-) | 34 (20–57) | 5 (-) | 5 (-) |
| Post * (95% CI) | 58 (35–95) | 5 (-) | 5 (-) | NT | 145 (91–230) | 5 (-) | 5 (-) | 94 (62–143) | 5 (-) | 5 (5–6) | ||
| % with HI ≥ 40 Post * | 73 | 0 | 0 | NT | 90 | 0 | 0 | 83 | 0 | 0 | ||
| % ≥ 4 fold rise | 73% (22) | 0 | 0 | NT | 63% (19) | 0 | 0 | 27% (8) | 0 | 0 | ||
| Viruses | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | NT | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | H3N2 Vic361 | H7N9 AH1 | H7N9 SH2 | ||
| MN | GMT | Pre * (95% CI) | 8 (5–11) | 5 (-) | 5 (-) | NT | 34 (19–60) | 5 (-) | 5 (-) | 88 (49–153) | 5 (-) | 5 (-) |
| Post * (95% CI) | 78 (47–130) | 5 (-) | 5 (-) | NT | 298 (189–472) | 5 (-) | 5 (-) | 240 (159–362) | 5 (5–6) | 5 (-) | ||
| % with MN ≥ 40 Post | 70 | 0 | 0 | NT | 97 | 0 | 0 | 90 | 0 | 0 | ||
| % ≥ 4 fold rise (n) | 67% (20) | 0 | 0 | NT | 57% (17) | 0 | 0 | 30% (9) | 0 | 0 | ||
| NAI functional antibodies (ELLA) | Viruses | H6N1 CA07 | H6N9 SH2 | NT | H6N1CA07 | H6N2 Vic 361 | H6N9AH1 | H6N9SH2 | H6N1 CA07 | H6N9 SH2 | NT | |
| GMT | Pre * (95% CI) | 6 (5–8) | 5 (-) | NT | 27 (16–45) | 23 (17–32) | 13 (10–18) | 11 (8–15) | 228 (135–388) | 97 (68–138) | NT | |
| Post * (95% CI) | 19 (12–29) | 8(6–11) | NT | 124 (77–200) | 70 (56–87) | 44 (32–61) | 52 (33–81) | 347 (211–570) | 189 (122–292) | NT | ||
| % ≥2 fold rise % (N) | 63% (19) | 30% (9) | NT | 97% (29) | 80% (24) | 80% (24) | 90% (27) | 50% (15) | 63% (19) | NT | ||
| % ≥4 fold rise %(N) | 50% (15) | 20% (6) | NT | 73% (22) | 57% (17) | 57% (17) | 70% (21) | 7% (2) | 17% (5) | NT | ||
| NA binding antibodies (ELISA) | rNAs | rN1 CA07 | rN9 AH1 | NT | rN1 CA07 | NT | rN9 AH1 | NT | rN1 CA07 | rN9 AH1 | NT | |
| GMT | Pre * (95% CI) | 4783 (3477–6581) | 3676 (2589–5218) | NT | 6475 (4590–9134) | NT | 3469 (2751–4376) | NT | 11,273 (7812–16,267) | 3200 (2314–4425) | NT | |
| Post * (95% CI) | 7558 (5781–9882) | 4050 (2928–5603) | NT | 12362 (8105–18856) | NT | 4371 (3314–5765) | NT | 13,878 (10,227–18,832) | 3313 (2403–4568) | NT | ||
| % ≥2 fold rise %(N) | 52% (13) | 24%(6) | NT | 63% (19) | NT | 27% (8) | NT | 20% (6) | 7% (2) | NT | ||
| % ≥4 fold rise %(N) | 16%(4) | 0% (0) | NT | 27% (8) | NT | 20% (6) | NT | 3% (1) | 0% (0) | NT | ||
| Assay | Antigen | Subtype | GMT | % ≥4 Fold Rise %(N) | |
|---|---|---|---|---|---|
| Pre * (95% CI) | Post * (95% CI) | ||||
| MN | A/Switzerland/9715293/2013# | A(H3N2) | 22 (12-43) | 542 (364-806) | 87% (20) |
| HI | A/Switzerland/9715293/2013# | A(H3N2) | 16 (9-28) | 330 (211-517) | 91% (21) |
| ADCC | A/Switzerland/9715293/2013# | A(H3N2) | 94 (59-152) | 453 (331-619) | 61% (14) |
| ADCC | A/Shanghai/2/2013 | A(H7N9) | 37 (24-58) | 48 (30-76) | 4% (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levine, M.Z.; Holiday, C.; Bai, Y.; Zhong, W.; Liu, F.; Jefferson, S.; Gross, F.L.; Tzeng, W.-p.; Fries, L.; Smith, G.; et al. Influenza A(H7N9) Pandemic Preparedness: Assessment of the Breadth of Heterologous Antibody Responses to Emerging Viruses from Multiple Pre-Pandemic Vaccines and Population Immunity. Vaccines 2022, 10, 1856. https://doi.org/10.3390/vaccines10111856
Levine MZ, Holiday C, Bai Y, Zhong W, Liu F, Jefferson S, Gross FL, Tzeng W-p, Fries L, Smith G, et al. Influenza A(H7N9) Pandemic Preparedness: Assessment of the Breadth of Heterologous Antibody Responses to Emerging Viruses from Multiple Pre-Pandemic Vaccines and Population Immunity. Vaccines. 2022; 10(11):1856. https://doi.org/10.3390/vaccines10111856
Chicago/Turabian StyleLevine, Min Z., Crystal Holiday, Yaohui Bai, Weimin Zhong, Feng Liu, Stacie Jefferson, F. Liaini Gross, Wen-pin Tzeng, Louis Fries, Gale Smith, and et al. 2022. "Influenza A(H7N9) Pandemic Preparedness: Assessment of the Breadth of Heterologous Antibody Responses to Emerging Viruses from Multiple Pre-Pandemic Vaccines and Population Immunity" Vaccines 10, no. 11: 1856. https://doi.org/10.3390/vaccines10111856
APA StyleLevine, M. Z., Holiday, C., Bai, Y., Zhong, W., Liu, F., Jefferson, S., Gross, F. L., Tzeng, W.-p., Fries, L., Smith, G., Boutet, P., Friel, D., Innis, B. L., Mallett, C. P., Davis, C. T., Wentworth, D. E., York, I. A., Stevens, J., Katz, J. M., & Tumpey, T. (2022). Influenza A(H7N9) Pandemic Preparedness: Assessment of the Breadth of Heterologous Antibody Responses to Emerging Viruses from Multiple Pre-Pandemic Vaccines and Population Immunity. Vaccines, 10(11), 1856. https://doi.org/10.3390/vaccines10111856





