SARS-CoV-2 mRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response
Abstract
:1. Introduction
2. Materials and Methods
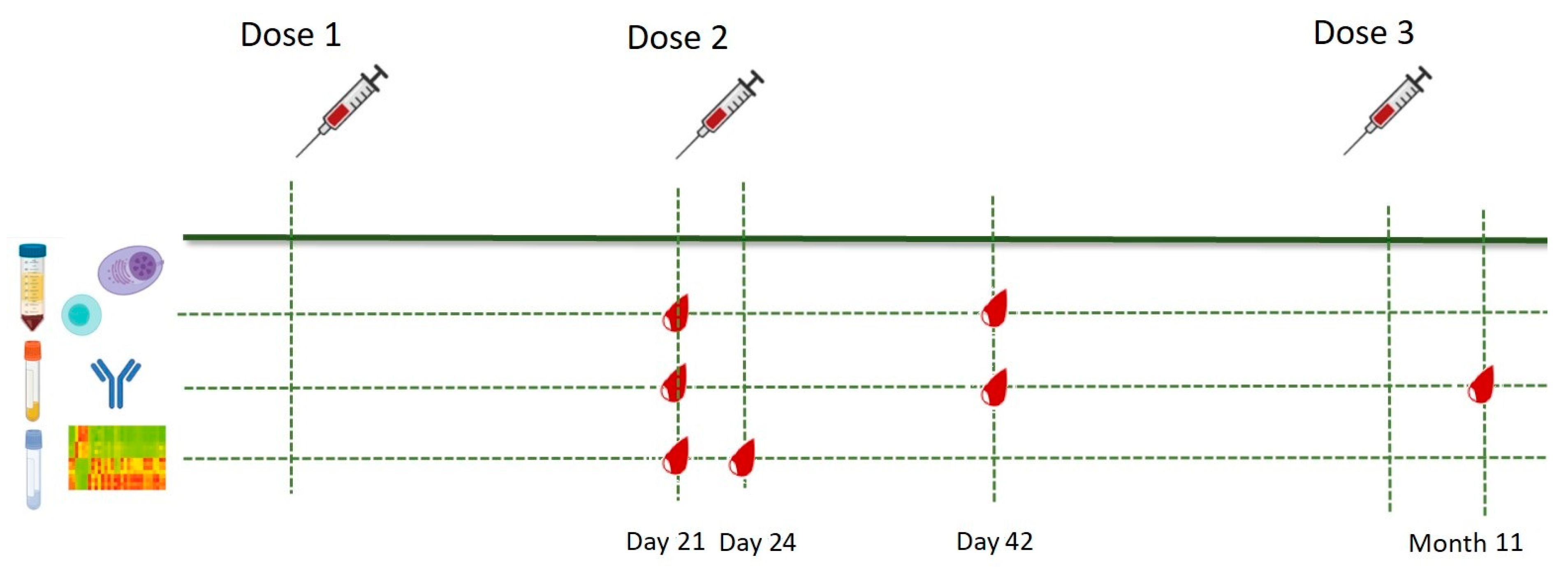
2.1. Study Population and Study Design
2.2. Sample Collection and Storage
2.3. Whole RNA Sequencing and Differential Expression Gene (DEG) Analysis
2.4. Ex Vivo Spike (S)-Protein Stimulation of PBMCs and Flow Cytometry Analysis
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Statistical Analysis
3. Results
3.1. Transcriptional Signature of the Immune Response to BNT162b2
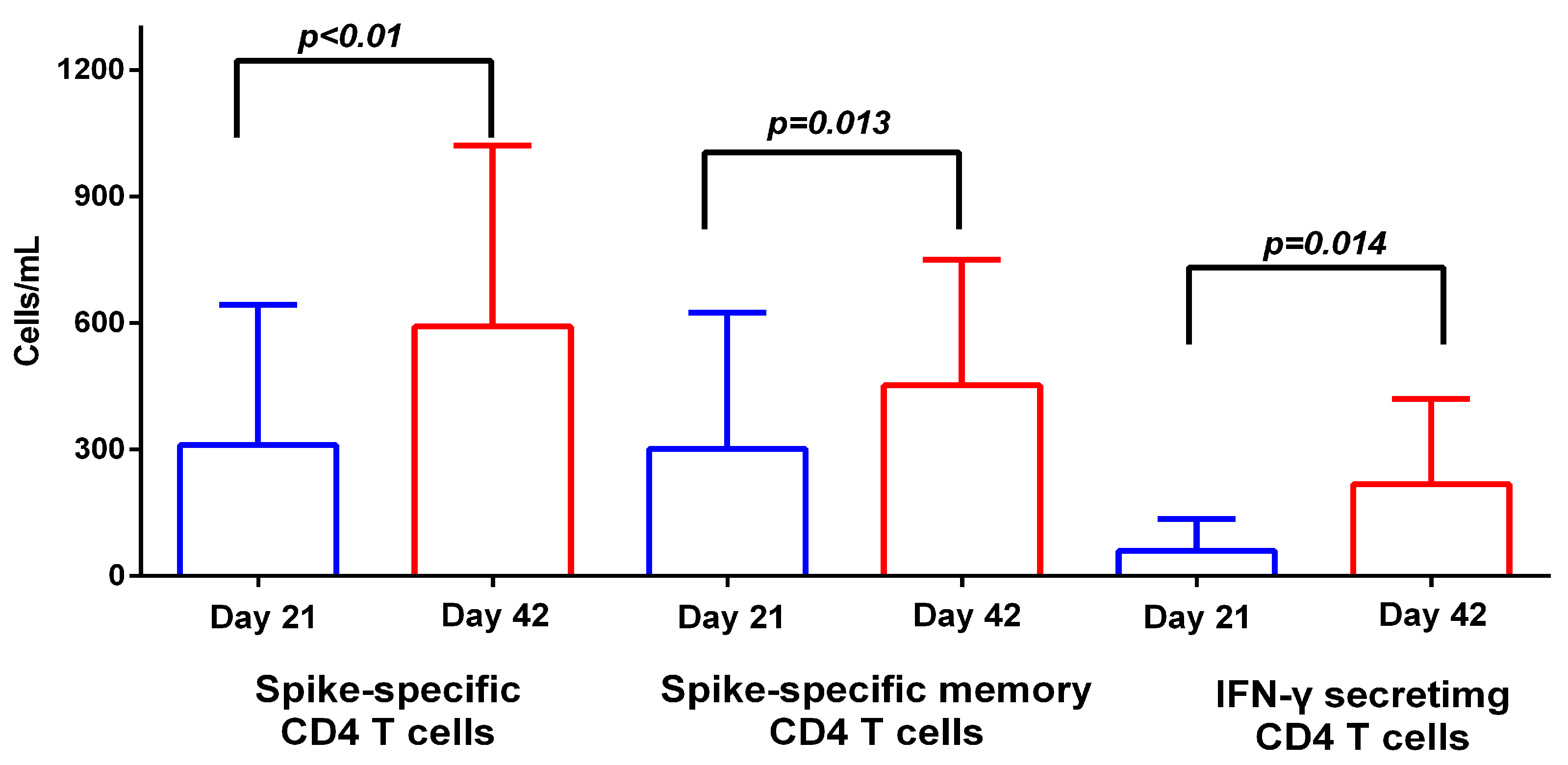
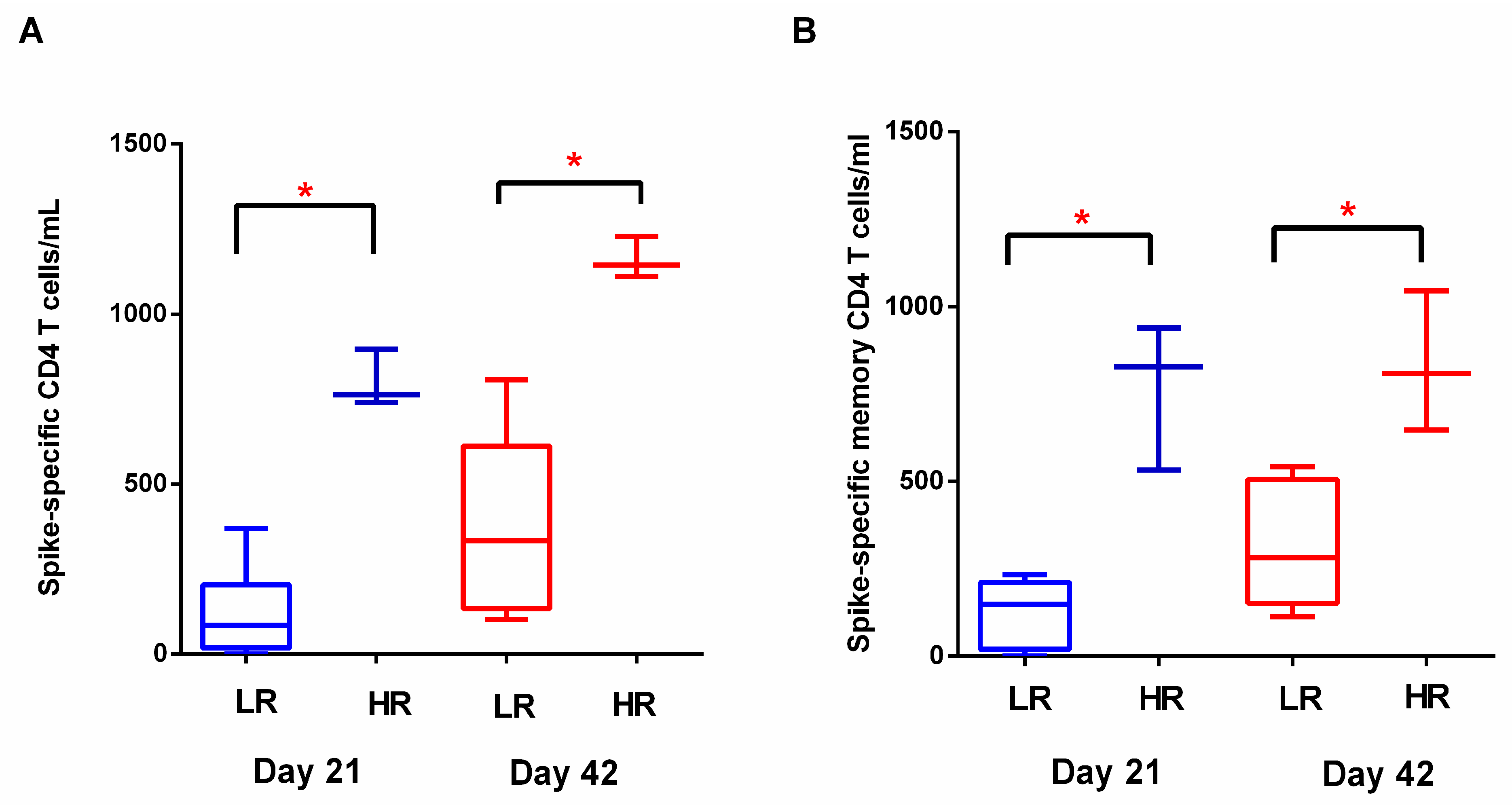
3.2. T Cell Response to BNT162b2
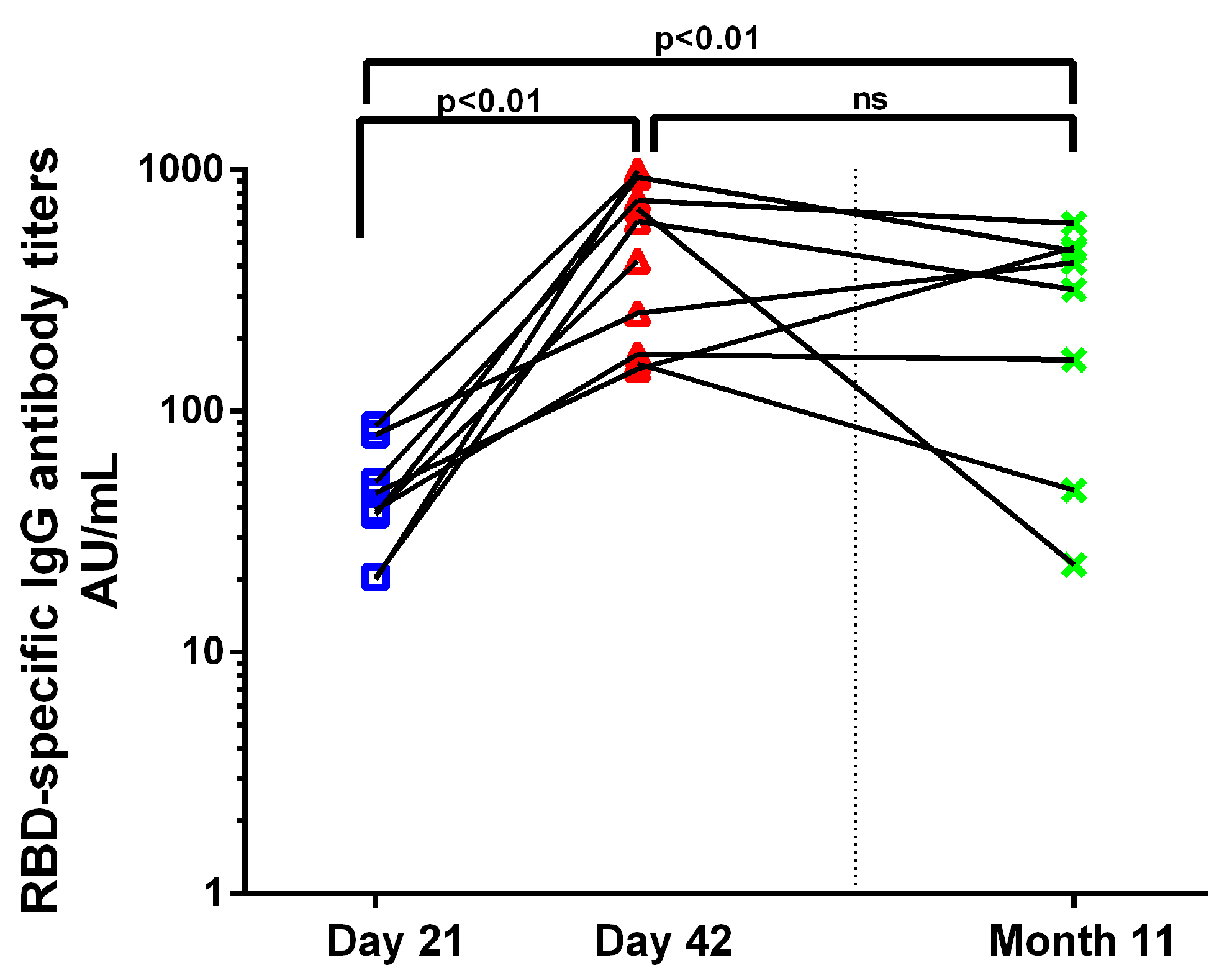
3.3. Antibody Response to BNT162b2
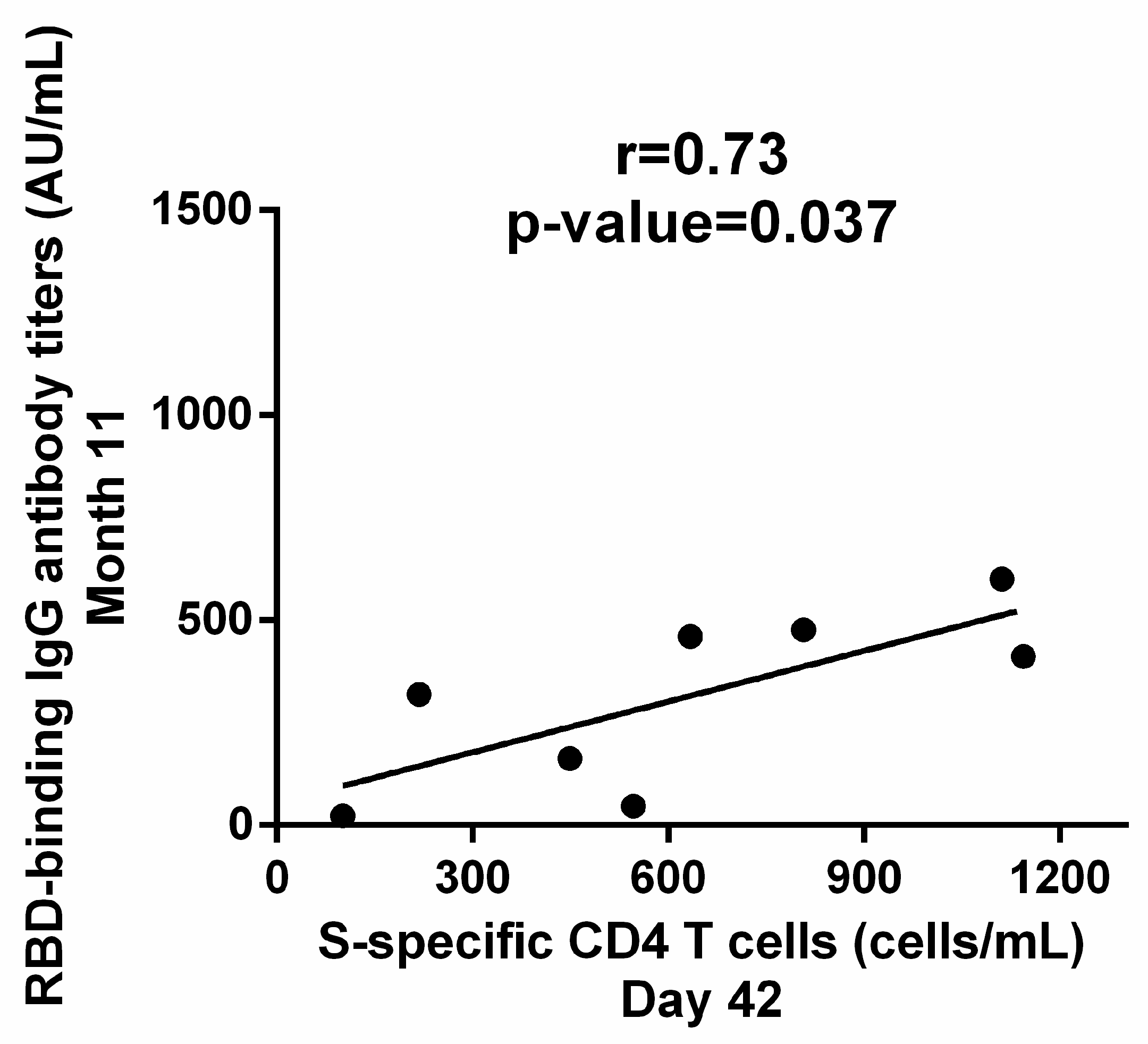
3.4. Correlation between Cellular and Humoral Response to BNT162b2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). COVID-19 Weekly Epidemiological Update. Available online: https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update (accessed on 8 December 2022).
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 COVID-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 384, e84. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; Ferreira, I.A.T.M.; Kotagiri, P.; Datir, R.P.; Lim, E.Y.; Touizer, E.; Meng, B.; Abdullahi, A.; Baker, S.; Dougan, G.; et al. Age-Related Immune Response Heterogeneity to SARS-CoV-2 Vaccine BNT162b2. Nature 2021, 596, 417–422. [Google Scholar] [CrossRef]
- Kobashi, Y.; Kawamura, T.; Shimazu, Y.; Zhao, T.; Sugiyama, A.; Nakayama, A.; Kaneko, Y.; Nishikawa, Y.; Omata, F.; Takita, M.; et al. Humoral Immunity after Second Dose of BNT162b2 Vaccine in Japanese Communities: An Observational Cross-Sectional Study, Fukushima Vaccination Community Survey. Sci. Rep. 2022, 12, 18929. [Google Scholar] [CrossRef]
- Lagousi, T.; Routsias, J.; Spoulou, V. Development of an Enzyme-Linked Immunosorbent Assay (ELISA) for Accurate and Prompt Coronavirus Disease 2019 (COVID-19) Diagnosis Using the Rational Selection of Serological Biomarkers. Diagnostics 2021, 11, 1970. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Fanidis, D.; Moulos, P. Integrative, Normalization-Insusceptible Statistical Analysis of RNA-Seq Data, with Improved Differential Expression and Unbiased Downstream Functional Analysis. Brief. Bioinform. 2021, 22, bbaa156. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachim, M.Y.; Al Heialy, S.; Hachim, I.Y.; Halwani, R.; Senok, A.C.; Maghazachi, A.A.; Hamid, Q. Interferon-Induced Transmembrane Protein (IFITM3) Is Upregulated Explicitly in SARS-CoV-2 Infected Lung Epithelial Cells. Front. Immunol. 2020, 11, 1372. [Google Scholar] [CrossRef]
- Sajid, M.; Ullah, H.; Yan, K.; He, M.; Feng, J.; Shereen, M.A.; Hao, R.; Li, Q.; Guo, D.; Chen, Y.; et al. The Functional and Antiviral Activity of Interferon Alpha-Inducible IFI6 against Hepatitis B Virus Replication and Gene Expression. Front. Immunol. 2021, 12, 634937. [Google Scholar] [CrossRef]
- Liu, G.Q.; Lee, J.H.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; van Gent, M.; Riedl, W.; Davis-Gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-Dependent Activation of the Sensor MDA5 Is Antagonized by the SARS-CoV-2 Papain-like Protease to Evade Host Innate Immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Farber, D.L. COVID-19 Vaccines: Modes of Immune Activation and Future Challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Scott, M.K.D.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L.; et al. Systems Vaccinology of the BNT162b2 MRNA Vaccine in Humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef]
- Lee, H.K.; Knabl, L.; Moliva, J.I.; Knabl, L.; Werner, A.P.; Boyoglu-Barnum, S.; Kapferer, S.; Pateter, B.; Walter, M.; Sullivan, N.J.; et al. MRNA Vaccination in Octogenarians 15 and 20 Months after Recovery from COVID-19 Elicits Robust Immune and Antibody Responses That Include Omicron. Cell Rep. 2022, 39, 110680. [Google Scholar] [CrossRef]
- Kishnani, P.S.; del Angel, G.; Zhou, S.; Rush, E.T. Investigation of ALPL Variant States and Clinical Outcomes: An Analysis of Adults and Adolescents with Hypophosphatasia Treated with Asfotase Alfa. Mol. Genet. Metab. 2021, 133, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, M.; Ransohoff, R.M. Chemokine Receptor CXCR2: Physiology Regulator and Neuroinflammation Controller? J Neuroimmunol. 2012, 246, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audrito, V.; Messana, V.G.; Deaglio, S. NAMPT and NAPRT: Two Metabolic Enzymes with Key Roles in Inflammation. Front. Oncol. 2020, 10, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diana, P.; Carvalheira, G.M.G. NIBAN1, Exploring Its Roles in Cell Survival Under Stress Context. Front. Cell Dev. Biol. 2022, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Aillaud, C.; Bosc, C.; Peris, L.; Bosson, A.; Heemeryck, P.; Van Dijk, J.; Le Friec, J.; Boulan, B.; Vossier, F.; Sanman, L.E.; et al. Vasohibins/SVBP Are Tubulin Carboxypeptidases (TCPs) That Regulate Neuron Differentiation. Science 2017, 358, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, O.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of MRNA BNT162b2 COVID-19 Vaccine up to 6 Months in a Large Integrated Health System in the USA: A Retrospective Cohort Study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Tang, P.; Hasan, M.R.; AlMukdad, S.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 Infection in Qatar. N. Engl. J. Med. 2021, 385, e83. [Google Scholar] [CrossRef] [PubMed]
- Akondy, R.S.; Fitch, M.; Edupuganti, S.; Yang, S.; Kissick, H.T.; Li, K.W.; Youngblood, B.A.; Abdelsamed, H.A.; McGuire, D.J.; Cohen, K.W.; et al. Origin and Differentiation of Human Memory CD8 T Cells after Vaccination. Nature 2017, 552, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund, E.; Lewis, M.W.; Hansen, S.G.; Strelow, L.I.; Nelson, J.A.; Sexton, G.J.; Hanifin, J.M.; Slifka, M.K. Duration of Antiviral Immunity after Smallpox Vaccination. Nat. Med. 2003, 9, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.R.; Painter, M.M.; Apostolidis, S.A.; Mathew, D.; Meng, W.; Rosenfeld, A.M.; Lundgreen, K.A.; Reynaldi, A.; Khoury, D.S.; Pattekar, A.; et al. MRNA Vaccines Induce Durable Immune Memory to SARS-CoV-2 and Variants of Concern. Science 2021, 374, abm0829. [Google Scholar] [CrossRef]
- Griffin, J.B.; Haddix, M.; Dana, P.; Fisher, R.; Koo, T.H.; Traub, E.; Gounder, P.; Jarashow, C.; Balter, S. SARS-CoV-2 Infections and Hospitalisations among Persons Aged ≥ 16 Years, by Vaccination Status—Los Angeles County, California, May 1–July 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 1170–1176. [Google Scholar] [CrossRef]
- Woldemeskel, B.A.; Garliss, C.C.; Aytenfisu, T.Y.; Johnston, T.S.; Cox, A.L.; Karaba, A.H.; Blankson, J.N. Discordant Antibody and T Cell Responses to the SARS-CoV-2 Omicron Variant in COVID-19 MRNA Vaccine Recipient. Clin. Infect. Dis. 2022, 75, 1652–1654. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and Immunological Assessment of Asymptomatic SARS-CoV-2 Infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef]
- Casado, J.L.; Vizcarra, P.; Haemmerle, J.; Velasco, H.; Martín-hondarza, A.; Rodríguez-domínguez, M.J.; Velasco, T.; Martín, S.; Romero-hernández, B.; Fernández-escribano, M.; et al. Pre-Existing T Cell Immunity Determines the Frequency and Magnitude of Cellular Immune Response to Two Doses of MRNA Vaccine Against. Vaccine X 2022, 11, 2–6. [Google Scholar] [CrossRef]
- Campagna, R.; Mazzuti, L.; Guerrizio, G.; Nonne, C.; Migliara, G.; De Vito, C.; Mezzaroma, I.; Chiaretti, S.; Fimiani, C.; Pistolesi, V.; et al. Humoral and T-Cell Mediated Response after Administration of MRNA Vaccine BNT162b2 in Frail Populations. Vaccine X 2022, 12, 100246. [Google Scholar] [CrossRef]
- Atmar, R.L.; Lyke, K.E.; Deming, M.E.; Jackson, L.A.; Branche, A.R.; El Sahly, H.M.; Rostad, C.A.; Martin, J.M.; Johnston, C.; Rupp, R.E.; et al. Homologous and Heterologous COVID-19 Booster Vaccinations. N. Engl. J. Med. 2022, 386, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Go, J.; Sung, H.; Kim, S.W.; Walter, M.; Knabl, L.; Furth, P.A.; Hennighausen, L.; Huh, J.W. Heterologous ChAdOx1-BNT162b2 Vaccination in Korean Cohort Induces Robust Immune and Antibody Responses That Includes Omicron. iScience 2022, 25, 104473. [Google Scholar] [CrossRef] [PubMed]

| N | 18 |
| Age | |
| Mean (in years) | 41.46 |
| SD (in years) | 12.36 |
| Gender | |
| Female (%) | 13 (72.2%) |
| Male (%) | 5 (27.8%) |
| Body Mass Index (BMI) | |
| Mean (SD) | 25.3 (4.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadatou, I.; Geropeppa, M.; Verrou, K.-M.; Tzanoudaki, M.; Lagousi, T.; Liatsis, E.; Spoulou, V. SARS-CoV-2 mRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response. Vaccines 2023, 11, 103. https://doi.org/10.3390/vaccines11010103
Papadatou I, Geropeppa M, Verrou K-M, Tzanoudaki M, Lagousi T, Liatsis E, Spoulou V. SARS-CoV-2 mRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response. Vaccines. 2023; 11(1):103. https://doi.org/10.3390/vaccines11010103
Chicago/Turabian StylePapadatou, Ioanna, Maria Geropeppa, Kleio-Maria Verrou, Marianna Tzanoudaki, Theano Lagousi, Emmanouil Liatsis, and Vana Spoulou. 2023. "SARS-CoV-2 mRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response" Vaccines 11, no. 1: 103. https://doi.org/10.3390/vaccines11010103
APA StylePapadatou, I., Geropeppa, M., Verrou, K.-M., Tzanoudaki, M., Lagousi, T., Liatsis, E., & Spoulou, V. (2023). SARS-CoV-2 mRNA Dual Immunization Induces Innate Transcriptional Signatures, Establishes T-Cell Memory and Coordinates the Recall Response. Vaccines, 11(1), 103. https://doi.org/10.3390/vaccines11010103






