VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dot Blot Assay
2.2. NTA Analysis
2.3. Quantification of S, E, and M by ELISA
2.4. Production of PRAK-03202 at 5 Liter Scale
- (i.)
- Upstream processing
- (ii.)
- Downstream processing
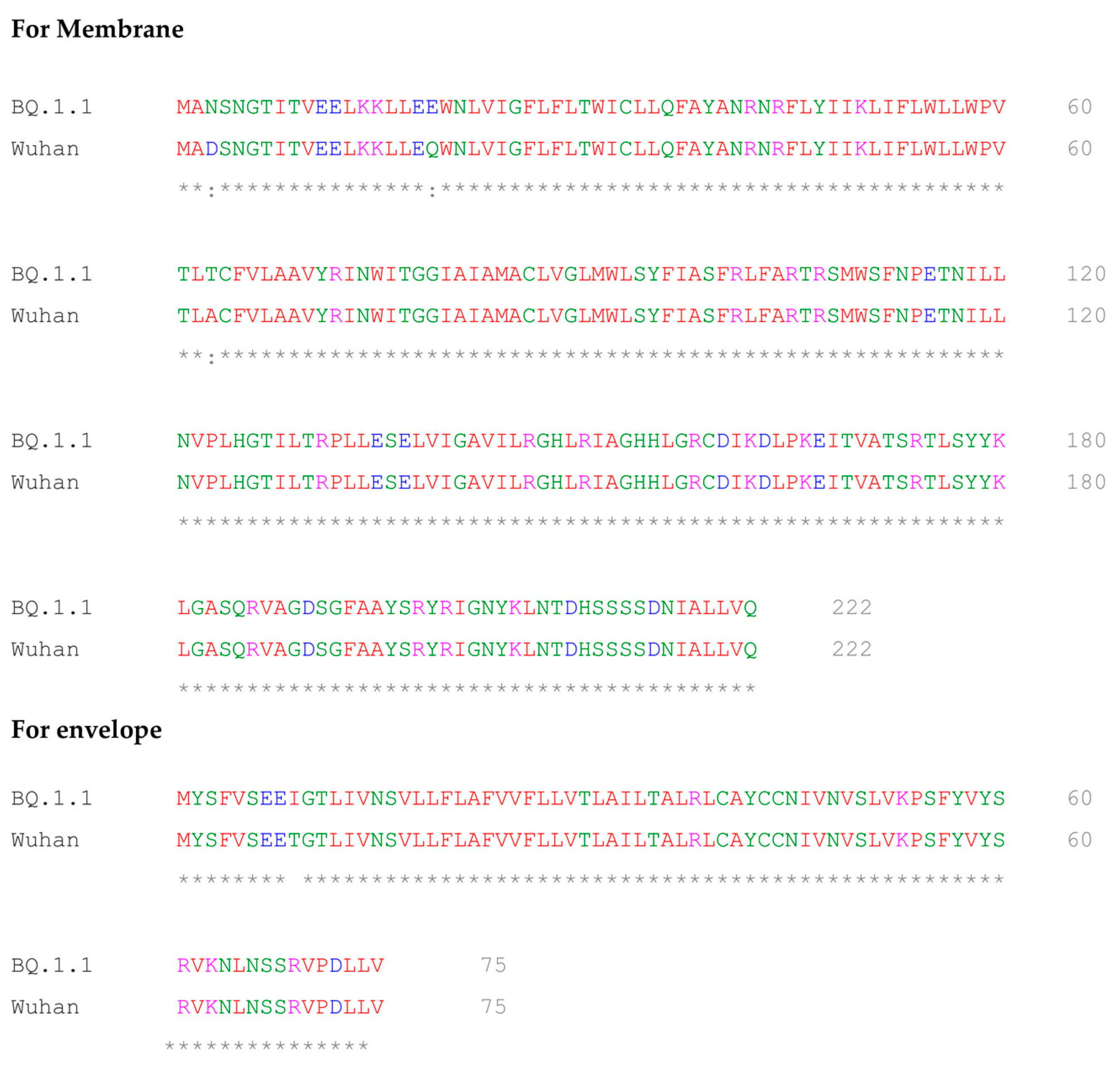
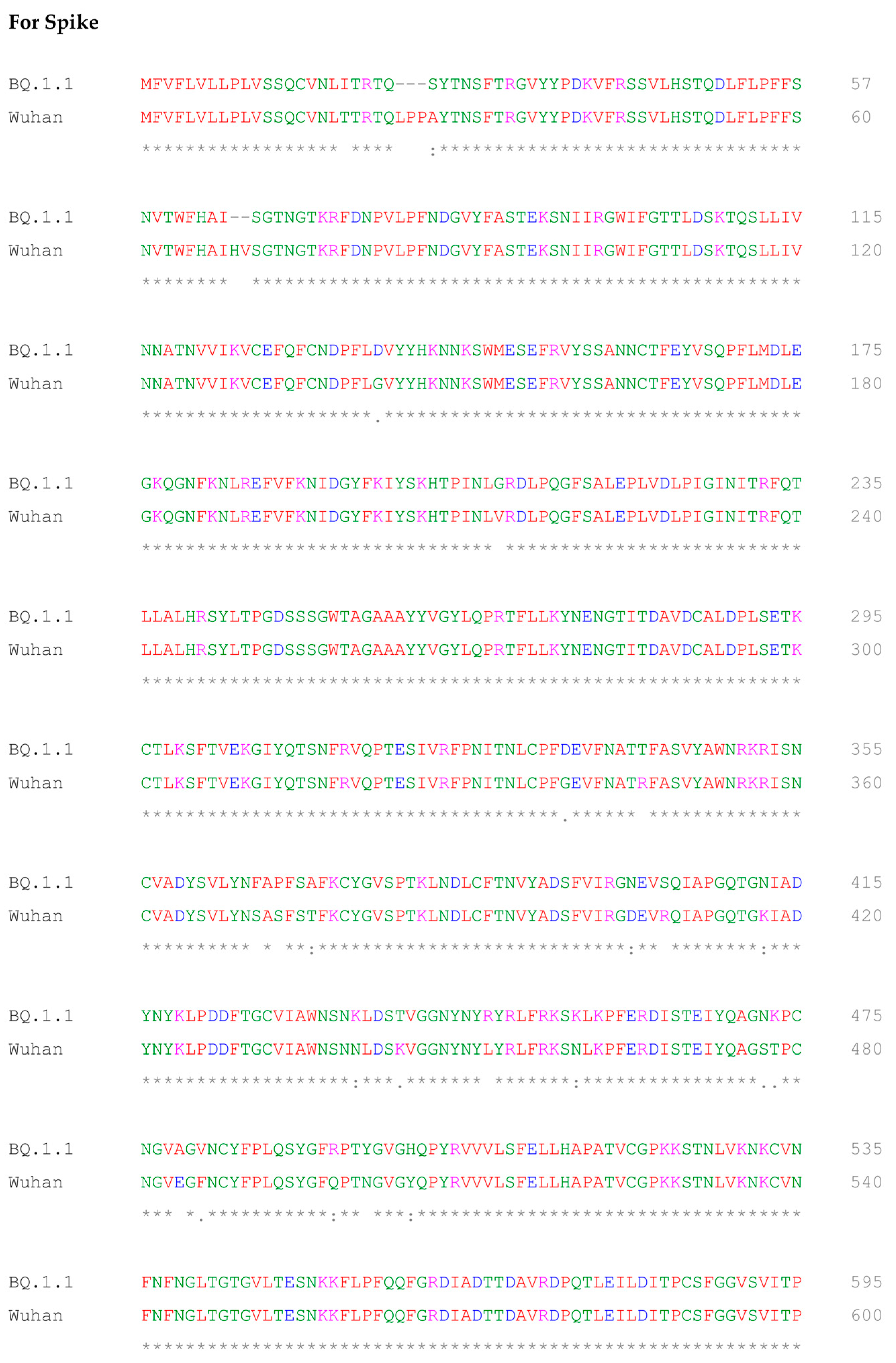
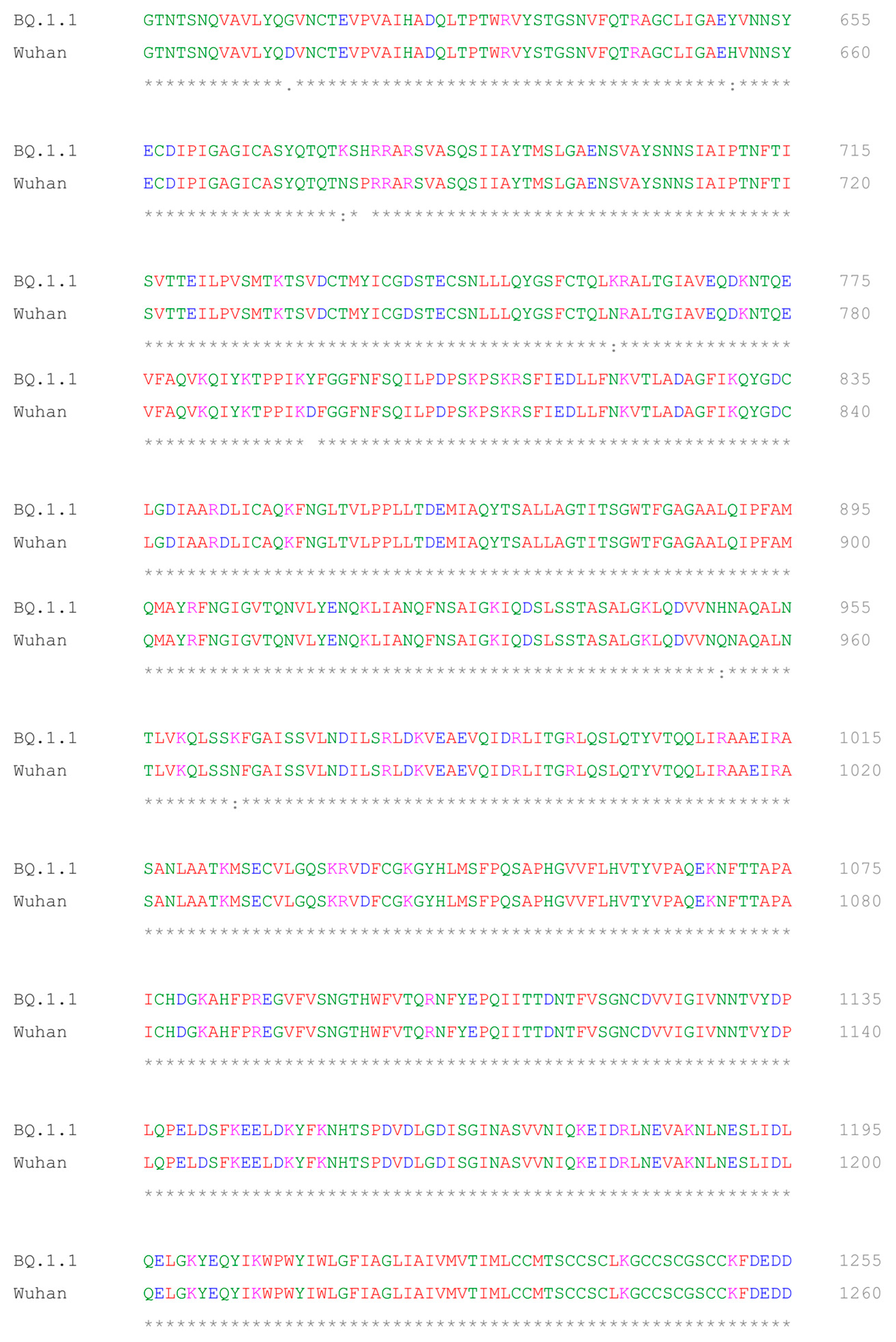
2.5. Multiple Sequence Alignment of BQ.1.1 and Wuhan Variant of SARS-CoV-2
2.6. Human Serum
2.7. ELISA
2.8. Calculation of Cut-Off, Sensitivity, Specificity, and Accuracy
3. Results
3.1. PRAK-03202 Production and Characterization
Dot Blot Assay and Particle Analysis of PRAK-03202
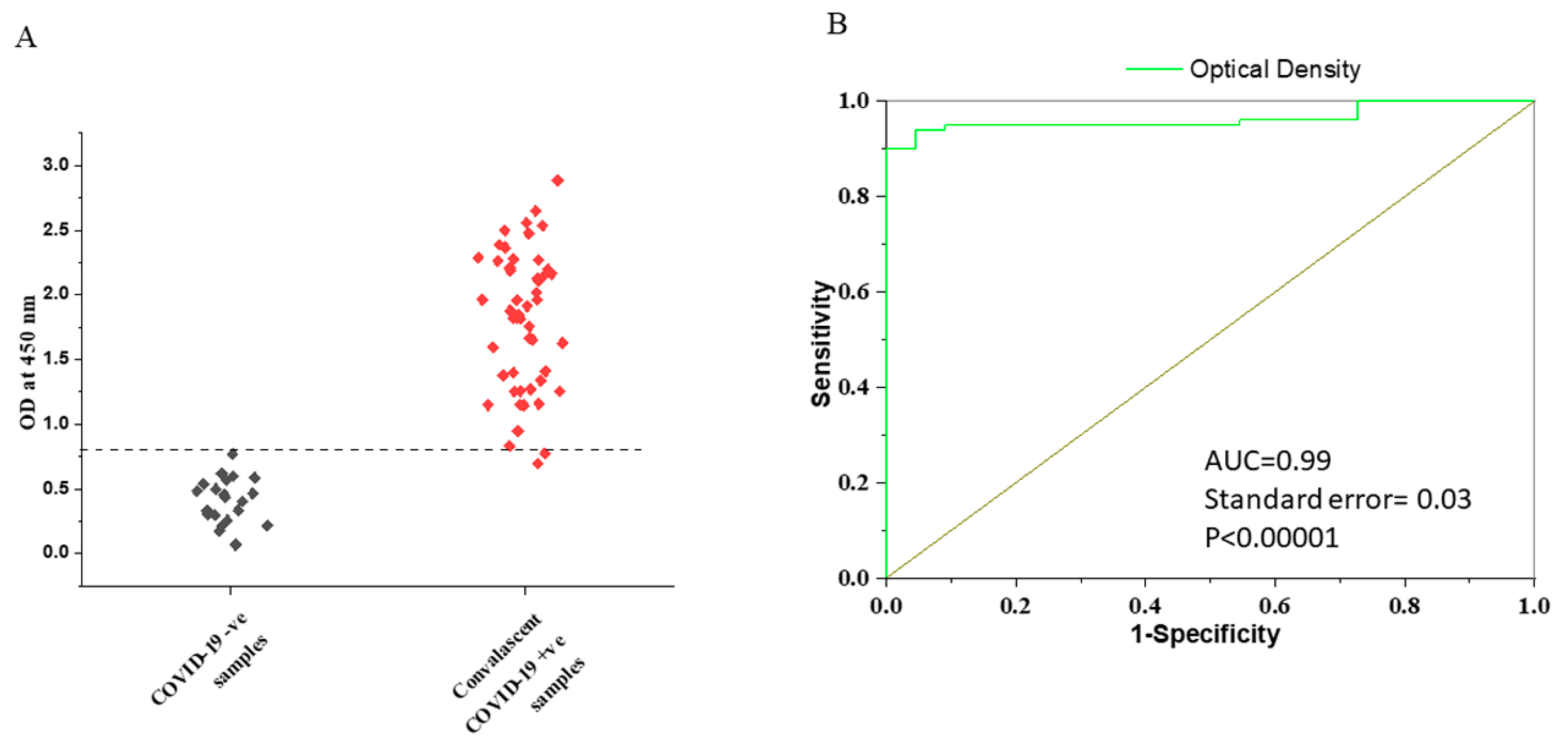
3.2. Determination of the Cut-Off Value for VLP-ELISA Assay
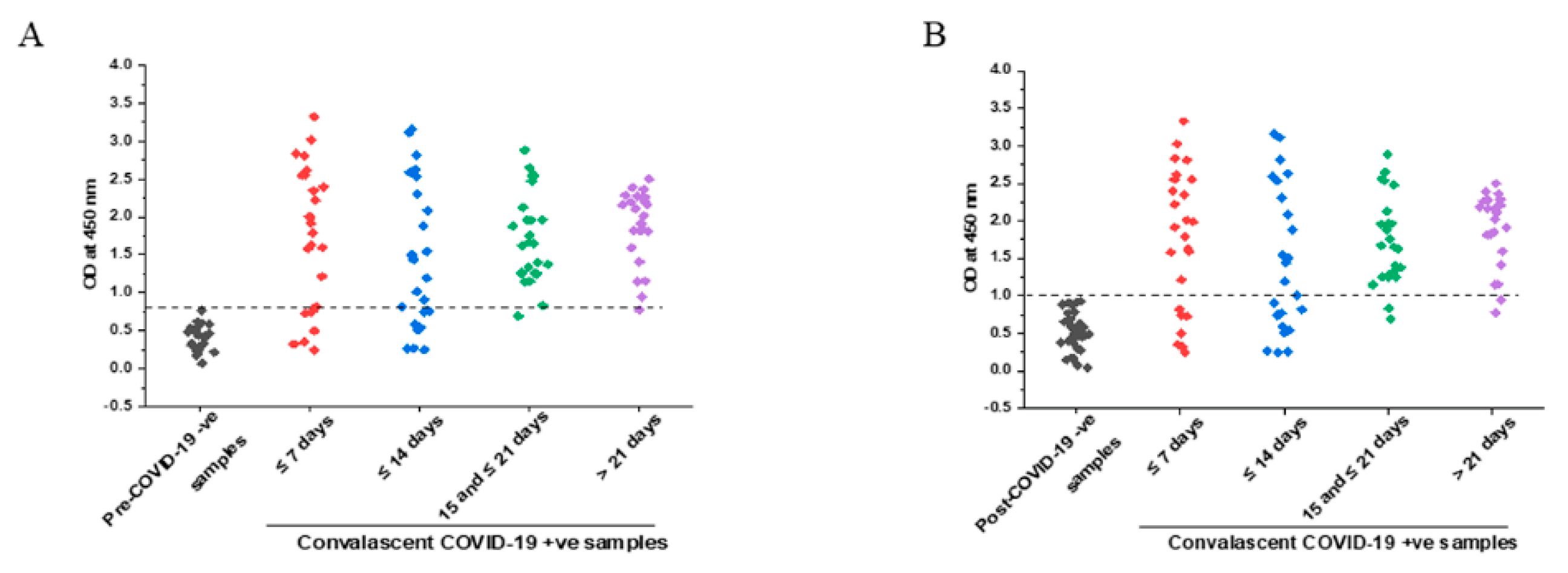
3.3. IgG Response with Respect to Time after Symptoms Onset
3.4. Comparison of Specificity in Pre- and Post-COVID Negative Controls
3.5. Comparison of IgG Titers in Early and Later Phases of COVID-19 Symptoms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saluzzo, F.; Mantegani, P.; Poletti De Chaurand, V.; Quaresima, V.; Cugnata, F.; Di Serio, C.; Macé, A.; De Vos, M.; Sacks, J.A.; Cirillo, D.M. SARS-CoV-2 Antibody Rapid Tests: Valuable Epidemiological Tools in Challenging Settings. Microbiol. Spectr. 2021, 9, e0025021. [Google Scholar] [CrossRef] [PubMed]
- Naushin, S.; Sardana, V.; Ujjainiya, R.; Bhatheja, N.; Kutum, R.; Bhaskar, A.K.; Pradhan, S.; Prakash, S.; Khan, R.; Rawat, B.S.; et al. Insights from a Pan India Sero-Epidemiological survey (Phenome-India Cohort) for SARS-CoV2. Elife 2021, 10, e66537. [Google Scholar] [CrossRef] [PubMed]
- Espejo, A.P.; Akgun, Y.; Al Mana, A.; Tjendra, Y.; Millan, N.C.; Gomez-Fernandez, C.; Cray, C. Review of Current Advances in Serologic Testing for COVID-19. Am. J. Clin. Pathol. 2020, 154, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Ichinose, N.; Okada, Y. False-negative rate of SARS-CoV-2 RT-PCR tests and its relationship to test timing and illness severity: A case series. IDCases 2022, 28, e01496. [Google Scholar] [CrossRef] [PubMed]
- Jalali Nadoushan, M.; Ahmadi, S.; Jalali Nadoushan, P. Serology Testing for SARS-CoV-2: Benefits and Challenges. Iran. J. Pathol. 2020, 15, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Bellizzi, S.; Alsawalha, L.; Ali, S.S.; Sharkas, G.; Muthu, N.; Ghazo, M.; Aly, E.; Rashidian, A.; Al Ariqi, L.; Hayajneh, W.; et al. A three-phase population based sero-epidemiological study: Assessing the trend in prevalence of SARS-CoV-2 during COVID-19 pandemic in Jordan. One Health 2021, 13, 100292. [Google Scholar] [CrossRef]
- Ayoub, H.H.; Chemaitelly, H.; Makhoul, M.; Al Kanaani, Z.; Al Kuwari, E.; Butt, A.A.; Coyle, P.; Jeremijenko, A.; Kaleeckal, A.H.; Latif, A.N.; et al. Epidemiological impact of prioritising SARS-CoV-2 vaccination by antibody status: Mathematical modelling analyses. BMJ Innov. 2021, 7, 327–336. [Google Scholar] [CrossRef]
- Ayoub, H.H.; Chemaitelly, H.; Makhoul, M.; Al Kanaani, Z.; Al Kuwari, E.; Butt, A.A.; Coyle, P.; Jeremijenko, A.; Kaleeckal, A.H.; Latif, A.N.; et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020, 129, 104512. [Google Scholar]
- Van Elslande, J.; Decru, B.; Jonckheere, S.; Van Wijngaerden, E.; Houben, E.; Vandecandelaere, P.; Indevuyst, C.; Depypere, M.; Desmet, S.; André, E.; et al. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin. Microbiol. Infect. 2020, 26, 1557.e1–1557.e7. [Google Scholar] [CrossRef]
- Theel, E.S.; Harring, J.; Hilgart, H.; Granger, D. Performance Characteristics of Four High-Throughput Immunoassays for Detection of IgG Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01243-20. [Google Scholar] [CrossRef]
- Perera, R.A.; Tso, E.; Tsang, O.T.; Tsang, D.N.; Fung, K.; Leung, Y.W.; Chin, A.W.; Chu, D.K.; Cheng, S.M.; Poon, L.L.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Eurosurveillance 2020, 25, 2000421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayouba, A.; Thaurignac, G.; Morquin, D.; Tuaillon, E.; Raulino, R.; Nkuba, A.; Lacroix, A.; Vidal, N.; Foulongne, V.; Le Moing, V.; et al. Multiplex detection and dynamics of IgG antibodies to SARS-CoV2 and the highly pathogenic human coronaviruses SARS-CoV and MERS-CoV. J. Clin. Virol. 2020, 129, 104521. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Heslan, C.; Jégou, G.; Eriksson, L.A.; Le Gallo, M.; Thibault, V.; Chevet, E.; Godey, F.; Avril, T. SARS-CoV-2 integral membrane proteins shape the serological responses of patients with COVID-19. iScience 2021, 24, 103185. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.K.; Hegde, S.T. The important role of serology for COVID-19 control. Lancet Infect. Dis. 2020, 20, 758–759. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M. SARS-CoV-2 antibodies titration: A reappraisal. Ann. Transl. Med. 2020, 8, 1032. [Google Scholar] [CrossRef]
- Gillot, C.; Douxfils, J.; Cadrobbi, J.; Laffineur, K.; Dogné, J.M.; Elsen, M.; Eucher, C.; Melchionda, S.; Modaffarri, É.; Tré-Hardy, M. An Original ELISA-Based Multiplex Method for the Simultaneous Detection of 5 SARS-CoV-2 IgG Antibodies Directed against Different Antigens. J. Clin. Med. 2020, 9, 3752. [Google Scholar] [CrossRef]
- Osterman, A.; Baldauf, H.M.; Eletreby, M.; Wettengel, J.M.; Afridi, S.Q.; Fuchs, T.; Holzmann, E.; Maier, A.; Döring, J.; Grzimek-Koschewa, N.; et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Med. Microbiol. Immunol. 2021, 210, 65–72. [Google Scholar] [CrossRef]
- Fox, T.; Geppert, J.; Dinnes, J.; Scandrett, K.; Bigio, J.; Sulis, G.; Hettiarachchi, D.; Mathangasinghe, Y.; Weeratunga, P.; Wickramasinghe, D.; et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2022, 11, CD013652. [Google Scholar]
- Jörrißen, P.; Schütz, P.; Weiand, M.; Vollenberg, R.; Schrempf, I.M.; Ochs, K.; Frömmel, C.; Tepasse, P.R.; Schmidt, H.; Zibert, A. Antibody Response to SARS-CoV-2 Membrane Protein in Patients of the Acute and Convalescent Phase of COVID-19. Front. Immunol. 2021, 12, 679841. [Google Scholar] [CrossRef]
- Szymczak, A.; Jędruchniewicz, N.; Torelli, A.; Kaczmarzyk-Radka, A.; Coluccio, R.; Kłak, M.; Konieczny, A.; Ferenc, S.; Witkiewicz, W.; Montomoli, E.; et al. Antibodies specific to SARS-CoV-2 proteins N, S and E in COVID-19 patients in the normal population and in historical samples. J. Gen. Virol. 2021, 102, 001692. [Google Scholar] [CrossRef]
- van Tol, S.; Mögling, R.; Li, W.; Godeke, G.J.; Swart, A.; Bergmans, B.; Brandenburg, A.; Kremer, K.; Murk, J.L.; van Beek, J.; et al. Accurate serology for SARS-CoV-2 and common human coronaviruses using a multiplex approach. Emerg. Microbes Infect. 2020, 9, 1965–1973. [Google Scholar] [CrossRef]
- Mazumder, S.; Rastogi, R.; Undale, A.; Arora, K.; Arora, N.M.; Pratim, B.; Kumar, D.; Joseph, A.; Mali, B.; Arya, V.B.; et al. PRAK-03202: A triple antigen virus-like particle vaccine candidate against SARS CoV-2. Heliyon 2021, 7, e08124. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Roessler, J.; Pich, D.; Albanese, M.; Wratil, P.R.; Krähling, V.; Hellmuth, J.C.; Scherer, C.; von Bergwelt-Baildon, M.; Becker, S.; Keppler, O.T.; et al. Quantitation of SARS-CoV-2 neutralizing antibodies with a virus-free, authentic test. PNAS Nexus 2022, 1, pgac045. [Google Scholar] [CrossRef] [PubMed]
- Munitz, A.; Edry-Botzer, L.; Itan, M.; Tur-Kaspa, R.; Dicker, D.; Marcoviciu, D.; Goren, M.G.; Mor, M.; Lev, S.; Gottesman, T.; et al. Rapid seroconversion and persistent functional IgG antibodies in severe COVID-19 patients correlates with an IL-12p70 and IL-33 signature. Sci. Rep. 2021, 11, 3461. [Google Scholar] [CrossRef] [PubMed]
- Döhla, M.; Boesecke, C.; Schulte, B.; Diegmann, C.; Sib, E.; Richter, E.; Eschbach-Bludau, M.; Aldabbagh, S.; Marx, B.; Eis-Hübinger, A.M.; et al. Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public. Health 2020, 182, 170–172. [Google Scholar] [CrossRef]
- Hoffman, T.; Nissen, K.; Krambrich, J.; Rönnberg, B.; Akaberi, D.; Esmaeilzadeh, M.; Salaneck, E.; Lindahl, J.; Lundkvist, Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect. Ecol. Epidemiol. 2020, 10, 1754538. [Google Scholar] [CrossRef] [Green Version]
- Tré-Hardy, M.; Wilmet, A.; Beukinga, I.; Dogné, J.M.; Douxfils, J.; Blairon, L. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020, 92, 1724–1727. [Google Scholar]
- Tré-Hardy, M.; Wilmet, A.; Beukinga, I.; Dogné, J.M.; Douxfils, J.; Blairon, L. Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin. Chem. Lab. Med. 2020, 58, 1357–1364. [Google Scholar] [CrossRef]
- Stock da Cunha, T.; Goma-Garces, E.; Avello, A.; Pereira-Garcia, M.; Mas-Fontao, S.; Ortiz, A.; Gonzalez-Parra, E. The Spectrum of Clinical and Serological Features of COVID-19 in Urban Hemodialysis Patients. J. Clin. Med. 2020, 9, 2264. [Google Scholar] [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef] [PubMed]
- Mardian, Y.; Kosasih, H.; Karyana, M.; Neal, A.; Lau, C.-Y. Review of Current COVID-19 Diagnostics and Opportunities for Further Development. Front. Med. 2021, 8, 615099. [Google Scholar] [CrossRef] [PubMed]
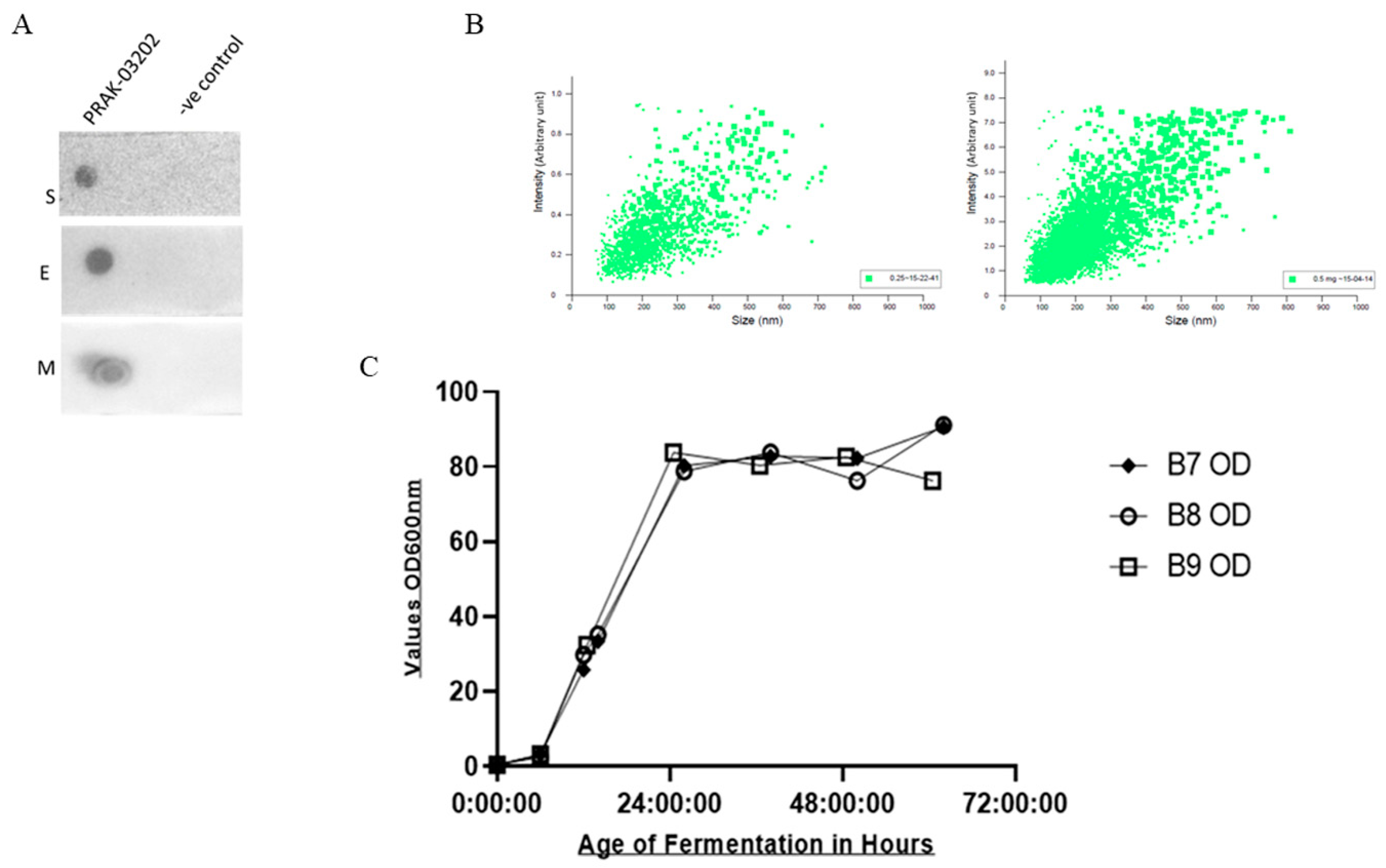

| Antibody | Amount of Protein (ng) | Mol. Wt. (kDa) | Number of Molecules |
|---|---|---|---|
| RBD | 105 ± 16 | 420 (trimer) | 1.4 × 1011 ± 0.32 |
| E-specific | 10.25 ± 2.12 | 54 (pentamer) | 1.1 × 1011 ± 0.3 |
| M-specific | 62.5 ± 1 | 25 | 1.51 × 1012 ± 0.05 |
| SN | Test | Method | Specification |
|---|---|---|---|
| 1 | Appearance | Visual | Whitish suspension to clear liquid, free from foreign particles, and in which the mineral carrier tends to settle down on keeping |
| 2 | pH | pH meter | 6.5–7.5 |
| 3 | Total protein content | Bradford | Not less than (NLT) 10 µg/dose |
| 4 | Identity test | ELISA | Positive for S, M and E antibodies |
| 5 | Purity | Size exclusion chromatography–HPLC | >98% Pure |
| 6 | Endotoxin | Kinetic chromogenic method | Not more than (NMT) 10 EU/µg |
| 7 | Sterility | Membrane Filtration | No evidence of microbial growth |
| 8 | Abnormal toxicity test | Test method as per IP 2018 | None of the animals die or show signs of ill health in 7 days following injection |
| 9 | Extractable volume | By pipetting | Not less than nominal volume of 0.5 mL |
| 10 | Sucrose estimation | Enzymatic method | NMT 15% w/v |
| 11 | Aluminum content (Al3+) | ICP-MS | NMT 800 µg/dose |
| Days after COVID-19 Symptoms Onset (Pre-COVID Era) | ||||
| PRAK-03202 | 0–7 Days (95% CI) | 8–14 Days (95% CI) | 15–21 Days (95% CI) | ≥22 Days (95% CI) |
| True-positive patients (n = 25/each group) | 18 | 15 | 24 | 24 |
| True-negative (n = 23) | 22 | |||
| Sensitivity | 72% (51–88%) | 60% (38–78%) | 96% (80–99%) | 96% (80–99%) |
| Specificity | 96% (78–99%) | 96% (78–99%) | 96% (78–99%) | 96% (78–99%) |
| Accuracy | 83% (70–92%) | 77% (63–88%) | 97% (86–99%) | 97% (86–99%) |
| Positive predictive value | 94% (72–99%) | 94% (68–99%) | 96% (78–99%) | 96% (78–99%) |
| Negative predictive value | 76% (62–86%) | 69% (57–78%) | 97% (76–99%) | 97% (76–99%) |
| Days after COVID-19 Symptoms Onset (Post-COVID Era) | ||||
| PRAK-03202 | 0–7 Days (95% CI) | 8–14 Days (95% CI) | 15–21 Days (95% CI) | ≥22 Days (95% CI) |
| True-negative (n = 32) | 32 | |||
| Sensitivity | 72% (51–88%) | 60% (38–78%) | 92% (89–100%) | 92% (89–100%) |
| Specificity | 100% (89–100%) | 100% (89–100%) | 100% (74–99%) | 100% (74–99%) |
| Accuracy | 88% (76–94%) | 82% (70–91%) | 96% (88–98%) | 96% (88–98%) |
| Positive predictive value | 100% | 100% | 100% | 100% |
| Negative predictive value | 82% (76–94%) | 76% (66–84%) | 94% (80–98%) | 94% (80–98%) |
| Serial Number | Sample ID | Time Since Symptom Onset | IgG Reciprocal Titer PRAK-03202 |
|---|---|---|---|
| 1. | A1002 | ≤14 Days | 50,000 |
| 2. | A1003 | ≤14 Days | 10,000 |
| 3. | A1004 | ≤14 Days | 10,000 |
| 4. | A1005 | ≤14 Days | 10,000 |
| 5. | A1007 | ≤14 Days | 50,000 |
| 6. | A1012 | ≤14 Days | 10,000 |
| 7. | A1021 | ≤14 Days | 10,000 |
| 8. | A1030 | ≤14 Days | 10,000 |
| 9. | A1039 | ≤14 Days | 10,000 |
| 10. | A1041 | ≤14 Days | 10,000 |
| 11. | A1045 | ≤14 Days | 10,000 |
| 12. | A1051 | >14 Days | 10,000 |
| 13. | A1052 | >14 Days | 10,000 |
| 14. | A1075 | >14 Days | 50,000 |
| 15. | A1076 | >14 Days | 50,000 |
| 16. | A1078 | >14 Days | 50,000 |
| 17. | A1079 | >14 Days | 10,000 |
| 18. | A1091 | >14 Days | 50,000 |
| 19. | A1092 | >14 Days | 50,000 |
| 20. | A1093 | >14 Days | 50,000 |
| 21. | A1094 | >14 Days | 10,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, D.; Roy, S.S.; Rastogi, R.; Arora, K.; Undale, A.; Gupta, R.; Arora, N.M.; Kundu, P.K. VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population. Vaccines 2023, 11, 743. https://doi.org/10.3390/vaccines11040743
Kumar D, Roy SS, Rastogi R, Arora K, Undale A, Gupta R, Arora NM, Kundu PK. VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population. Vaccines. 2023; 11(4):743. https://doi.org/10.3390/vaccines11040743
Chicago/Turabian StyleKumar, Dilip, Sourav Singha Roy, Ruchir Rastogi, Kajal Arora, Avinash Undale, Reeshu Gupta, Nupur Mehrotra Arora, and Prabuddha K. Kundu. 2023. "VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population" Vaccines 11, no. 4: 743. https://doi.org/10.3390/vaccines11040743
APA StyleKumar, D., Roy, S. S., Rastogi, R., Arora, K., Undale, A., Gupta, R., Arora, N. M., & Kundu, P. K. (2023). VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population. Vaccines, 11(4), 743. https://doi.org/10.3390/vaccines11040743





