Length-Dependent Modulation of B Cell Activating Factor Transcripts in Chicken Macrophage by Viral Double-Stranded RNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Macrophages
2.2. Activation of Macrophages
2.3. RT-PCR Array
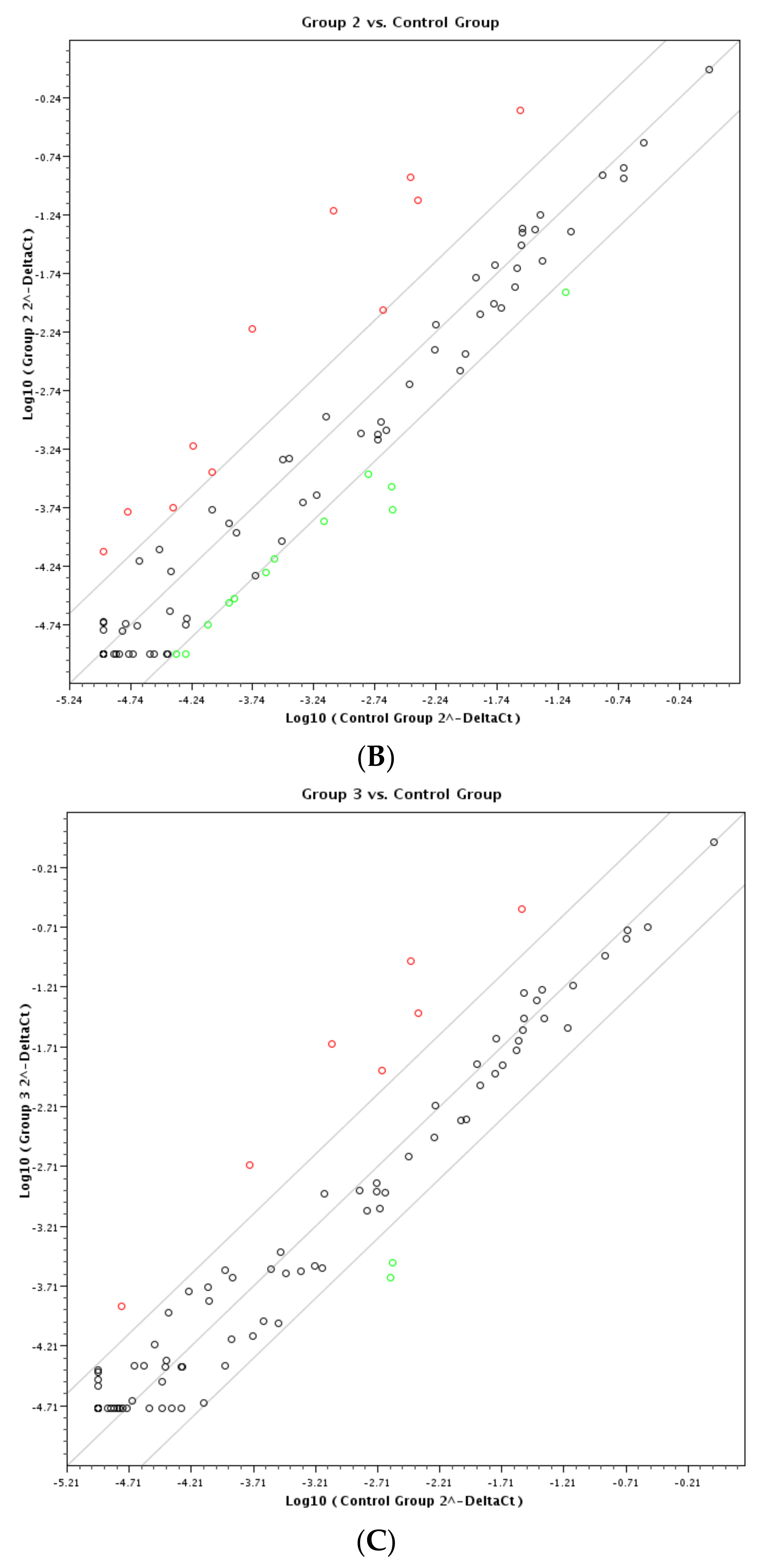
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Broz, P.; Monack, D.M. Newly described pattern recognition receptors team up against intracellular pathogens. Nat. Rev. Immunol. 2013, 13, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, S. Macrophages: Plastic solutions to environmental heterogeneity. Inflamm. Res. 2013, 62, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Kc, M.; Ngunjiri, J.M.; Ghorbani, A.; Lee, K. TLR3 and MDA5 Knockout DF-1 cells Enhance Replication of Avian Orthoavulavirus 1. Avian Dis. 2023, 67, 94–101. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; e Sousa, C.R. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Im, J.H.; Duic, I.; Yoshimura, S.H.; Onomoto, K.; Yoneyama, M.; Kato, H.; Fujita, T. Mechanisms of length-dependent recognition of viral double-stranded RNA by RIG-I. Sci. Rep. 2023, 13, 6318. [Google Scholar] [CrossRef]
- Cornelissen, J.B.W.J.; Post, J.; Peeters, B.; Vervelde, L.; Rebel, J.M.J. Differential innate responses of chickens and ducks to low-pathogenic avian influenza. Avian Pathol. 2012, 41, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Watanabe, C.; Suzuki, Y.; Tanikawa, T.; Uchida, Y.; Saito, T. Chicken MDA5 senses short double-stranded RNA with implications for antiviral response against avian influenza viruses in chicken. J. Innate Immun. 2013, 6, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, J.B.W.J.; Vervelde, L.; Post, J.; Rebel, J.M.J. Differences in highly pathogenic avian influenza viral pathogenesis and associated early inflammatory response in chickens and ducks. Avian Pathol. 2013, 42, 347–364. [Google Scholar] [CrossRef]
- Boone, A.C.; Kulkarni, R.R.; Cortes, A.L.; Villalobos, T.; Esandi, J.; Gimeno, I.M. In ovo HVT vaccination enhances cellular responses at hatch and addition of poly I:C offers minimal adjuvant effects. Vaccine 2023, 41, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Klasing, K.C. Avian macrophages: Regulators of local and systemic immune responses. Poult. Sci. 1998, 77, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Ciraci, C.; Tuggle, C.K.; Wannemuehler, M.J.; Nettleton, D.; Lamont, S.J. Unique genome-wide transcriptome profiles of chicken macrophages exposed to Salmonella-derived endotoxin. BMC Genom. 2010, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Barchet, W.; Gilfillan, S.; Cella, M.; Beutler, B.; Flavell, R.A.; Diamond, M.S.; Colonna, M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 2006, 103, 8459–8464. [Google Scholar] [CrossRef]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar] [CrossRef]
- Schlee, M.; Hartmann, E.; Coch, C.; Wimmenauer, V.; Janke, M.; Barchet, W.; Hartmann, G. Approaching the RNA ligand for RIG-I? Immunol. Rev. 2008, 227, 66–74. [Google Scholar] [CrossRef]
- Peisley, A.; Jo, M.H.; Lin, C.; Wu, B.; Orme-Johnson, M.; Walz, T.; Hohng, S.; Hur, S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. USA 2012, 109, E3340–E3349. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Kothlow, S.; Schneider, P.; Tardivel, A.; Göbel, T.; Kaspers, B.; Staeheli, P. Chicken BAFF--a highly conserved cytokine that mediates B cell survival. Int. Immunol. 2004, 16, 139–148. [Google Scholar] [CrossRef]
- Kothlow, S.; Schenk-Weibhauser, K.; Ratcliffe, M.J.; Kaspers, B. Prolonged effect of BAFF on chicken B cell development revealed by RCAS retroviral gene transfer in vivo. Mol. Immunol. 2010, 47, 1619–1628. [Google Scholar] [CrossRef]
- Zheng, Y.; Gallucci, S.; Gaughan, J.P.; Gross, J.A.; Monestier, M. A role for B cell-activating factor of the TNF family in chemically induced autoimmunity. J. Immunol. 2005, 175, 6163–6168. [Google Scholar] [CrossRef]
- Biscarini, F.; Bovenhuis, H.; van Arendonk, J.A.M.; Parmentier, H.K.; Jungerius, A.P.; van der Poel, J.J. Across-line SNP association study of innate and adaptive immune response in laying hens. Anim. Genet. 2010, 41, 26–38. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, J.; Dai, D.; Liu, J.; Xu, J.; Miao, X.; Wang, H.; Mao, C.; Xiao, Y. Poly IC pretreatment suppresses B cell-mediated lupus-like autoimmunity through induction of Peli1. Acta Biochim. Biophys. Sin. 2018, 50, 862–868. [Google Scholar] [CrossRef]
- Caskey, M.; Lefebvre, F.; Filali-Mouhim, A.; Cameron, M.J.; Goulet, J.-P.; Haddad, E.K.; Breton, G.; Trumpfheller, C.; Pollak, S.; Shimeliovich, I.; et al. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J. Exp. Med. 2011, 208, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Nagato, T.; Lee, Y.-R.; Harabuchi, Y.; Celis, E. Combinatorial immunotherapy of polyinosinic-polycytidylic acid and blockade of programmed death-ligand 1 induce effective CD8 T-cell responses against established tumors. Clin. Cancer Res. 2014, 20, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Zinzula, L.; Tramontano, E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: Hide, mask, hit. Antivir. Res. 2013, 100, 615–635. [Google Scholar] [CrossRef] [PubMed]

| Cytokine Gene | Fold Increase in mRNA Expression Following Treatment | ||
|---|---|---|---|
| LMW Poly(I:C) | HMW Poly(I:C) | LMW + HMW Poly(I:C) | |
| CCL1 | <4 | 34 | 11 |
| CCL4 | 11 | 34 | 28 |
| CCL5 | <4 | 4 | 6 |
| CSF3 | 6 | 74 | 25 |
| IFNG | <4 | 10 | 8 |
| IL1B | <4 | 19 | 9 |
| IL8 | <4 | 16 | 10 |
| IL6 | <4 | 7 | <4 |
| IL12B | <4 | 4 | <4 |
| CSF2 | <4 | 5 | <4 |
| TNSF13B (BAFF) | <4 | 10 | <4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abo-Samaha, M.I.; Sharaf, M.M.; El-Nahas, A.F.; Odemuyiwa, S.O. Length-Dependent Modulation of B Cell Activating Factor Transcripts in Chicken Macrophage by Viral Double-Stranded RNA. Vaccines 2023, 11, 1561. https://doi.org/10.3390/vaccines11101561
Abo-Samaha MI, Sharaf MM, El-Nahas AF, Odemuyiwa SO. Length-Dependent Modulation of B Cell Activating Factor Transcripts in Chicken Macrophage by Viral Double-Stranded RNA. Vaccines. 2023; 11(10):1561. https://doi.org/10.3390/vaccines11101561
Chicago/Turabian StyleAbo-Samaha, Magda I., Mohammed M. Sharaf, Abeer F. El-Nahas, and Solomon O. Odemuyiwa. 2023. "Length-Dependent Modulation of B Cell Activating Factor Transcripts in Chicken Macrophage by Viral Double-Stranded RNA" Vaccines 11, no. 10: 1561. https://doi.org/10.3390/vaccines11101561
APA StyleAbo-Samaha, M. I., Sharaf, M. M., El-Nahas, A. F., & Odemuyiwa, S. O. (2023). Length-Dependent Modulation of B Cell Activating Factor Transcripts in Chicken Macrophage by Viral Double-Stranded RNA. Vaccines, 11(10), 1561. https://doi.org/10.3390/vaccines11101561






