Mycobacterium smegmatis, a Promising Vaccine Vector for Preventing TB and Other Diseases: Vaccinomics Insights and Applications
Abstract
1. Introduction:
2. System Biology Understanding of M.sm
3. Why Is M.sm an Effective Vaccine Vector for Development of Recombinant Mycobacterium Vaccines?
3.1. Omics-Based Antigen Discovery in Mycobacterium sp.
3.2. Preliminary Application of M.sm Vector Vaccine
4. Broad Spectrum Applications of M.sm Vector Vaccines
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- McCune, R.M.; Feldmann, F.M.; Lambert, H.P.; McDermott, W. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J. Exp. Med. 1966, 123, 445–468. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.E.; Smith, M.M. Rapidly growing, acid fast bacteria. I. Species’ descriptions of Mycobacterium phlei Lehmann and Neumann and M. sm (Trevisan) Lehmann and Neumann. J. Bacteriol. 1953, 66, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, E.K.; de Souza, N.; Santos, D.S.; Blanchard, J.S.; Basso, L.A. Drugs that inhibit mycolic acid biosynthesis in M. tb. Curr. Pharm. Biotechnol. 2002, 3, 197–225. [Google Scholar] [CrossRef] [PubMed]
- Ideker, T. Systems biology 101—What you need to know. Nat. Biotechnol. 2004, 22, 473–475. [Google Scholar] [CrossRef]
- Veenstra, T.D. Omics in Systems Biology: Current Progress and Future Outlook. Proteomics 2021, 21, 2000235. [Google Scholar] [CrossRef]
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef]
- Tavassoly, I.; Goldfarb, J.; Iyengar, R. Systems biology primer: The basic methods and approaches. Essays Biochem. 2018, 62, EBC20180003. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; van Riet, E.; Meiring, H.D.; Metz, B.; Kersten, G.F.A. Systems vaccinology and big data in the vaccine development chain. Immunology 2019, 156, 33–46. [Google Scholar] [CrossRef]
- Russo, G.; Reche, P.; Pennisi, M.; Pappalardo, F. The combination of artificial intelligence and systems biology for intelligent vaccine design. Expert. Opin. Drug Discov. 2020, 15, 1267–1281. [Google Scholar] [CrossRef]
- Bercovier, H.; Vincent, V. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. sm, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. Rev. Sci. Tech. Int. Off. Epizoot. 2001, 20, 265–290. [Google Scholar] [CrossRef]
- Pelicic, V.; Reyrat, J.M.; Gicquel, B. Genetic advances for studying M. tb pathogenicity. Mol. Microbiol. 1998, 28, 413–420. [Google Scholar] [CrossRef]
- Bange, F.C.; Collins, F.M.; Jacobs, W.R. Survival of mice infected with M. sm containing large DNA fragments from M. tb. Tuber. Lung Dis. 1999, 79, 171–180. [Google Scholar] [CrossRef]
- Lagier, B.; Pelicic, V.; Lecossier, D.; Prod’Hom, G.; Rauzier, J.; Guilhot, C.; Gicquel, B.; Hance, A.J. Identification of genetic loci implicated in the survival of M. sm in human mononuclear phagocytes. Mol. Microbiol. 1998, 29, 465–475. [Google Scholar] [CrossRef]
- Judd, J.A.; Canestrari, J.; Clark, R.; Joseph, A.; Lapierre, P.; Lasek-Nesselquist, E.; Mir, M.; Palumbo, M.; Smith, C.; Stone, M.; et al. A Mycobacterial Systems Resource for the Research Community. mBio 2021, 12, e02401–e02420. [Google Scholar] [CrossRef]
- Malhotra, S.; Vedithi, S.C.; Blundell, T.L. Decoding the similarities and differences among mycobacterial species. PLoS Negl. Trop. Dis. 2017, 11, e0005883. [Google Scholar] [CrossRef]
- Dragset, M.S.; Ioerger, T.R.; Zhang, Y.J.; Mærk, M.; Ginbot, Z.; Sacchettini, J.C.; Flo, T.H.; Rubin, E.J.; Steigedal, M. Genome-wide Phenotypic Profiling Identifies and Categorizes Genes Required for Mycobacterial Low Iron Fitness. Sci. Rep. 2019, 9, 11394. [Google Scholar] [CrossRef]
- Tyagi, J.S.; Sharma, D.M. M. sm and tuberculosis. Trends Microbiol. 2002, 10, 68–69. [Google Scholar] [CrossRef]
- Wu, Q.L.; Kong, D.; Lam, K.; Husson, R.N. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 1997, 179, 2922–2929. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- Li, X.; Mei, H.; Chen, F.; Tang, Q.; Yu, Z.; Cao, X.; Andongma, B.T.; Chou, S.-H.; He, J. Transcriptome Landscape of M. sm. Front. Microbiol. 2017, 8, 2505. [Google Scholar] [CrossRef] [PubMed]
- Maarsingh, J.D.; Yang, S.; Park, J.G.; Haydel, S.E. Comparative transcriptomics reveals PrrAB-mediated control of metabolic, respiration, energy-generating, and dormancy pathways in M. sm. BMC Genom. 2019, 20, 942. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Xu, M.; Wang, R.; Li, Q.; Yu, Z.; Xie, J. Overexpression of Rv2788 increases mycobacterium stresses survival. Microbiol. Res. 2017, 195, 51–59. [Google Scholar] [CrossRef]
- Zhu, J.; Wolf, I.D.; Dulberger, C.L.; Won, H.I.; Kester, J.C.; Judd, J.A.; Wirth, S.E.; Clark, R.R.; Li, Y.; Luo, Y.; et al. Spatiotemporal localization of proteins in mycobacteria. Cell Rep. 2021, 37, 110154. [Google Scholar] [CrossRef] [PubMed]
- De Wet, T.J.; Winkler, K.R.; Mhlanga, M.; Mizrahi, V.; Warner, D.F. Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes. eLife 2020, 9, e60083. [Google Scholar] [CrossRef]
- Faludi, I.; Szabó, A.M.; Burián, K.; Endrész, V.; Miczák, A. Recombinant M. sm vaccine candidates. Acta Microbiol. Immunol. Hung. 2011, 58, 13–22. [Google Scholar] [CrossRef]
- Long, Q.; Zhou, Q.; Ji, L.; Wu, J.; Wang, W.; Xie, J.M. M. sm genomic characteristics associated with its saprophyte lifestyle. J. Cell Biochem. 2012, 113, 3051–3055. [Google Scholar] [CrossRef]
- Houben, E.N.G.; Korotkov, K.V.; Bitter, W. Take five—Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta BBA Mol. Cell Res. 2014, 1843, 1707–1716. [Google Scholar] [CrossRef]
- Abdallah, A.M.; Van Pittius, N.C.; Champion, P.A.; Cox, J.; Luirink, J.; Vandenbroucke-Grauls, C.M.; Appelmelk, B.J.; Bitter, W. Type VII secretion—Mycobacteria show the way. Nat. Rev. Microbiol. 2007, 5, 883–891. [Google Scholar] [CrossRef]
- Lagune, M.; Petit, C.; Sotomayor, F.V.; Johansen, M.D.; Beckham, K.S.H.; Ritter, C.; Girard-Misguich, F.; Wilmanns, M.; Kremer, L.; Maurer, F.P.; et al. Conserved and specialized functions of Type VII secretion systems in non-tuberculous mycobacteria. Microbiology 2021, 167, 001054. [Google Scholar] [CrossRef]
- Coros, A.; Callahan, B.; Battaglioli, E.; Derbyshire, K.M. The specialized secretory apparatus ESX-1 is essential for DNA transfer in M. sm. Mol. Microbiol. 2008, 69, 794–808. [Google Scholar] [CrossRef]
- Fang, Z.; Newton-Foot, M.; Sampson, S.L.; Gey van Pittius, N.C. Two promoters in the esx-3 gene cluster of M. sm respond inversely to different iron concentrations in vitro. BMC Res. Notes 2017, 10, 426. [Google Scholar] [CrossRef]
- Clark, R.R.; Judd, J.; Lasek-Nesselquist, E.; Montgomery, S.A.; Hoffmann, J.G.; Derbyshire, K.M.; Gray, T.A. Direct cell-cell contact activates SigM to express the ESX-4 secretion system in M. sm. Proc. Natl. Acad. Sci. USA 2018, 115, E6595–E6603. [Google Scholar] [CrossRef]
- Mi, Y.; Bao, L.; Gu, D.; Luo, T.; Sun, C.; Yang, G.M. M. tb PPE25 and PPE26 proteins expressed in M. sm modulate cytokine secretion in mouse macrophages and enhance mycobacterial survival. Res. Microbiol. 2017, 168, 234–243. [Google Scholar] [CrossRef]
- Houben, E.N.G.; Bestebroer, J.; Ummels, R.; Wilson, L.; Piersma, S.R.; Jiménez, C.R.; Ottenhoff, T.H.M.; Luirink, J.; Bitter, W. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 2012, 86, 472–484. [Google Scholar] [CrossRef]
- Bashiri, G.; Baker, E.N. Production of recombinant proteins in M. sm for structural and functional studies. Protein Sci. Publ. Protein Soc. 2015, 24, 1–10. [Google Scholar] [CrossRef]
- Dhiman, N.; Khuller, G.K. Immunoprophylactic properties of 71-kDa cell wall-associated protein antigen of M. tb H37Ra. Med. Microbiol. Immunol. 1997, 186, 45–51. [Google Scholar] [CrossRef]
- Dhiman, N.; Khuller, G.K. Protective efficacy of mycobacterial 71-kDa cell wall associated protein using poly (DL-lactide-co-glycolide) microparticles as carrier vehicles. FEMS Immunol. Med. Microbiol. 1998, 21, 19–28. [Google Scholar] [CrossRef]
- Skeiky, Y.A.W.; Ovendale, P.J.; Jen, S.; Alderson, M.R.; Dillon, D.C.; Smith, S.; Wilson, C.B.; Orme, I.M.; Reed, S.G.; Campos-Neto, A. T cell expression cloning of a M. tb gene encoding a protective antigen associated with the early control of infection. J. Immunol. 2000, 165, 7140–7149. [Google Scholar] [CrossRef]
- Dillon, D.C.; Alderson, M.R.; Day, C.H.; Lewinsohn, D.M.; Coler, R.; Bement, T.; Campos-Neto, A.; Skeiky, Y.A.W.; Orme, I.M.; Roberts, A.; et al. Molecular characterization and human T-cell responses to a member of a novel M. tb mtb39 gene family. Infect. Immun. 1999, 67, 2941–2950. [Google Scholar] [CrossRef]
- Kunnath-Velayudhan, S.; Porcelli, S. Recent Advances in Defining the Immunoproteome of M. tb. Front. Immunol. 2013, 4, 335. [Google Scholar] [CrossRef] [PubMed]
- Arlehamn, C.S.L.; Gerasimova, A.; Mele, F.; Henderson, R.; Swann, J.; Greenbaum, J.A.; Kim, Y.; Sidney, J.; James, E.A.; Taplitz, R.; et al. Memory T cells in latent M. tb infection are directed against three antigenic islands and largely contained in a CXCR3 + CCR6 + Th1 subset. PLoS Pathog. 2013, 9, e1003130. [Google Scholar] [CrossRef]
- Kunnath-Velayudhan, S.; Salamon, H.; Wang, H.-Y.; Davidow, A.L.; Molina, D.M.; Huynh, V.T.; Cirillo, D.M.; Michel, G.; Talbot, E.A.; Perkins, M.D.; et al. Dynamic antibody responses to the M. tb proteome. Proc. Natl. Acad. Sci. USA 2010, 107, 14703–14708. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.S.; Bosma, A.; Chinai, N.; Frost, J.; Jesdale, B.M.; Gonzalez, M.A.; Martin, W.; Saint-Aubin, C. From genome to vaccine: In silico predictions, ex vivo verification. Vaccine 2001, 19, 4385–4395. [Google Scholar] [CrossRef]
- Dockrell, H.M.; Brahmbhatt, S.; Robertson, B.D.; Britton, S.; Fruth, U.; Gebre, N.; Hunegnaw, M.; Hussain, R.; Manandhar, R.; Murillo, L.; et al. A postgenomic approach to identification of Mycobacterium leprae-specific peptides as T-cell reagents. Infect. Immun. 2000, 68, 5846–5855. [Google Scholar] [CrossRef]
- Martin, W.; Sbai, H.; De Groot, A.S. Bioinformatics tools for identifying class I-restricted epitopes. Methods 2003, 29, 289–298. [Google Scholar] [CrossRef]
- De Groot, A.S.; Bishop, E.A.; Khan, B.; Lally, M.; Marcon, L.; Franco, J.; Mayer, K.H.; Carpenter, C.C.; Martin, W. Engineering immunogenic consensus T helper epitopes for a cross-clade HIV vaccine. Methods 2004, 34, 476–487. [Google Scholar] [CrossRef]
- Mollenkopf, H.J.; Grode, L.; Mattow, J.; Stein, M.; Mann, P.; Knapp, B.; Ulmer, J.; Kaufmann, S.H.E. Application of mycobacterial proteomics to vaccine design: Improved protection by Mycobacterium bovis BCG prime-Rv3407 DNA boost vaccination against tuberculosis. Infect. Immun. 2004, 72, 6471–6479. [Google Scholar] [CrossRef]
- Rodriguez, L.; Tirado, Y.; Reyes, F.; Puig, A.; Kadir, R.; Borrero, R.; Fernandez, S.; Reyes, G.; Alvarez, N.; Garcia, M.A.; et al. Proteoliposomes from M. sm induce immune cross-reactivity against M. tb antigens in mice. Vaccine 2011, 29, 6236–6241. [Google Scholar] [CrossRef]
- García, M.d.L.A.; Borrero, R.; Marrón, R.; Lanio, M.E.; Canet, L.; Otero, O.; Kadir, R.; Suraiya, S.; Zayas, C.; López, Y.; et al. Evaluation of specific humoral immune response and cross reactivity against M. tb antigens induced in mice immunized with liposomes composed of total lipids extracted from M. sm. BMC Immunol. 2013, 14 (Suppl. 1), S11. [Google Scholar] [CrossRef]
- Borrero, R.; García, M.D.L.A.; Canet, L.; Zayas, C.; Reyes, F.; Prieto, J.L.; Infante, J.F.; E Lanio, M.; Kadir, R.; López, Y.; et al. Evaluation of the humoral immune response and cross reactivity against M. tb of mice immunized with liposomes containing glycolipids of M. sm. BMC Immunol. 2013, 14 (Suppl. 1), S13. [Google Scholar] [CrossRef]
- Harth, G.; Lee, B.Y.; Horwitz, M.A. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major M. tb extracellular proteins considered to be leading vaccine candidates and drug targets. Infect. Immun. 1997, 65, 2321–2328. [Google Scholar] [CrossRef]
- Xu, M.; Wen, C.B.; Bing, S.X.; Su, C.; Ai, L.Y.; Wang, G.Z. Effects of M. sm vaccine on cytokines production and Th1/Th2 responses in mice. Zhonghua Jie He He Hu Xi Za Zhi Zhonghua Jiehe He Huxi Zazhi Chin. J. Tuberc. Respir. Dis. 2005, 28, 781–784. [Google Scholar]
- Sweeney, K.A.; Dao, D.N.; Goldberg, M.F.; Hsu, T.; Venkataswamy, M.M.; Henao-Tamayo, M.; Ordway, D.; Sellers, R.S.; Jain, P.; Chen, B.; et al. A recombinant M. sm induces potent bactericidal immunity against M. tb. Nat. Med. 2011, 17, 1261–1268. [Google Scholar] [CrossRef]
- Ali, K.; Zhen, G.; Nzungize, L.; Stojkoska, A.; Duan, X.; Li, C.; Duan, W.; Xu, J.; Xie, J.M. M. tb PE31 (Rv3477) Attenuates Host Cell Apoptosis and Promotes Recombinant, M. sm Intracellular Survival via Up-regulating GTPase Guanylate Binding Protein-1. Front. Cell Infect. Microbiol. 2020, 10, 40. [Google Scholar] [CrossRef]
- Yang, W.; Deng, W.; Zeng, J.; Ren, S.; Ali, K.; Gu, Y.; Li, Y.; Xie, J.M. M. tb PE_PGRS18 enhances the intracellular survival of M. sm via altering host macrophage cytokine profiling and attenuating the cell apoptosis. Apoptosis Int. J. Program. Cell Death 2017, 22, 502–509. [Google Scholar] [CrossRef]
- Deng, G.; Zhang, F.; Yang, S.; Kang, J.; Sha, S.; Ma, Y.M. M. tb Rv0431 expressed in M. sm, a potentially mannosylated protein, mediated the immune evasion of RAW 264.7 macrophages. Microb. Pathog. 2016, 100, 285–292. [Google Scholar] [CrossRef]
- Garbe, T.; Harris, D.; Vordermeier, M.; Lathigra, R.; Ivanyi, J.; Young, D. Expression of the M. tb 19-kilodalton antigen in M. sm: Immunological analysis and evidence of glycosylation. Infect. Immun. 1993, 61, 260–267. [Google Scholar] [CrossRef]
- Sha, S.; Shi, X.; Deng, G.; Chen, L.; Xin, Y.; Ma, Y.M. M. tb Rv1987 induces Th2 immune responses and enhances M. sm survival in mice. Microbiol. Res. 2017, 197, 74–80. [Google Scholar] [CrossRef]
- Li, Y.; Bao, L.; Zhang, H.D.; Li, Y.S.; Zhu, H.L. Construction of recombinant M. sm expressing ESAT-6 and its effects on macrophages. Nan Fang. Yi Ke Da Xue Xue Bao 2006, 26, 923–926. [Google Scholar]
- Zhang, H.; Peng, P.; Miao, S.; Zhao, Y.; Mao, F.; Wang, L.; Bai, Y.; Xu, Z.; Wei, S.; Shi, C. Recombinant, M. sm expressing an ESAT6-CFP10 fusion protein induces anti-mycobacterial immune responses and protects against M. tb challenge in mice. Scand. J. Immunol. 2010, 72, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Kadir, N.A.; Sarmiento, M.E.; Acosta, A.; Norazmi, M.N. Cellular and humoral immunogenicity of recombinant M. sm expressing Ag85B epitopes in mice. Int. J. Mycobacteriol. 2016, 5, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, D.R.; Dhandayuthapani, S.; Jagannath, C. Anti-tuberculosis immunity induced in mice by vaccination with M. sm over-expressing Antigen 85B is due to the increased influx of IFNgamma-positive CD4 T cells into the lungs. Tuberculosis 2009, 89 (Suppl. 1), S46–S48. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.W.; Winter, N.; Triccas, J.A.; Feng, C.G.; Britton, W.J. Expression of M. tb MPT64 in recombinant Myco. smegmatis: Purification, immunogenicity and application to skin tests for tuberculosis. Clin. Exp. Immunol. 1996, 103, 226–232. [Google Scholar] [CrossRef]
- Kannan, N.; Haug, M.; Steigedal, M.; Flo, T.H.M. M. sm Vaccine Vector Elicits CD4+ Th17 and CD8+ Tc17 T Cells With Therapeutic Potential to Infections With Mycobacterium avium. Front. Immunol. 2020, 11, 1116. [Google Scholar] [CrossRef]
- Falcone, V.; Bassey, E.; Jacobs, W.; Collins, F. The immunogenicity of recombinant M. sm bearing BCG genes. Microbiology 1995, 141 Pt 5, 1239–1245. [Google Scholar] [CrossRef]
- Jagadeb, M.; Pattanaik, K.P.; Rath, S.N.; Sonawane, A. Identification and evaluation of immunogenic MHC-I and MHC-II binding peptides from M. tb. Comput. Biol. Med. 2021, 130, 104203. [Google Scholar] [CrossRef]
- Tsolaki, A.G.; Nagy, J.; Leiva, S.; Kishore, U.; Rosenkrands, I.; Robertson, B.D.M. M. tb antigen 85B and ESAT-6 expressed as a recombinant fusion protein in M. sm elicits cell-mediated immune response in a murine vaccination model. Mol. Immunol. 2013, 54, 278–283. [Google Scholar] [CrossRef][Green Version]
- Wang, P.; Wang, L.; Zhang, W.; Bai, Y.; Kang, J.; Hao, Y.; Luo, T.; Shi, C.; Xu, Z. Immunotherapeutic efficacy of recombinant M. sm expressing Ag85B-ESAT6 fusion protein against persistent tuberculosis infection in mice. Hum. Vaccines Immunother. 2014, 10, 150–158. [Google Scholar] [CrossRef]
- Guo, X.Q.; Wei, Y.M.; Yu, B. Recombinant M. sm expressing Hsp65–hIL–2 fusion protein and its influence on lymphocyte function in mice. Asian Pac. J. Trop. Med. 2012, 5, 347–351. [Google Scholar] [CrossRef]
- Hao, S.; Zhao, Y.; Mao, F.; Zhang, C.; Bai, B.; Zhang, H.; Shi, C.; Xu, Z. Protective and therapeutic efficacy of M. sm expressing HBHA-hIL12 fusion protein against M. tb in mice. PLoS ONE 2012, 7, e31908. [Google Scholar] [CrossRef]
- Yang, C.; He, Y.L.; Zhang, L.; Xu, L.; Yi, Z.; Wang, Y.; Zhu, D. GLS/IL-12-modified M. sm as a novel anti-tuberculosis immunotherapeutic vaccine. Int. J. Tuberc. Lung Dis. 2009, 13, 1360–1366. [Google Scholar]
- Yi, Z.; Fu, Y.; Yang, C.; Li, J.; Luo, X.; Chen, Q.; Zeng, W.; Jiang, S.; Jiang, Y.; He, Y.; et al. Recombinant, M. sm vaccine targeted delivering IL-12/GLS into macrophages can induce specific cellular immunity against M. tb in BALB/c mice. Vaccine 2007, 25, 638–648. [Google Scholar] [CrossRef]
- Junqueira-Kipnis, A.P.; de Oliveira, F.M.; Trentini, M.M.; Tiwari, S.; Chen, B.; Resende, D.P.; Silva, B.D.S.; Chen, M.; Tesfa, L.; Jacobs, W.R., Jr.; et al. Prime-boost with M. sm recombinant vaccine improves protection in mice infected with M. tb. PLoS ONE 2013, 8, e78639. [Google Scholar] [CrossRef]
- Lü, L.; Zeng, H.-Q.; Wang, P.-L.; Shen, W.; Xiang, T.-X.; Mei, Z.-C. Oral immunization with recombinant M. sm expressing the outer membrane protein 26-kilodalton antigen confers prophylactic protection against Helicobacter pylori infection. Clin. Vaccine Immunol. 2011, 18, 1957–1961. [Google Scholar] [CrossRef]
- Lü, L.; Cao, H.-D.; Zeng, H.-Q.; Wang, P.-L.; Wang, L.-J.; Liu, S.-N.; Xiang, T.-X. Recombinant, M. sm mc(2)155 vaccine expressing outer membrane protein 26 kDa antigen affords therapeutic protection against Helicobacter pylori infection. Vaccine 2009, 27, 972–978. [Google Scholar] [CrossRef]
- Vasconcellos, H.L.F.; Scaramuzzi, K.; Nascimento, I.P.; Ferreira, J.M.D.C.; Abe, C.M.; Piazza, R.M.; Kipnis, A.; da Silva, W.D. Generation of recombinant bacillus Calmette-Guérin and M. sm expressing BfpA and intimin as vaccine vectors against enteropathogenic Escherichia coli. Vaccine 2012, 30, 5999–6005. [Google Scholar] [CrossRef]
- Yue, Q.; Hu, X.; Yin, W.; Xu, X.; Wei, S.; Lei, Y.; Lü, X.; Yang, J.; Su, M.; Xu, Z.; et al. Immune responses to recombinant M. sm expressing fused core protein and preS1 peptide of hepatitis B virus in mice. J. Virol. Methods 2007, 141, 41–48. [Google Scholar] [CrossRef]
- Skerry, C.; Klinkenberg, L.G.; Page, K.R.; Karakousis, P.C. TLR2-Modulating Lipoproteins of the M. tb Complex Enhance the HIV Infectivity of CD4+ T Cells. PLoS ONE 2016, 11, e0147192. [Google Scholar] [CrossRef]
- Cayabyab, M.J.; Hovav, A.-H.; Hsu, T.; Krivulka, G.R.; Lifton, M.A.; Gorgone, D.A.; Fennelly, G.J.; Haynes, B.F., Jr.; Letvin, N.L. Generation of CD8+ T-cell responses by a recombinant nonpathogenic M. sm vaccine vector expressing human immunodeficiency virus type 1 Env. J. Virol. 2006, 80, 1645–1652. [Google Scholar] [CrossRef]
- Yu, J.-S.; Peacock, J.W.; Vanleeuwen, S.; Hsu, T.; Jacobs, W.R.; Cayabyab, M.J.; Letvin, N.L.; Frothingham, R.; Staats, H.F.; Liao, H.-X.; et al. Generation of mucosal anti-human immunodeficiency virus type 1 T-cell responses by recombinant M. sm. Clin. Vaccine Immunol. 2006, 13, 1204–1211. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Gong, J.R.; Kim, G.N.; Kim, B.R.; Lee, S.Y.; Kook, Y.H.; Kim, B.J. Recombinant, M. sm with a pMyong2 vector expressing Human Immunodeficiency Virus Type I Gag can induce enhanced virus-specific immune responses. Sci. Rep. 2017, 7, 44776. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, S.; Wu, L.; Fan, X.; Ma, H.; Wu, K.; Wu, J.; Zhang, J. IL-17A Autoantibody Induced by Recombinant, M. sm Expressing Ag85A-IL-17A Fusion Protein. Appl. Biochem. Biotechnol. 2015, 176, 2018–2026. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, W.; Mao, S.; Zhu, R.; Zhang, J. Autoantibody of interleukin-17A induced by recombinant M. sm attenuates airway inflammation in mice with neutrophilic asthma. J. Asthma 2022, 59, 2117–2126. [Google Scholar] [CrossRef]
- Nicolò, C.; Sali, M.; Di Sante, G.; Geloso, M.C.; Signori, E.; Penitente, R.; Uniyal, S.; Rinaldi, M.; Ingrosso, L.; Fazio, V.M.; et al. M. sm expressing a chimeric protein MPT64-proteolipid protein (PLP) 139–151 reorganizes the PLP-specific T cell repertoire favoring a CD8-mediated response and induces a relapsing experimental autoimmune encephalomyelitis. J. Immunol. 2010, 184, 222–235. [Google Scholar] [CrossRef]
- Deshpande, V.; Krishnan, R.; Philip, S.; Faludi, I.; Ponnusamy, T.; Thota, L.N.R.; Endresz, V.; Lu, X.; Kakkar, V.V.; Mundkur, L.A. Oral administration of recombinant M. sm expressing a tripeptide construct derived from endogenous and microbial antigens prevents atherosclerosis in ApoE (−/−) mice. Cardiovasc. Ther. 2016, 34, 314–324. [Google Scholar] [CrossRef]
- Haley, J.L.; Young, D.G.; Alexandroff, A.; James, K.; Jackson, A.M. Enhancing the immunotherapeutic potential of mycobacteria by transfection with tumour necrosis factor-alpha. Immunology 1999, 96, 114–121. [Google Scholar] [CrossRef]
- Young, S.L.; Murphy, M.; Zhu, X.W.; Harnden, P.; O’Donnell, M.A.; James, K.; Patel, P.M.; Selby, P.J.; Jackson, A.M. Cytokine-modified M. sm as a novel anticancer immunotherapy. Int. J. Cancer 2004, 112, 653–660. [Google Scholar] [CrossRef]
- Jian, W.; Li, X.; Kang, J.; Lei, Y.; Bai, Y.; Xue, Y. Antitumor effect of recombinant M. sm expressing MAGEA3 and SSX2 fusion proteins. Exp. Ther. Med. 2018, 16, 2160–2166. [Google Scholar] [CrossRef]
- Jeong, H.; Lee, S.Y.; Seo, H.; Kim, B.J. Recombinant M. sm delivering a fusion protein of human macrophage migration inhibitory factor (MIF) and IL-7 exerts an anticancer effect by inducing an immune response against MIF in a tumor-bearing mouse model. J. Immunother. Cancer 2021, 9, e003180. [Google Scholar] [CrossRef]

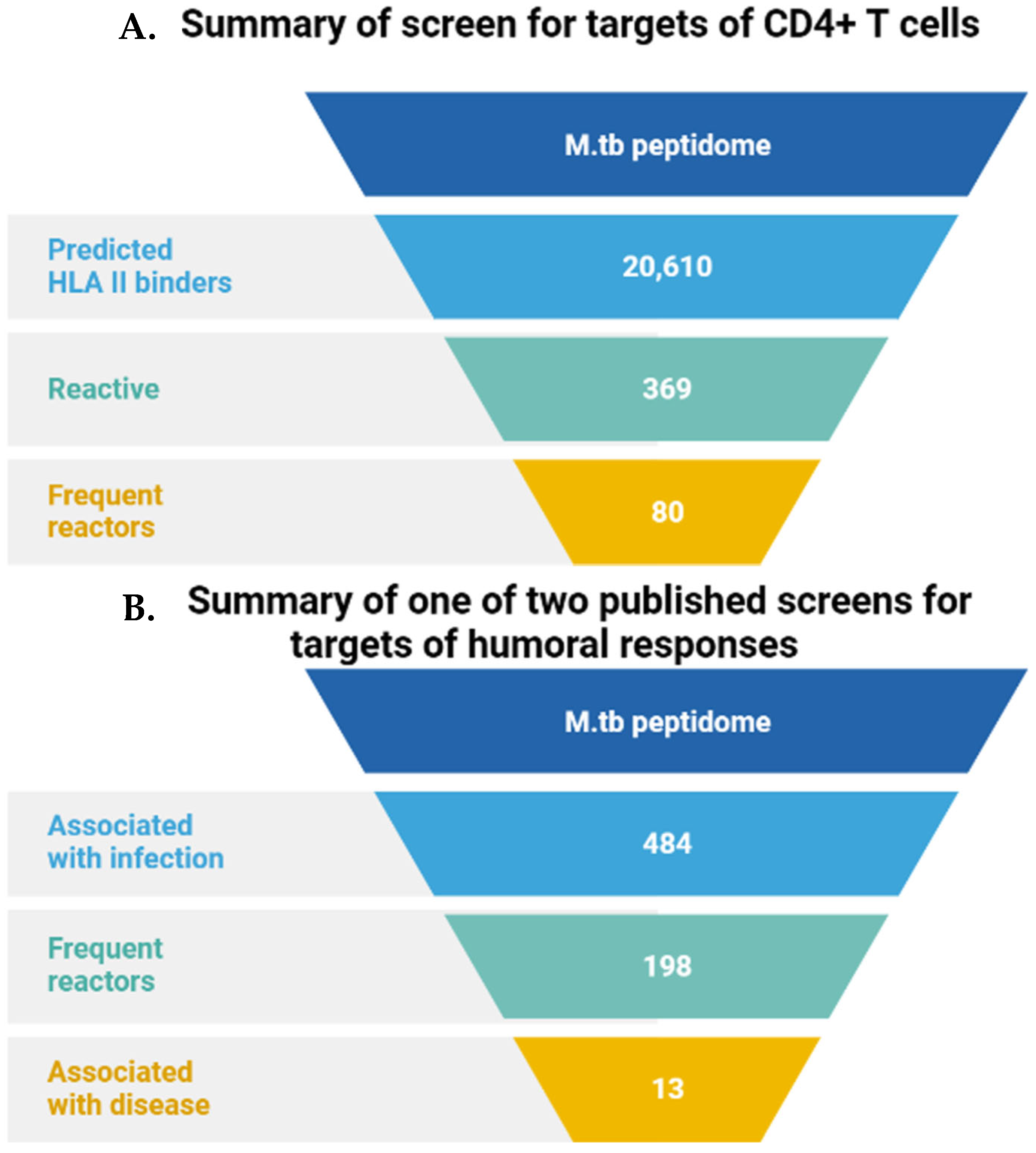
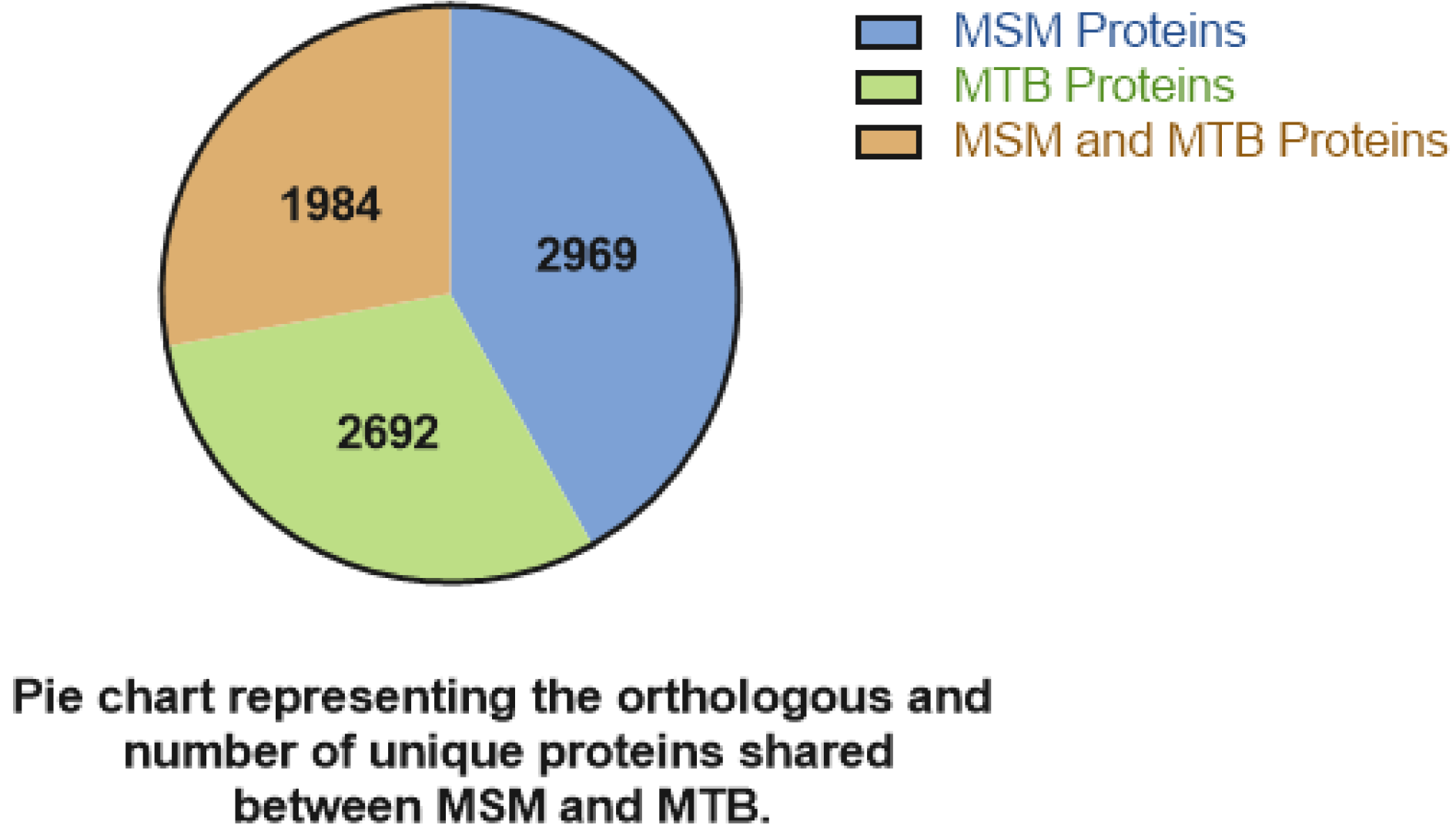
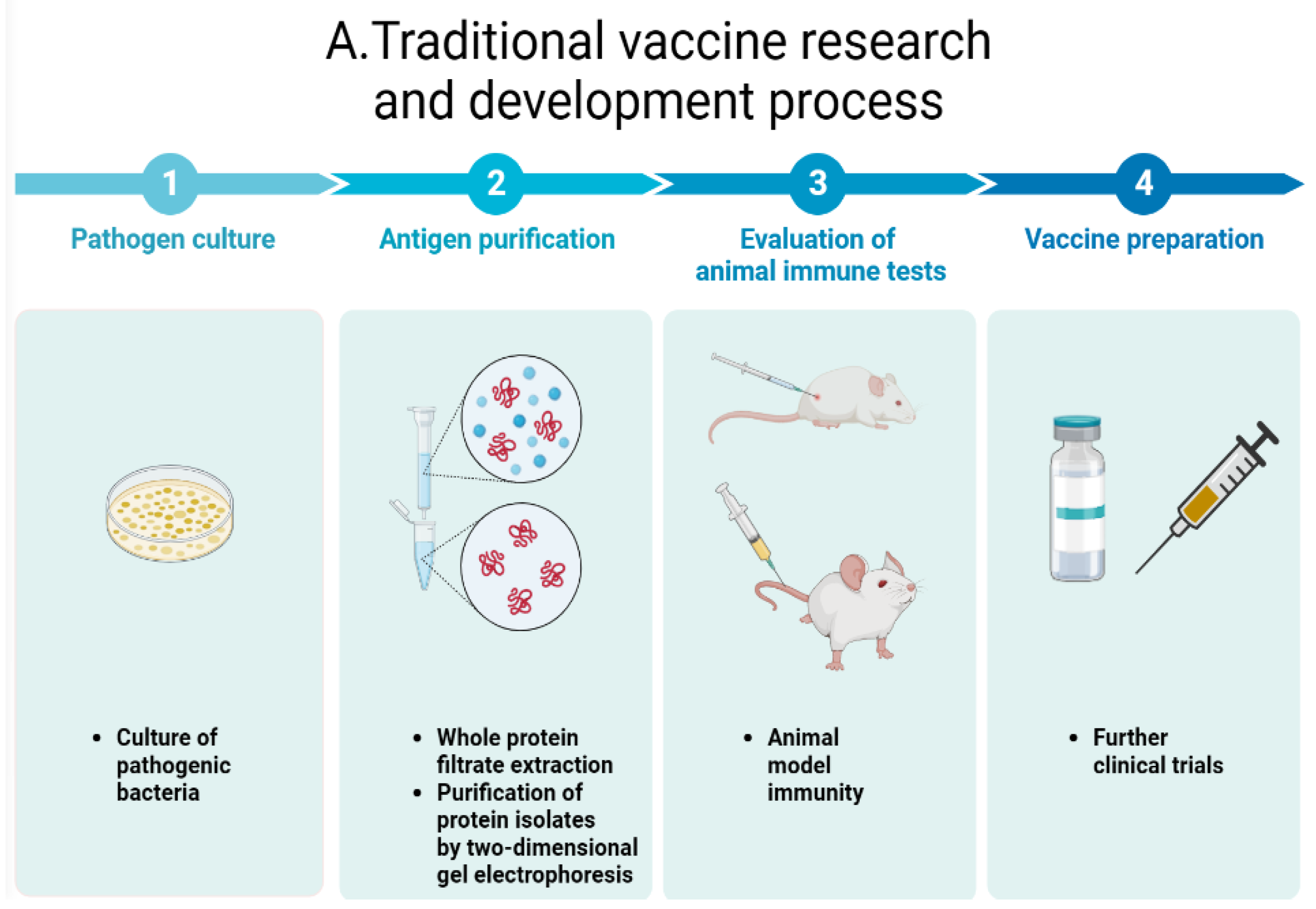

| Species | ESX-1 | ESX-2 | ESX-3 | ESX-4 | ESX-5 | Pathogenicity | |
|---|---|---|---|---|---|---|---|
| RGM | M.smegmatis | ● | ● | ● | saprophyte | ||
| SGM | M.tuberculosis | ● | ▲ | ● | ● | ▲ | pathogen |
| Antigen | Sanger ID | Methods of Identification | Present in CF | Theoretical Mass (kDa) | PATRIC Annotation | Homologous Genes Sanger ID in M.sm (Identity) |
|---|---|---|---|---|---|---|
| ESAT-6 | Rv3875 | Biochemical purification | + | 9.9 | Early secretory antigenic target EsxA | MSMEG0066 (71.56%) |
| Ag85A | Rv3804c | Biochemical purification | + | 35.7 | Secreted antigen 85-A FbpA | MSMEG2078 (69.21%) |
| Ag85B | Rv1886c | Biochemical purification | + | 34.6 | Secreted antigen 85-B FbpB | MSMEG2078 (71.08%) |
| MPT51 | Rv3803c | Biochemical purification | + | 28.7 | Secreted MPT51/MPB51 antigen protein FbpD | MSMEG6396 (67.00%) |
| MPT64 | Rv1980c | Biochemical purification | + | 24.8 | Immunogenic protein | MSMEG2331, MSMEG1051 (42.73%) |
| CFP10 a | Rv3874 | Molecular cloning serological expression cloning | + | 10.8 | ESAT-6-like protein EsxB | MSMEG0065 (61.00%) |
| TB10.4 a | Rv0288 | Antibody expression cloning | + | 10.4 | Antigen 7 EsxH | MSMEG0621 (75.79%) |
| M.tb8.4 | Rv1174c | Biochemical purification | + | 10.9 | T cell antigen | MSMEG4804 (52.29%) |
| HspX | Rv2031c | Biochemical purification | – | 15.4 | Heat shock protein | MSMEG3932 (60.84%) |
| CFP6 | Rv3004 | Biochemical purification | + | 12.2 | Antigen 6 | MSMEG2371 (60.71%) |
| M.tb12 | Rv2376c | Biochemical purification | + | 12.5 | Antigen CFP2 | MSMEG3903 (38.81%) |
| M.tb9.9 a antigens | Rv1793, MT3721, Rv1198, Rv1037c, Rv3619c | T cell expression cloning | + | 9.8–9.9 | ESAT-6 like protein | / |
| M.tb32A | Rv0125 | Antibody expression cloning | + | 32 | Serine protease | MSMEG6289 (49.83%) |
| PstS-1 | Rv0934 | Affinity purification | + | 38.2 | Phosphate uptake surface protein | pstS (34.23%) |
| PstS-2 | Rv0932c | Antibody expression cloning | + | 37.8 | Phosphate uptake surface protein | pstS (48.47%) |
| PstS-3 | Rv0928 | Antibody expression cloning | + | 37.9 | Phosphate uptake surface protein | pstS (48.42%) |
| MPT63 | Rv1926c | Biochemical purification | + | 16.5 | Immunogenic protein | MSMEG5412 (54.40%) |
| M.tb39 | Rv1196 | Serological expression cloning | – | 39.1 | PPE family protein | MSMEG0619 (41.18%) |
| M.tb41 | Rv0915c | T cell expression cloning | – | 41.4 | PPE family protein | MSMEG0619 (42.00%) |
| MPT83 | Rv2873 | Antibody expression cloning | – | 22.1 | Cell surface lipoprotein | MSMEG5196 (40.37%) |
| 71-kDa | Unknown | Biochemical purification | – | 71 | Unknown surface protein | / |
| PPE 68 | Rv3873 | Comparative genomics | – | 37.3 | PPE family protein | MSMEG0064 (62.57%) |
| LppX | Rv2945c | Comparative genomics | + | 24 | lipoprotein | / |
| Rv3878 | Comparative genomics | ? | ESX-1 secretion-associated protein EspJ | / | ||
| Rv3407 | In silico prediction | ? | 10 | Antitoxin | / | |
| Rv1818c | Molecular cloning | – | 40.7 | PE-PGRS family protein | / |
| Main Author | Antigen | Results/Influences | Reference | |
|---|---|---|---|---|
| Mycobacteriosis | G. Harth et al., 1997. | M.tb four major extracellular proteins (the 30-, 32-, 16-, and 23.5-kDa proteins) | Extracellular protein recombination of M.tb first achieved | [52] |
| Miao Xu et al., 2005. | M.sm autologous vaccine | Strong immunogenicity promoted Th1 responses and inhibited Th2 response in mice | [53] | |
| Sweeney et al., 2011 | M.tb Esx-3 | Superior protection to BCG observed (intravenous administration trial) | [54] | |
| Md Kaisar Ali et al., 2020 | M.tb PE subfamily member PE31 (Rv3477) | PE31 identified as a potential vaccine target for ESX-5 | [55] | |
| Wenmin Yang et al., 2017 | M.tb PE_PGRS18 | Increased host interleukin production and survival in infected macrophages | [56] | |
| Guoying Deng et al., 2016 | M.tb Rv0431 | Implications in mycobacterial immunity | [57] | |
| T. Garbe et al., 1993 | M.tb 19 Dalton glycosylation-associated protein | Stimulation of T cell proliferation and differentiation | [58] | |
| Shanshan Sha et al., 2017 | M.tb Rv1987 encoded by the region of difference (RD)-2 gene | Stimulation of T cell proliferation and differentiation | [59] | |
| Yan Li et al., 2006 | M.tb ESAT-6 protein | Greater immunogenicity achieved | [60] | |
| H. Zhang et al., 2010 | M.tb ESAT-6 and CFP10 protein | Protective efficacy was found similar to BCG | [61] | |
| Ayuni Kadir et al., 2016 | M.tb Ag85B | Resistance of serum IgG and its subclasses to Ag85B epitopes significantly enhanced in immunized mice | [62] | |
| Devin R. Lindsey et al., 2009 | M.tb Ag85B(overexpressed) | Effective stimulation and increased numbers of CD4+ IFN-γ + T cells observed in lung | [63] | |
| P. W. Roche et al., 1996 | M.tb MPT64 | Recombinant vaccine’s T cell reactivity is demonstrated by the ability to stimulate human rMPB64 T cell line | [64] | |
| Nisha Kannan et al., 2020 | M. avium MPT64 | BCG-like protection was observed with strong induction of IL-17; producing CD4+ and CD8+ T cells | [65] | |
| Valeria Falcone et al., 1995 | Genomic library of BCG | Superior splenic survival rate observed | [66] | |
| Manaswini Jagadeb et al., 2021 | M.tb Pep-9 and Pep-15 | Secretion of pro-inflammatory cytokines induced in stimulated macrophages | [67] | |
| Anthony G. et al., 2013 | M.tb Ag85B and ESAT-6 | Proof for effective expression of major antigens (with dimer variation) secreted by M.tb | [68] | |
| Ping Wang et al., 2014 | M.tb Ag85B and ESAT-6 | Strong stimulation of spleen cells producing IFN-γ- and il-2 and increased activity of antigen-specific cytotoxic T lymphocytes (CTL) to produce TH1 type immune response | [69] | |
| Xiao-Qing Guo et al., 2012 | A fusion protein of heat shock protein 65 (Hsp65) and human interleukin 2 (IL-2) | Lymphocyte function markedly enhanced | [70] | |
| Shanmin Zhao et al., 2012 | A fusion protein of heparin-binding hemagglutinin (HBHA) and human interleukin 12 | Enhanced Th1-type cellular responses (IFN-γ and IL-2) in mice and reduced bacterial burden in lungs compared to BCG vaccination | [71] | |
| C. Yang et al., 2009 | A co-expression plasmid encoding human granulysin (GLS) and mouse interleukin-12 (IL-12) | Immunotherapeutic effect found to be related to stimulation of Th1 response and GLS antimicrobial activity | [72] | |
| Zhengjun Yi et al., 2007 | IL-12/GLS (granulolysin) | Strong induction of specific Th1 responses against M.tb | [73] | |
| Ana Paula Junqueira-Kipnis et al., 2013 | M.tb Ag85c, MPT51, and HspX | Significant immune responses induced | [74] |
| Main Author | Antigen | Results/Influences | References | |
|---|---|---|---|---|
| Gastrointestinal Diseases | Lin Lü et al., 2009, 2011 | Helicobacter pylori (H. pylori) outer membrane protein 26kDa antigen (Omp26) | Inducing protection and a significant reduction in bacterial colonization in the stomach | [75,76] |
| Vasconcellos et al., 2012 | (enteropathgenic) E. coli BfpA or intimin | Effectively stimulate the production of TNF-α and INF-γ | [77] | |
| Viral Diseases | Qiaohong Yue et al., 2007 | A fusion protein consisting of the HBV truncated core protein (amino acids 1–155) and the preS1 peptide (amino acids 1–55) | A stronger cellular immune response and a longer duration of humoral immune response were induced | [78] |
| Ciaran Skerry et al., 2016 | Mycobacterium bovis BCG lipoproteins | Determining the role of potential TLR2-stimulated lipoproteins on mycobacterial mediated HIV infection in CD4 + T cells | [79] | |
| Mark J Cayabyab et al., 2006 | HIV-1 HXBc2 gp120 envelope protein | Inducing both effector and memory T lymphocytes and generating a stable virus-specific central memory pool | [80] | |
| Jae-Sung Yu et al., 2006 | HIV-1 group M consensus envelope protein | Insufficient expression of insertional protein can still effectively generate immune response against HIV | [81] | |
| Byoung-Jun Kim et al., 2017 | HIV gag proteins | Eliciting more potent immunity | [82] | |
| Autoimmune Diseases | Ling Chen et al., 2015 | A fusion protein Ag85a-IL-17A | Inducing the production of interleukin-17A autoantibodies that reduced airway inflammation in mice with neutrophil asthma | [83] |
| Wanting Xu et al., 2022 | A fusion protein Ag85A-IL-17A | Obtaining in the immune test of asthmatic mouse model established by ovalbumin | [84] | |
| Chiara Nicolò et al., 2010 | A chimeric protein containing the self-epitope of proteolipid protein 139–151 (p139) fused to MPT64 | The disease severity was significantly reduced by comparing with p139 alone | [85] | |
| Vrushali Deshpande et al., 2016 | Tripeptide constructs (AHC; peptides from Apolipoprotein B, Heat-shock protein 60 and Chlamydia pneumoniae outer membrane protein) | Inducing regulatory immune responses and reducing the development of atherosclerosis in a mouse mode | [86] | |
| Cancer | J. L. Haley et al., 1999 | Human tumor necrosis factor-alpha (TNF-alpha) | The transgenic mycobacterium was more effective in inducing or upregulating a range of anticancer cytokines | [87] |
| Sarah L. Young et al., 2004 | Human tumor necrosis factor-alpha (TNF-alpha) | Further demonstrating that the expression of mammalian cytokines significantly increased the antitumor properties | [88] | |
| Wen Jian et al., 2018 | MAGEA3 and SSX2 | Activating the immune system and enhancing anti-tumor effects | [89] | |
| Hyein Jeong et al., 2021 | A fusion protein of human macrophage migration inhibitory factor (MIF) and interleukin 7 | Exerting a strong antitumor immune response in mice model | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, W.; Wang, L.; Luo, D.; Soni, V.; Rosenn, E.H.; Wang, Z. Mycobacterium smegmatis, a Promising Vaccine Vector for Preventing TB and Other Diseases: Vaccinomics Insights and Applications. Vaccines 2023, 11, 1302. https://doi.org/10.3390/vaccines11081302
Xie W, Wang L, Luo D, Soni V, Rosenn EH, Wang Z. Mycobacterium smegmatis, a Promising Vaccine Vector for Preventing TB and Other Diseases: Vaccinomics Insights and Applications. Vaccines. 2023; 11(8):1302. https://doi.org/10.3390/vaccines11081302
Chicago/Turabian StyleXie, Weile, Longlong Wang, Dan Luo, Vijay Soni, Eric H. Rosenn, and Zhe Wang. 2023. "Mycobacterium smegmatis, a Promising Vaccine Vector for Preventing TB and Other Diseases: Vaccinomics Insights and Applications" Vaccines 11, no. 8: 1302. https://doi.org/10.3390/vaccines11081302
APA StyleXie, W., Wang, L., Luo, D., Soni, V., Rosenn, E. H., & Wang, Z. (2023). Mycobacterium smegmatis, a Promising Vaccine Vector for Preventing TB and Other Diseases: Vaccinomics Insights and Applications. Vaccines, 11(8), 1302. https://doi.org/10.3390/vaccines11081302







