NK Cell Subset Redistribution and Antibody Dependent Activation after Ebola Vaccination in Africans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants and Samples
2.2. NK Cell Assays
2.3. Flow Cytometry
2.4. Statistics
3. Results
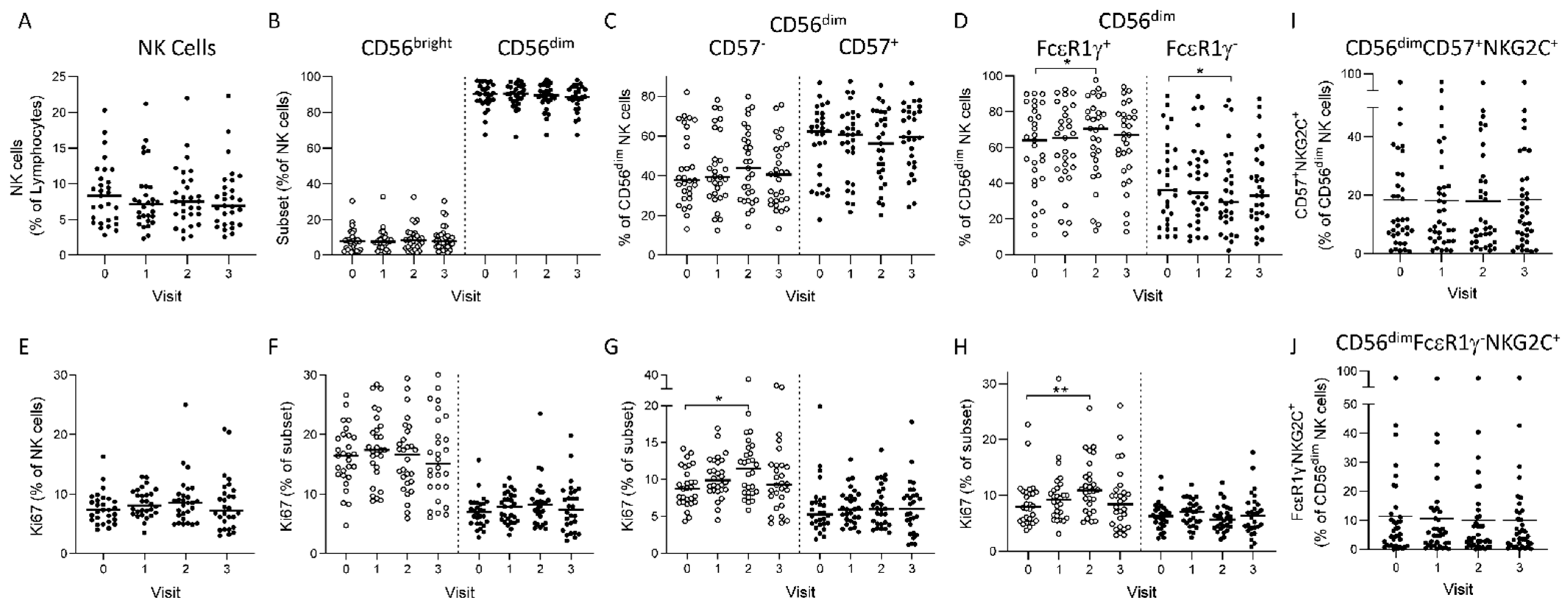
3.1. Longitudinal NK Cell Subset Changes after Heterologous 2 Dose Ebola Vaccination
3.2. Post-Ad26.ZEBOV, MVA-BN-Filo Vaccination-Induced Antibody-Dependent NK Cell Activation at 21 Days Post-Dose 2
3.3. Reduced Ebola Glycoprotein Specific Antibody-Dependent Responses in Post-Vaccination NK Cells
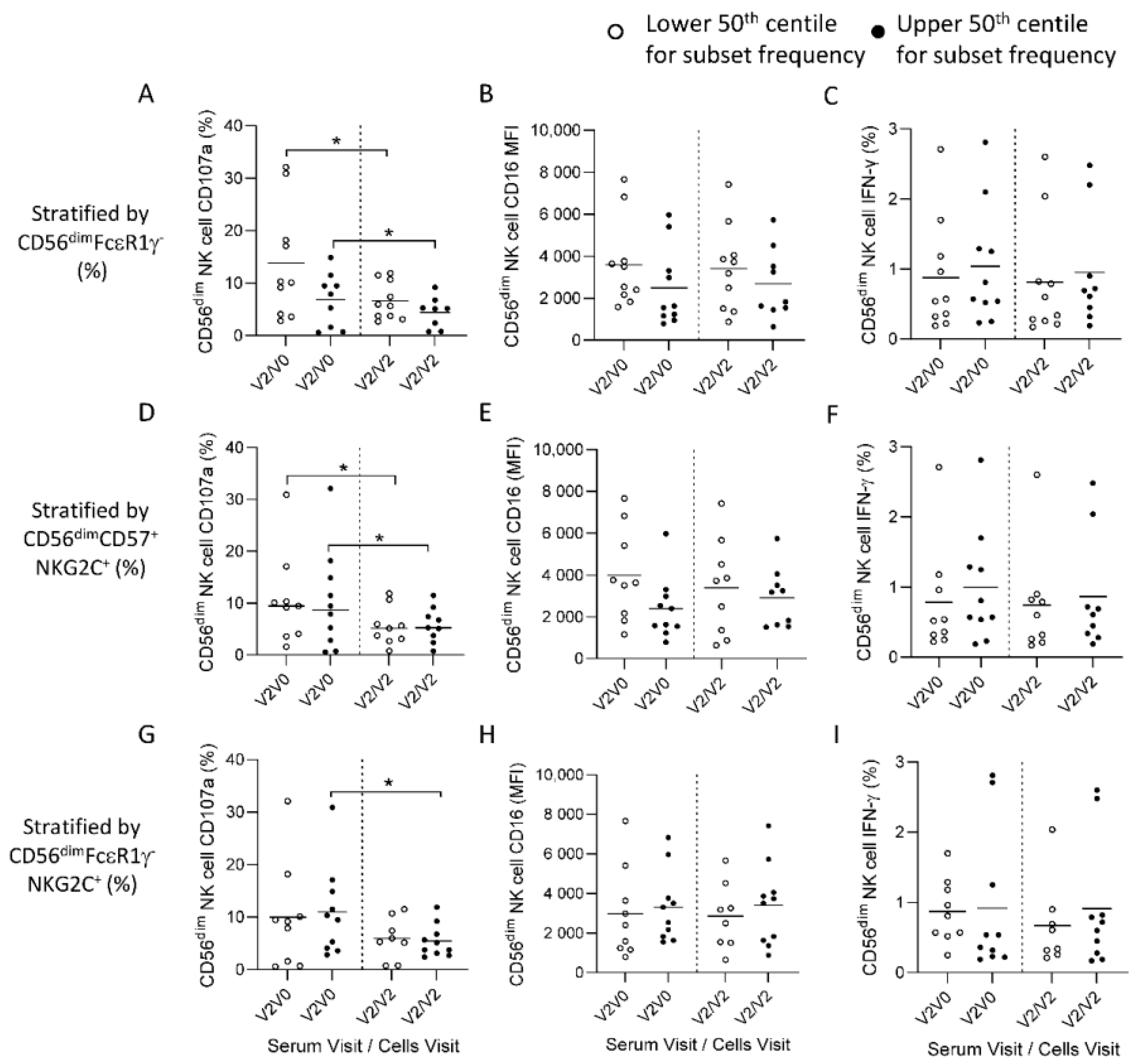
3.4. HCMV Associated NK Cell Expansions do Not Influence Antibody-Dependent Responses to Ebolavirus Glycoprotein
3.5. Anti-Ebolavirus GP Antibody Concentration does not Correlate with the ADNKA Response of Autologous NK Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centres for disease control and prevetion (CDC), Ebola Virus Disease—Democratic Republic of the Congo. Disease Outbreak News. Available online: https://www.cdc.gov/vhf/ebola/outbreaks/drc/2022-apr.html (accessed on 30 March 2022).
- Mutua, G.; Anzala, O.; Luhn, K.; Robinson, C.; Bockstal, V.; Anumendem, D.; Douoguih, M. Randomized clinical trial ex-amining safety and immunogenicity of heterologous prime-boost Ebola vaccines, Ad26.ZEBOV and MVA-BN-Filo: 12-month data from Nairobi, Kenya. J. Infect. Dis. 2019, 220, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Anywaine, Z.; Whitworth, H.; Kaleebu, P.; Praygod, G.; Shukarev, G.; Manno, D.; Kapiga, S.; Grosskurth, H.; Kalluvya, S.; Bockstal, V.; et al. Randomized clinical trial examining safety and immunogenicity of heterologous prime-boost Ebola vaccines, Ad26.ZEBOV and MVA-BN-Filo: 12-month data from Uganda and Tanzania. J. Infect. Dis. 2019, 220, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winslow, R.L.; Milligan, I.D.; Voysey, M.; Luhn, K.; Shukarev, G.; Douoguih, M.; Snape, M. Immune Responses to Novel Adenovirus Type 26 and Modified Vaccinia Virus Ankara–Vectored Ebola Vaccines at 1 Year. JAMA J. Am. Med. Assoc. 2017, 317, 1075–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukarev, G.; Callendret, B.; Luhn, K.; Douoguih, M. A two-dose heterologous prime-boost vaccine regimen eliciting sus-tained immune responses to Ebola Zaire could support a preventive strategy for future outbreaks. Hum. Vaccines Immunother. 2017, 13, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Milligan, I.D.; Gibani, M.M.; Sewell, R.; Clutterbuck, E.A.; Campbell, D.; Plested, E.; Nuthall, E.; Voysey, M.; Silva-Reyes, L.; McElrath, M.J.; et al. Safety and Immunogenicity of Novel Adenovirus Type 26- and Modified Vaccinia Anka-ra-Vectored Ebola Vaccines: A Randomized Clinical Trial. JAMA 2016, 315, 1610–1623. [Google Scholar] [CrossRef]
- Barry, H.; Mutua, G.; Kibuuka, H.; Anywaine, Z.; Sirima, S.B.; Meda, N.; Anzala, O.; Eholie, S.; Bétard, C.; Richert, L.; et al. Safety and immunogenicity of 2-dose heterologous Ad26.ZEBOV, MVA-BN-Filo Ebola vaccination in healthy and HIV-infected adults: A randomised, placebo-controlled Phase II clinical trial in Africa. PLoS Med. 2021, 18, e1003813. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Launay, O.; Lelievre, J.-D.; Lacabaratz, C.; Grande, S.; Goldstein, N.; Robinson, C.; Gaddah, A.; Bockstal, V.; Wiedemann, A.; et al. Safety and immunogenicity of a two-dose heterologous Ad26.ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): A randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 2020, 21, 493–506. [Google Scholar] [CrossRef]
- Gunn, B.M.; Yu, W.-H.; Karim, M.M.; Brannan, J.M.; Herbert, A.S.; Wec, A.Z.; Halfmann, P.J.; Fusco, M.L.; Schendel, S.; Gangavarapu, K.; et al. A Role for Fc Function in Therapeutic Monoclonal Antibody-Mediated Protection against Ebola Virus. Cell Host Microbe 2018, 24, 221–233. [Google Scholar] [CrossRef]
- Liu, Q.; Fan, C.; Li, Q.; Zhou, S.; Huang, W.; Wang, L.; Sun, C.; Wang, M.; Wu, X.; Ma, J.; et al. Anti-body-dependent-cellular-cytotoxicity-inducing antibodies significantly affect the post-exposure treatment of Ebola virus infection. Sci. Rep. 2017, 7, 45552. [Google Scholar] [CrossRef] [Green Version]
- Bornholdt, Z.A.; Herbert, A.S.; Mire, C.E.; He, S.; Cross, R.W.; Wec, A.Z.; Abelson, D.M.; Geisbert, J.B.; James, R.M.; Rahim, M.N.; et al. A Two-Antibody Pan-Ebolavirus Cocktail Confers Broad Ther-apeutic Protection in Ferrets and Nonhuman Primates. Cell Host Microbe 2019, 25, 49–58.e5. [Google Scholar] [CrossRef] [Green Version]
- Wec, A.Z.; Bornholdt, Z.A.; He, S.; Herbert, A.S.; Goodwin, E.; Wirchnianski, A.S.; Gunn, B.M.; Zhang, Z.; Zhu, W.; Liu, G.; et al. Development of a Human Antibody Cocktail that Deploys Multiple Functions to Confer Pan-Ebolavirus Protection. Cell Host Microbe 2019, 25, 39–48.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagstaffe, H.R.; Clutterbuck, E.A.; Bockstal, V.; Stoop, J.N.; Luhn, K.; Douoguih, M.; Shukarev, G.; Snape, M.D.; Pollard, A.J.; Riley, E.M.; et al. Antibody-Dependent Natural Killer Cell Activation After Ebola Vaccination. J. Infect. Dis. 2019, 223, 1171–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagstaffe, H.R.; Clutterbuck, E.A.; Bockstal, V.; Stoop, J.N.; Luhn, K.; Douoguih, M.J.; Shukarev, G.; Snape, M.D.; Pollard, A.J.; Riley, E.M.; et al. Ebola virus glycoprotein stimulates IL-18–dependent natural killer cell responses. J. Clin. Invest. 2020, 130, 3936–3946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagstaffe, H.R.; Susannini, G.; Thiébaut, R.; Richert, L.; Lévy, Y.; Bockstal, V.; Stoop, J.N.; Luhn, K.; Douoguih, M.; Riley, E.M.; et al. Durable natural killer cell responses after heterologous two-dose Ebola vaccination. NPJ Vaccines 2021, 6, 19. [Google Scholar] [CrossRef]
- Hill, D.L.; Carr, E.J.; Rutishauser, T.; Moncunill, G.; Campo, J.J.; Innocentin, S.; Mpina, M.; Nhabomba, A.; Tumbo, A.; Jairoce, C.; et al. Immune system development varies ac-cording to age, location, and anemia in African children. Sci. Transl. Med. 2020, 12, eaaw9522. [Google Scholar] [CrossRef]
- Goodier, M.R.; White, M.J.; Darboe, A.; Nielsen, C.M.; Goncalves, A.; Bottomley, C.; Moore, S.E.; Riley, E.M. Rapid NK cell differentiation in a population with near-universal human cytomegalovirus infection is attenuated by NKG2C deletions. Blood 2014, 124, 2213–2222. [Google Scholar] [CrossRef] [Green Version]
- Miles, D.J.C.; van der Sande, M.; Jeffries, D.; Kaye, S.; Ismaili, J.; Ojuola, O.; Sanneh, M.; Touray, E.S.; Waight, P.; Rowland-Jones, S.; et al. Cytomegalovirus Infection in Gambian Infants Leads to Profound CD8 T-Cell Differentiation. J. Virol. 2007, 81, 5766–5776. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic Modification and Antibody-Dependent Expansion of Memory-like NK Cells in Human Cytomegalovirus-Infected Individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Béziat, V.; Holmes, T.D.; Han, H.; Chiang, S.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus Infection Drives Adaptive Epigenetic Diversification of NK Cells with Altered Signaling and Effector Function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Stockdale, L.; Nash, S.; Nalwoga, A.; Gibson, L.; Painter, H.; Raynes, J.; Asiki, G.; Newton, R.; Fletcher, H. HIV, HCMV and mycobacterial antibody levels: A cross-sectional study in a rural Ugandan cohort. Trop. Med. Int. Health 2018, 24, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Bates, M.; Brantsaeter, A.B. Human cytomegalovirus (CMV) in Africa: A neglected but important pathogen. J. Virus Erad. 2016, 2, 136–142. [Google Scholar] [CrossRef]
- Logue, J.; Tuznik, K.; Follmann, D.; Grandits, G.; Marchand, J.; Reilly, C.; Sarro, Y.D.S.; Pettitt, J.; Stavale, E.J.; Fallah, M.; et al. Use of the Filovirus Animal Non-Clinical Group (FANG) Ebola virus immuno-assay requires fewer study participants to power a study than the Alpha Diagnostic International assay. J. Virol. Methods 2018, 255, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Pejoski, D.; de Rham, C.; Martinez-Murillo, P.; Santoro, F.; Auderset, F.; Medaglini, D.; Pozzi, G.; Vono, M.; Lambert, P.H.; Huttner, A.; et al. Rapid dose-dependent Natural Killer (NK) cell modulation and cytokine responses following human rVSV-ZEBOV Ebo-lavirus vaccination. NPJ Vaccines 2020, 5, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björkström, N.K.; Riese, P.; Heuts, F.; Andersson, S.; Fauriat, C.; Ivarsson, M.A.; Björklund, A.T.; Flodström-Tullberg, M.; Michaëlsson, J.; Rottenberg, M.E.; et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 2010, 116, 3853–3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, S.; Tomezsko, P.J.; Rands, K.; Toth, I.; Lichterfeld, M.; Gandhi, R.T.; Altfeld, M. CD4 + T-Cell Help Enhances NK Cell Function following Therapeutic HIV-1 Vaccination. J. Virol. 2014, 88, 8349–8354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, C.M.; White, M.J.; Bottomley, C.; Lusa, C.; Galan, A.R.; Turner, S.E.G.; Goodier, M.; Riley, E.M. Impaired NK Cell Responses to Pertussis and H1N1 Influenza Vaccine Antigens in Human Cytomegalovirus-Infected Individuals. J. Immunol. 2015, 194, 4657–4667. [Google Scholar] [CrossRef]
- White, M.J.; Nielsen, C.M.; McGregor, R.H.C.; Riley, E.H.C.; Goodier, M. Differential activation of CD 57-defined natural killer cell subsets during recall responses to vaccine antigens. Immunology 2014, 142, 140–150. [Google Scholar] [CrossRef]
- Darboe, A.; Danso, E.; Clarke, E.; Umesi, A.; Touray, E.; Wegmuller, R.; Moore, S.E.; Riley, E.M.; Goodier, M.R. Enhancement of cytokine-driven NK cell IFN-gamma production after vaccination of HCMV infected Africans. Eur. J. Immunol. 2017, 47, 1040–1050. [Google Scholar] [CrossRef] [Green Version]
- Goodier, M.R.; Rodriguez-Galan, A.; Lusa, C.; Nielsen, C.M.; Darboe, A.; Moldoveanu, A.L.; White, M.J.; Behrens, R.; Riley, E.M. Influenza Vaccination Generates Cytokine-Induced Memory-like NK Cells: Impact of Human Cytomegalovirus In-fection. J. Immunol. 2016, 197, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Della Chiesa, M.; Falco, M.; Podesta, M.; Locatelli, F.; Moretta, L.; Frassoni, F.; Moretta, A. Phenotypic and functional het-erogeneity of human NK cells developing after umbilical cord blood transplantation: A role for human cytomegalovirus? Blood 2012, 119, 399–410. [Google Scholar] [CrossRef]
- Wald, A.; Selke, S.; Magaret, A.; Boeckh, M. Impact of human cytomegalovirus (CMV) infection on immune response to pandemic 2009 H1N1 influenza vaccine in healthy adults. J. Med. Virol. 2013, 85, 1557–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furman, D.; Jojic, V.; Sharma, S.; Shen-Orr, S.S.; Angel, C.J.L.; Onengut-Gumuscu, S.; Kidd, B.A.; Maecker, H.T.; Concannon, P.; Dekker, C.L.; et al. Cytomegalovirus infection enhances the immune response to influenza. Sci. Transl. Med. 2015, 7, 281ra43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.; Adetifa, J.U.; Noho-Konteh, F.; Njie-Jobe, J.; Sanyang, L.C.; Drammeh, A.; Plebanski, M.; Whittle, H.C.; Rowland-Jones, S.L.; Robertson, I.; et al. Limited Impact of Human Cytomegalovirus Infection in African Infants on Vaccine-Specific Responses Following Diphtheria-Tetanus-Pertussis and Measles Vaccination. Front. Immunol. 2020, 11, 1083. [Google Scholar] [CrossRef]
- Sharpe, H.R.; Provine, N.M.; Bowyer, G.S.; Folegatti, P.M.; Belij-Rammerstorfer, S.; Flaxman, A.; Makinson, R.; Hill, A.V.; Ewer, K.J.; Pollard, A.J.; et al. CMV-associated T cell and NK cell terminal differentiation does not affect immunogenicity of ChAdOx1 vaccination. JCI Insight 2022, 7, e154187. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.M.; Wolf, A.-S.; Goodier, M.; Riley, E.M. Synergy between Common γ Chain Family Cytokines and IL-18 Potentiates Innate and Adaptive Pathways of NK Cell Activation. Front. Immunol. 2016, 7, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherratt, S.; Patel, A.; Baker, D.A.; Riley, E.M.; Goodier, M.R. Differential IL-18 Dependence of Canonical and Adaptive NK Cells for Antibody Dependent Responses to P. falciparum. Front. Immunol. 2020, 11, 533. [Google Scholar] [CrossRef]
- Reynard, S.; Journeaux, A.; Gloaguen, E.; Schaeffer, J.; Varet, H.; Pietrosemoli, N.; Mateo, M.; Baillet, N.; Laouenan, C.; Raoul, H.; et al. Immune parameters and outcomes during Ebola virus disease. JCI Insight 2019, 4, e125106. [Google Scholar] [CrossRef]
- Meyer, M.; Gunn, B.M.; Malherbe, D.C.; Gangavarapu, K.; Yoshida, A.; Pietzsch, C.; Kuzmina, N.A.; Saphire, E.O.; Collins, P.L.; Crowe, J.E.; et al. Ebola vaccine–induced protection in nonhuman primates correlates with antibody specificity and Fc-mediated effects. Sci. Transl. Med. 2021, 13, eabg6128. [Google Scholar] [CrossRef]



| Group 1 | Group 2 | |
|---|---|---|
| Vaccine Schedule | Ad26 1, MVA 2 28 Day Interval | Ad26, MVA 56 Day Interval |
| Age years: median (IQR) | 32, (24–46) | 25, (22–44) |
| Sex: n (m/f) | 12/2 | 10/5 |
| Baseline samples (Visit 0) (PBMC/Serum) | Day 1 (14/14) | Day 1 (15/15) |
| Post-dose 1 samples (Visit 1) 3 (PBMC/Serum) | Day 29 (14/0) | Day 57 (15/0) |
| 21 days post-dose 2 samples (Visit 2) (PBMC/Serum) | Day 50 (14/14) | Day 78 (15/15) |
| 180 days post-dose 2 samples (Visit 3) (PBMC/Serum) | Day 209 (14/0) | Day 237 (15/0) |
| Anti-Ebola GP IgG (Visit 2) concentration (EU/mL) (median, IQR) | 11,513 (3409–16,021) | 13,379 (3367–19,788) |
| Anti-HCMV IgG (Visit 0) concentration (IU/mL) (median, IQR) | 2879 (2492–3918) | 2677 (1258–4299) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagstaffe, H.R.; Anzala, O.; Kibuuka, H.; Anywaine, Z.; Sirima, S.B.; Thiébaut, R.; Richert, L.; Levy, Y.; Lacabaratz, C.; Bockstal, V.; et al. NK Cell Subset Redistribution and Antibody Dependent Activation after Ebola Vaccination in Africans. Vaccines 2022, 10, 884. https://doi.org/10.3390/vaccines10060884
Wagstaffe HR, Anzala O, Kibuuka H, Anywaine Z, Sirima SB, Thiébaut R, Richert L, Levy Y, Lacabaratz C, Bockstal V, et al. NK Cell Subset Redistribution and Antibody Dependent Activation after Ebola Vaccination in Africans. Vaccines. 2022; 10(6):884. https://doi.org/10.3390/vaccines10060884
Chicago/Turabian StyleWagstaffe, Helen R., Omu Anzala, Hannah Kibuuka, Zacchaeus Anywaine, Sodiomon B. Sirima, Rodolphe Thiébaut, Laura Richert, Yves Levy, Christine Lacabaratz, Viki Bockstal, and et al. 2022. "NK Cell Subset Redistribution and Antibody Dependent Activation after Ebola Vaccination in Africans" Vaccines 10, no. 6: 884. https://doi.org/10.3390/vaccines10060884
APA StyleWagstaffe, H. R., Anzala, O., Kibuuka, H., Anywaine, Z., Sirima, S. B., Thiébaut, R., Richert, L., Levy, Y., Lacabaratz, C., Bockstal, V., Luhn, K., Douoguih, M., & Goodier, M. R. (2022). NK Cell Subset Redistribution and Antibody Dependent Activation after Ebola Vaccination in Africans. Vaccines, 10(6), 884. https://doi.org/10.3390/vaccines10060884








