Surface-Exposed Protein Moieties of Burkholderia cenocepacia J2315 in Microaerophilic and Aerobic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Surface Shaving of Live B. cenocepacia Cells and Peptide Extraction
2.3. LC-MS/MS Analysis
2.4. Protein Identification by Database Searching
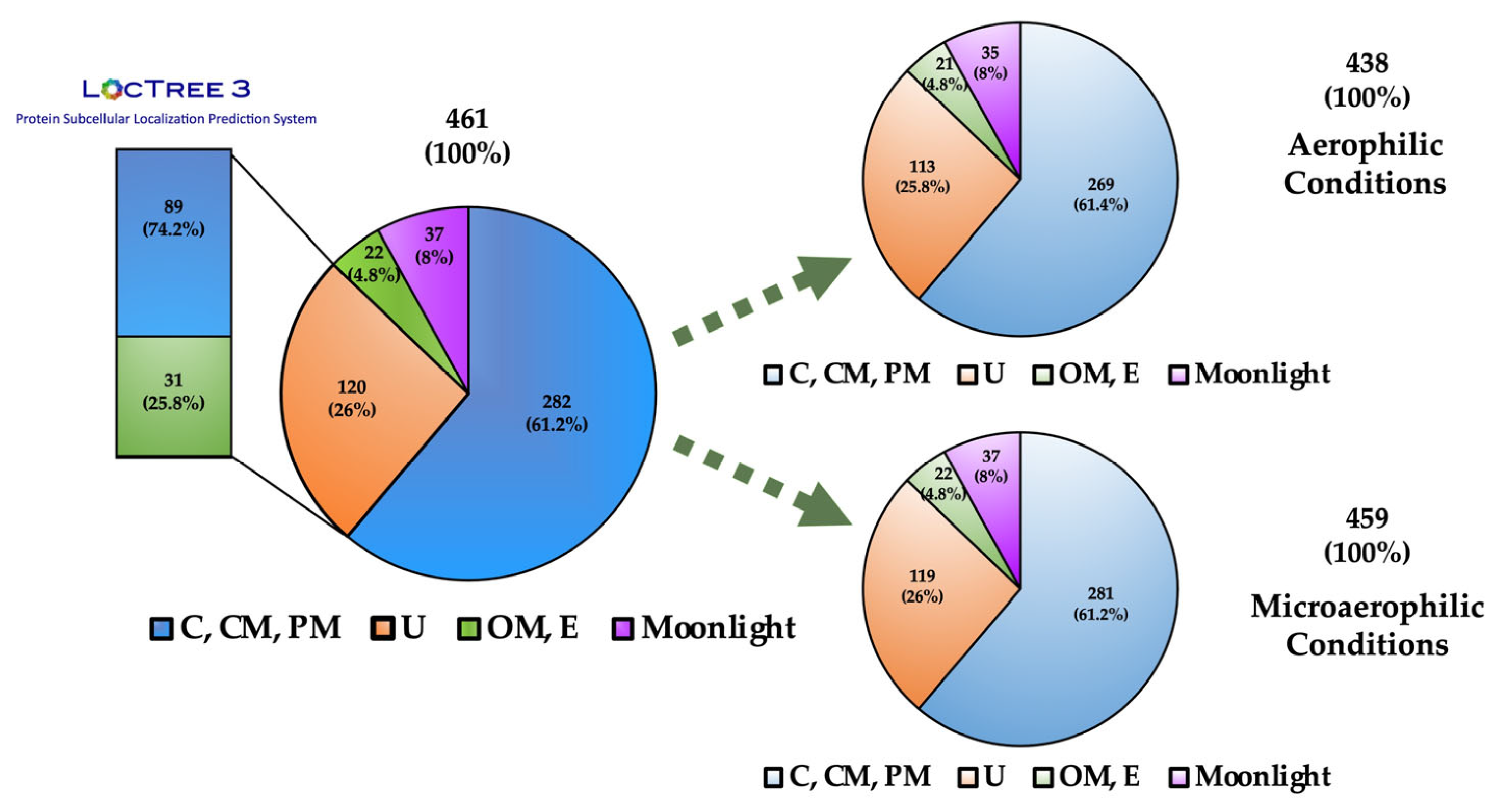
2.5. Bioinformatic Analysis of Protein Sequences
2.6. Production of Anti-Hfq2 and Anti-GroEL Polyclonal Antibodies
2.7. Fractionation of Cell Proteins
2.8. Western Blot Analyses
2.9. Cloning and Overexpression of B. cenocepacia J2315 bcal1985, bcal2645, and bcam1931
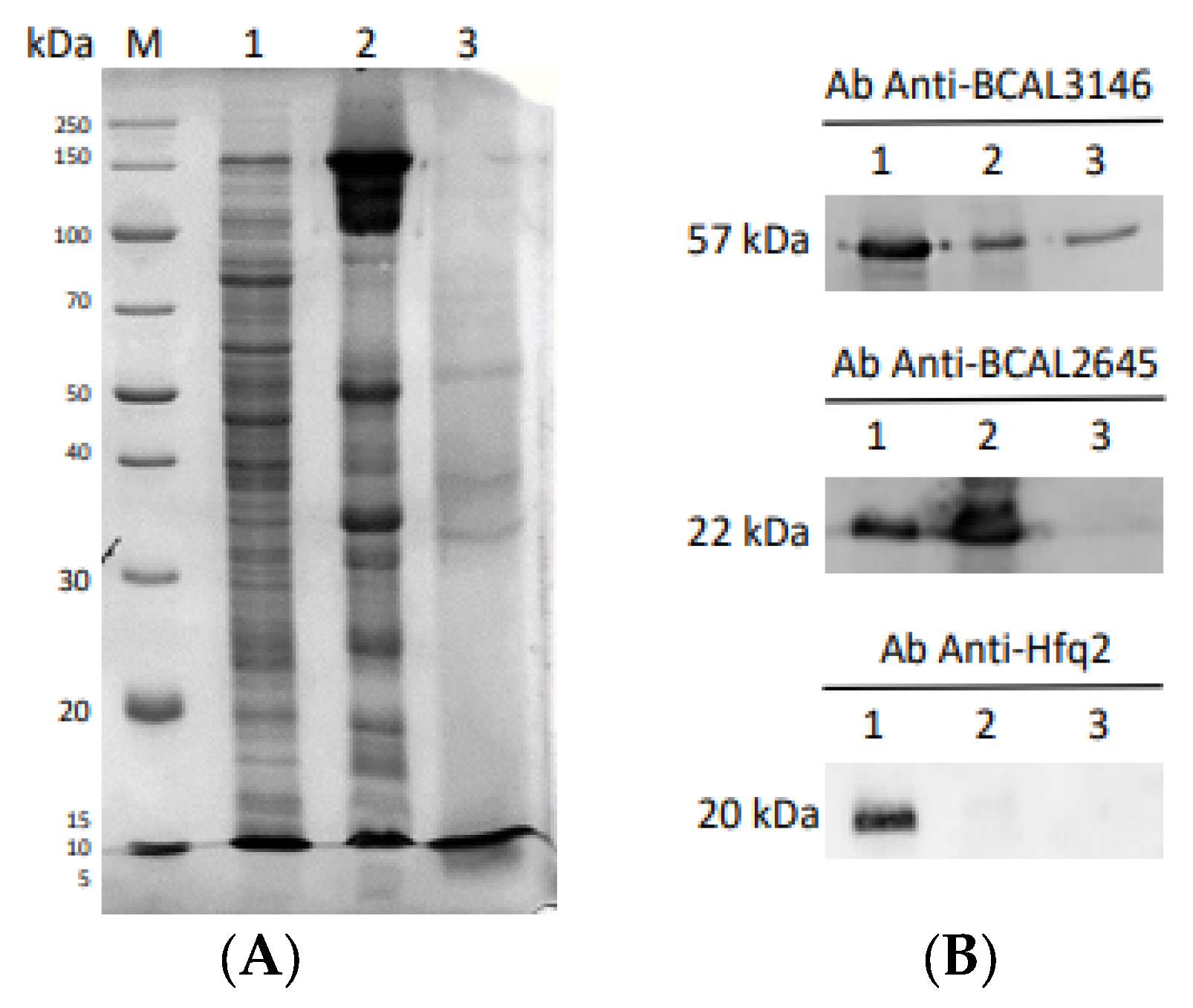
2.10. Purification of His-Tagged Proteins BCAL1985, BCAL2645, and BCAM1931
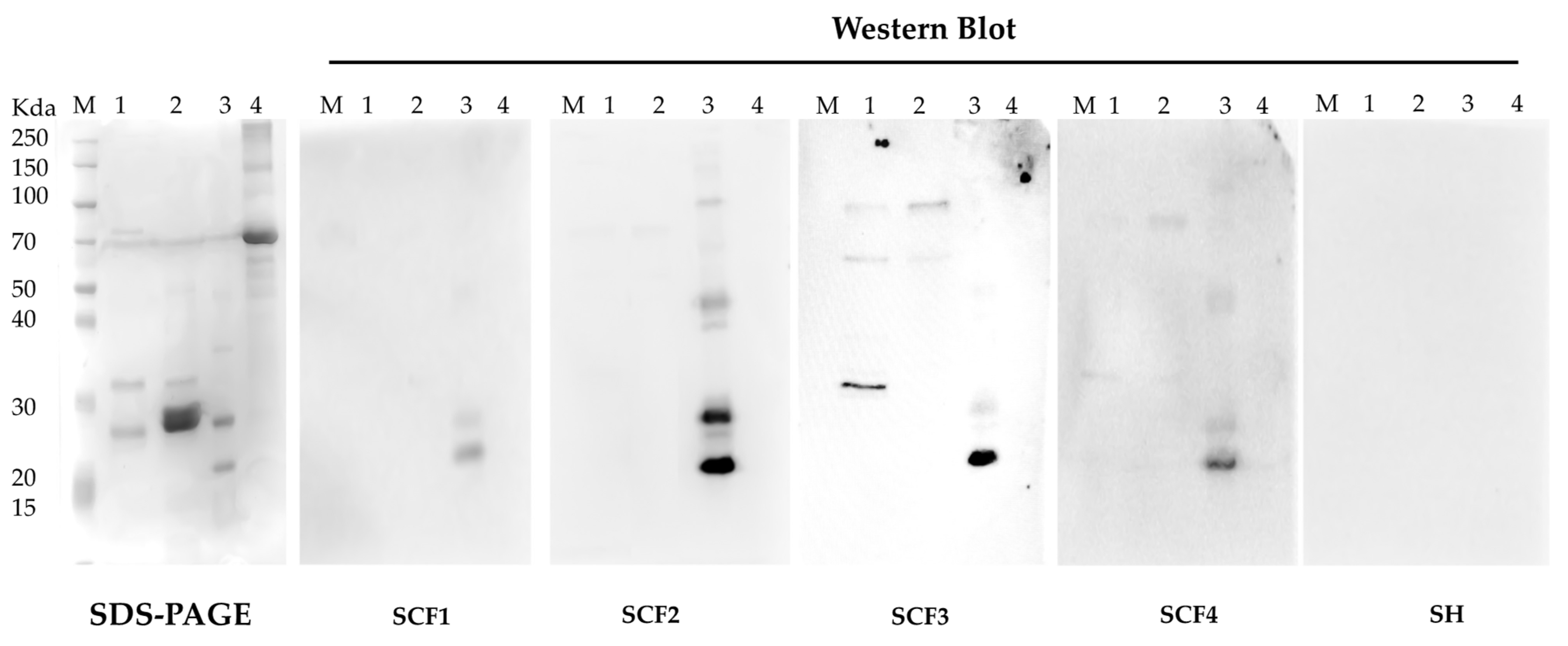
2.11. CF Patients’ Blood Sera Immunoreactivity Assay against BCAL1985, BCAL2645, and BCAM1931 Proteins
2.12. Data and Statistical Analysis
3. Results and Discussion
3.1. “Shaving” of Surface-Exposed Proteins of B. cenocepacia J2315 Live Cells
3.2. Surfome Proteins Predicted to be Located at the Outer Membrane
3.3. Surfome Proteins Predicted as Extracellular
3.4. Quantitative Analysis of Proteins Predicted as Outer Membrane or Extracellular Proteins
3.5. Characterization of B. cenocepacia BCAL1985, BCAL2645, and BCAM1931 Proteins’ Immunoreactivity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velez, L.S.; Aburjaile, F.F.; Farias, A.R.G.; Baia, A.D.B.; Oliveira, W.J.; Silva, A.M.F.; Benko-Iseppon, A.M.; Azevedo, V.; Brenig, B.; Ham, J.H.; et al. Burkholderia semiarida Sp. Nov. and Burkholderia sola Sp. Nov., Two Novel B. cepacia Complex Species Causing Onion Sour Skin. Syst. Appl. Microbiol. 2023, 46, 126415. [Google Scholar] [CrossRef] [PubMed]
- Zolin, A.; Orenti, A.; Jung, A.; van Rens, J.; Prasad, V.; Fox, A.; Krasnyk, M.; Mayor, S.L.; Naehrlich, L.; Gkolia, P.; et al. ECFSPR Annual Report 2021; European Cystic Fibrosis Society: Karup, Denmark, 2023. [Google Scholar]
- Tavares, M.; Kozak, M.; Balola, A.; Sá-Correia, I. Burkholderia cepacia Complex Bacteria: A Feared Contamination Risk in Water-Based Pharmaceutical Products. Clin. Microbiol. Rev. 2020, 33, e00139–e00219. [Google Scholar] [CrossRef]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in Cystic Fibrosis: Epidemiology and Molecular Mechanisms of Virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, K.M.; Green, A.E.; Connor, T.R.; Neill, D.R.; Mahenthiralingam, E. Identification of Two Distinct Phylogenomic Lineages and Model Strains for the Understudied Cystic Fibrosis Lung Pathogen Burkholderia multivorans. Microbiology 2023, 169, 1366. [Google Scholar] [CrossRef]
- Daccò, V.; Alicandro, G.; Consales, A.; Rosazza, C.; Sciarrabba, C.S.; Cariani, L.; Colombo, C. Cepacia Syndrome in Cystic Fibrosis: A Systematic Review of the Literature and Possible New Perspectives in Treatment. Pediatr. Pulmonol. 2023, 58, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia Infections in Cystic Fibrosis Patients: Drug Resistance and Therapeutic Approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Associated with Cystic Fibrosis Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Guzior, D.V.; Gonzalez, C.T.; Okros, M.; Mielke, J.; Padillo, L.; Querido, G.; Gil, M.; Thomas, R.; McClelland, M.; et al. Longitudinal Microbial and Molecular Dynamics in the Cystic Fibrosis Lung after Elexacaftor–Tezacaftor–Ivacaftor Therapy. Respir. Res. 2023, 24, 317. [Google Scholar] [CrossRef]
- Regan, K.H.; Bhatt, J. Eradication Therapy for Burkholderia cepacia Complex in People with Cystic Fibrosis. Cochrane Database Syst. Rev. 2019, 4, CD009876. [Google Scholar] [CrossRef]
- Seixas, A.M.M.M.; Sousa, S.A.; Leitão, J.H. Antibody-Based Immunotherapies as a Tool for Tackling Multidrug-Resistant Bacterial Infections. Vaccines 2022, 10, 1789. [Google Scholar] [CrossRef]
- Sousa, S.A.; Seixas, A.M.M.; Mandal, M.; Rodríguez-Ortega, M.J.; Leitão, J.H. Characterization of the Burkholderia cenocepacia J2315 Surface-Exposed Immunoproteome. Vaccines 2020, 8, 509. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving Live Organisms for a Fast Proteomic Identification of Surface Proteins. J. Proteomics 2014, 97, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Quintanilla, M.; Pulido, M.R.; Carretero-Ledesma, M.; McConnell, M.J. Vaccines for Antibiotic-Resistant Bacteria: Possibility or Pipe Dream? Trends Pharmacol. Sci. 2016, 37, 143–152. [Google Scholar] [CrossRef]
- Govan, J.R.; Deretic, V. Microbial Pathogenesis in Cystic Fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996, 60, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Worlitzsch, D.; Tarran, R.; Ulrich, M.; Schwab, U.; Cekici, A.; Meyer, K.C.; Birrer, P.; Bellon, G.; Berger, J.; Weiss, T.; et al. Effects of Reduced Mucus Oxygen Concentration in Airway Pseudomonas Infections of Cystic Fibrosis Patients. J. Clin. Investig. 2002, 109, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic Fibrosis Lung Environment and Pseudomonas aeruginosa Infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Tunney, M.M.; Field, T.R.; Moriarty, T.F.; Patrick, S.; Doering, G.; Muhlebach, M.S.; Wolfgang, M.C.; Boucher, R.; Gilpin, D.F.; McDowell, A.; et al. Detection of Anaerobic Bacteria in High Numbers in Sputum from Patients with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2008, 177, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Sass, A.M.; Schmerk, C.; Agnoli, K.; Norville, P.J.; Eberl, L.; Valvano, M.A.; Mahenthiralingam, E. The Unexpected Discovery of a Novel Low-Oxygen-Activated Locus for the Anoxic Persistence of Burkholderia cenocepacia. ISME J. 2013, 14, 1568–1581. [Google Scholar] [CrossRef]
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen Limitation Contributes to Antibiotic Tolerance of Pseudomonas aeruginosa in Biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664. [Google Scholar] [CrossRef]
- Leeper-Woodford, S.K.; Detmer, K. Acute Hypoxia Increases Alveolar Macrophage Tumor Necrosis Factor Activity and Alters NF-ΚB Expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 1999, 276, L909–L916. [Google Scholar] [CrossRef]
- Sriramulu, D.D.; Lünsdorf, H.; Lam, J.S.; Römling, U. Microcolony Formation: A Novel Biofilm Model of Pseudomonas aeruginosa for the Cystic Fibrosis Lung. J. Med. Microbiol. 2005, 54, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.; Hecht, M.; Hamp, T.; Karl, T.; Yachdav, G.; Ahmed, N.; Altermann, U.; Angerer, P.; Ansorge, S.; Balasz, K.; et al. LocTree3 Prediction of Localization. Nucleic Acids Res. 2014, 42, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Ong, E.; Cooke, M.F.; Huffman, A.; Xiang, Z.; Wong, M.U.; Wang, H.; Seetharaman, M.; Valdez, N.; He, Y. Vaxign2: The Second Generation of the First Web-Based Vaccine Design Program Using Reverse Vaccinology and Machine Learning. Nucleic Acids Res. 2021, 49, W671–W678. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, G.; Kumar, K.; Jain, P.; Ramachandran, S. SPAAN: A Software Program for Prediction of Adhesins and Adhesin-like Proteins Using Neural Networks. Bioinformatics 2005, 21, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Wickramasekara, S.; Neilson, J.; Patel, N.; Breci, L.; Hilderbrand, A.; Maier, R.M.; Wysocki, V. Proteomics Analyses of the Opportunistic Pathogen Burkholderia vietnamiensis Using Protein Fractionations and Mass Spectrometry. J. Biomed. Biotechnol. 2011, 2011, 701928. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, S.; Haigh, R.; Freestone, P. Fractionation by Ultracentrifugation of Gram Negative Cytoplasmic and Membrane Proteins. Bio-Protocol 2014, 4, e1287. [Google Scholar] [CrossRef]
- Seixas, A.M.M.; Sousa, S.A.; Feliciano, J.R.; Gomes, S.C.; Ferreira, M.R.; Moreira, L.M.; Leitão, J.H. A Polyclonal Antibody Raised against the Burkholderia cenocepacia OmpA-like Protein BCAL2645 Impairs the Bacterium Adhesion and Invasion of Human Epithelial Cells In Vitro. Biomedicines 2021, 9, 1788. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.G.; Seth-Smith, H.M.B.; Crossman, L.C.; Sebaihia, M.; Bentley, S.D.; Cerdeno-Tarraga, A.M.; Thomson, N.R.; Bason, N.; Quail, M.A.; Sharp, S.; et al. The Genome of Burkholderia cenocepacia J2315, an Epidemic Pathogen of Cystic Fibrosis Patients. J. Bacteriol. 2009, 191, 261–277. [Google Scholar] [CrossRef]
- Krishnan, S.; Prasadarao, N.V. Outer Membrane Protein A and OprF: Versatile Roles in Gram-Negative Bacterial Infections. FEBS J. 2012, 279, 919–993. [Google Scholar] [CrossRef]
- Feliciano, J.R.; Grilo, A.M.; Guerreiro, S.I.; Sousa, S.A.; Leitão, J.H. Hfq: A Multifaceted RNA Chaperone Involved in Virulence. Future Microbiol. 2016, 11, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK Stress Response Proteins as Antigens for Bacterial Diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Leung, P.K.; Wong, S.S.; Ho, P.L.; Yuen, K.Y. GroEL Encodes a Highly Antigenic Protein in Burkholderia pseudomallei. Clin. Diagn. Lab. Immunol. 2001, 8, 832–836. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shinoy, M.; Dennehy, R.; Coleman, L.; Carberry, S.; Schaffer, K.; Callaghan, M.; Doyle, S.; McClean, S. Immunoproteomic Analysis of Proteins Expressed by Two Related Pathogens, Burkholderia multivorans and Burkholderia cenocepacia, during Human Infection. PLoS ONE 2013, 8, e80796. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.A.; Soares-Castro, P.; Seixas, A.M.M.; Feliciano, J.R.; Balugas, B.; Barreto, C.; Pereira, L.; Santos, P.M.; Leitão, J.H. New Insights into the Immunoproteome of B. cenocepacia J2315 Using Serum Samples from Cystic Fibrosis Patients. N. Biotechnol. 2020, 54, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, J.B.; Ganesan, S.; Comstock, A.T.; Zhao, Y.; Sajjan, U.S. Cable Pili and the Associated 22 Kda Adhesin Contribute to Burkholderia cenocepacia Persistence In Vivo. PLoS ONE 2011, 6, e22435. [Google Scholar] [CrossRef] [PubMed]
- Badten, A.J.; Torres, A.G. Burkholderia pseudomallei Complex Subunit and Glycoconjugate Vaccines and Their Potential to Elicit Cross-Protection to Burkholderia Cepacia Complex. Vaccines 2024, 12, 313. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, G.C.; Deeraksa, A.; Qazi, O.; Judy, B.M.; Taylor, K.; Propst, K.L.; Duffy, A.J.; Johnson, K.; Kitto, G.B.; Brown, K.A.; et al. Protective Response to Subunit Vaccination against Intranasal Burkholderia mallei and B. pseudomallei Challenge. Procedia Vaccinol. 2010, 2, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Dyke, J.S.; Huertas-Diaz, M.C.; Michel, F.; Holladay, N.E.; Hogan, R.J.; He, B.; Lafontaine, E.R. The Peptidoglycan-Associated Lipoprotein Pal Contributes to the Virulence of Burkholderia mallei and Provides Protection against Lethal Aerosol Challenge. Virulence 2020, 11, 1024–1040. [Google Scholar] [CrossRef]
- Gerlach, R.G.; Hensel, M. Protein Secretion Systems and Adhesins: The Molecular Armory of Gram-Negative Pathogens. Int. J. Med. Microbiol. 2007, 297, 401–415. [Google Scholar] [CrossRef]
- Pimenta, A.I.; Mil-Homens, D.; Fialho, A.M. Burkholderia cenocepacia–Host Cell Contact Controls the Transcription Activity of the Trimeric Autotransporter Adhesin BCAM2418 Gene. Microbiologyopen 2020, 9, e998. [Google Scholar] [CrossRef]
- Pimenta, A.I.; Kilcoyne, M.; Bernardes, N.; Mil-Homens, D.; Joshi, L.; Fialho, A.M. Burkholderia cenocepacia BCAM2418 -induced Antibody Inhibits Bacterial Adhesion, Confers Protection to Infection and Enables Identification of Host Glycans as Adhesin Targets. Cell. Microbiol. 2021, 23, e13340. [Google Scholar] [CrossRef] [PubMed]
- Thibau, A.; Dichter, A.A.; Vaca, D.J.; Linke, D.; Goldman, A.; Kempf, V.A.J. Immunogenicity of Trimeric Autotransporter Adhesins and Their Potential as Vaccine Targets. Med. Microbiol. Immunol. 2020, 209, 243. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Holden, M.T.G.; Ge, Z.; Jones, A.M.; Ketchell, I.; Gill, R.T.; Mahenthiralingam, E. Gene Expression Changes Linked to Antimicrobial Resistance, Oxidative Stress, Iron Depletion and Retained Motility Are Observed When Burkholderia cenocepacia Grows in Cystic Fibrosis Sputum. BMC Infect. Dis. 2008, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Burtnick, M.N.; Brett, P.J.; Harding, S.V.; Ngugi, S.A.; Ribot, W.J.; Chantratita, N.; Scorpio, A.; Milne, T.S.; Dean, R.E.; Fritz, D.L.; et al. The Cluster 1 Type VI Secretion System Is a Major Virulence Determinant in Burkholderia pseudomallei. Infect. Immun. 2011, 79, 1512–1525. [Google Scholar] [CrossRef]
- Tapia, D.; Sanchez-Villamil, J.I.; Stevenson, H.L.; Torres, A.G. Multicomponent Gold-Linked Glycoconjugate Vaccine Elicits Antigen-Specific Humoral and Mixed Th1-Th17 Immunity, Correlated with Increased Protection against Burkholderia pseudomallei. MBio 2021, 12, 10.1128/mbio.01227-21. [Google Scholar] [CrossRef]
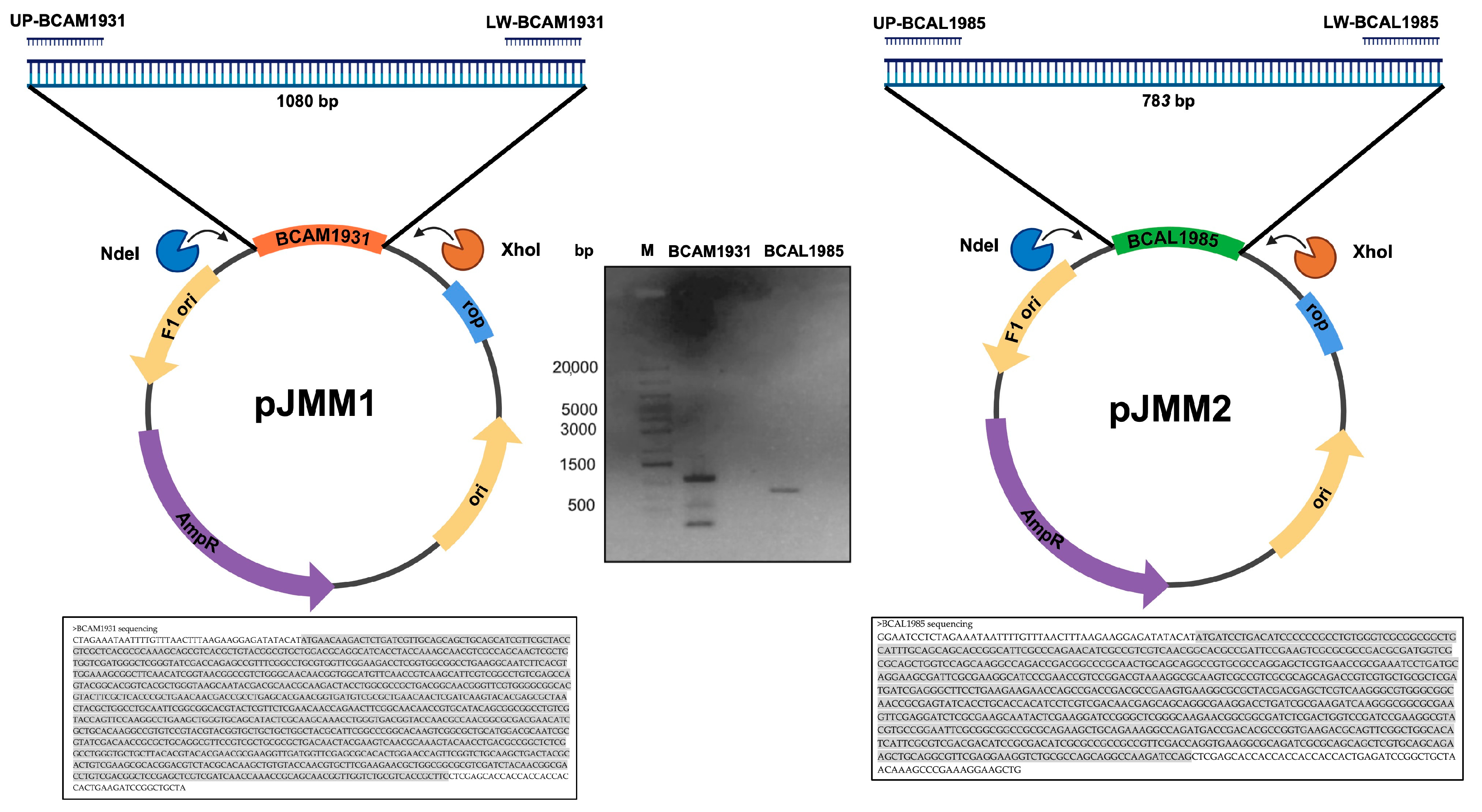

| ORF 1 | Product 1 | PSMs 2 | Unique Peptides 2 | Vaxign-ML Score 3 | Adhesin Probability (Pad) 4 | Surface-Exposed Protein Moieties with Predicted B-Cell Linear Epitopes 5 |
|---|---|---|---|---|---|---|
| BCAL0304 | VacJ-like lipoprotein | 9 | 2 | 90.9 | 0.513 | 195ANLLGAGDVLDAAALDK211 ○ |
| BCAL0349 * | Type VI secretion system-associated protein TagL | 5 | 2 | 94.8 | 0.756 | 195VSASEQGVLDQTLANR210 ○ 257TSNIALSQAR266 ○ |
| BCAL0565 | Flagellar basal body rod protein (FlgC) | 6 | 3 | 90.9 | 0.852 | 47QVVFATDPMGGARTASGQGVGGVR70 ● |
| BCAL0577 | Flagellar hook-associated protein (FlgL) | 18 | 4 | 94.3 | 0.846 | - |
| BCAL1893 | Family M23 peptidase | 7 | 3 | 91.3 | 0.645 | - |
| BCAL1985 | Putative exported isomerase | 26 | 6 | 90.9 | 0.517 | 77EGIPNRPDVK86 ◑ 235AQIAQQLVQQKLQAFEEGLR254 ◑ |
| BCAL2022 | PspA/IM30 family protein | 19 | 5 | 90.9 | 0.406 | 13GLLNDAADSVQDPS R27 ◑ 150DVAASALGGIGGKNLSEDFQK170 ◑ |
| BCAL2413 | HP | 17 | 3 | 90.9 | 0.639 | 70QMLFVDTVSASGAR83 ◑ 102DEIADPK108 ◑ |
| BCAL2645 | Putative OmpA family membrane protein | 22 | 3 | 90.9 | 0.623 | 79LAPSAAQTGTQVTEQPDGSLK99 ● 178LSAQGMGASNPIADNATEAGR198 ◑ |
| BCAL2820 | RND-4 efflux system outer membrane protein (oprM) | 6 | 3 | 94.9 | 0.537 | - |
| BCAL2958 | Putative ompA family protein | 78 | 9 | 92.8 | 0.581 | 93ITYQADALFDFDKATLKPLGKQKLDELASK122 ◑ 140IGSDKYNDR148 ◑ 203RVEVEVVGTQQVQK216 ◑ |
| BCAL3203 # | Tol-Pal system protein (TolB) | 16 | 4 | 93.7 | 0.832 | 176YQLQISDSDGQNAR189 ◑ |
| BCAL3204 | Putative OmpA family lipoprotein | 25 | 4 | 90.9 | 0.391 | 97HVLIQGNTDERGTSEYNLALGQK119 ◑ |
| BCAL3426 | Putative lipoprotein (SlyB) | 12 | 3 | 90.6 | 0.798 | 56IQSDGGGSAIGTLGGGALGAVAGSAIGGGK85 ○ 128SITQAASGEAFR139 ◑ |
| BCAM1931 | Putative porin | 62 | 10 | 92.5 | 0.915 | 44SLWSMGSGIDQSR56 ◑ 61GSEDLGGGLK70 ◑ 199LGAAYSQANLGDGTNANGATNIAAQGR225 ◑ |
| BCAM2418 # | Trimeric autotransporter adhesin | 26 | 3 | 93.05 | 0.898 | - |
| BCAM2549 | Multidrug efflux system outer membrane protein (OpcM) | 17 | 6 | 92.5 | 0.305 | 185ADQAQSEALFR195 ○ 253AKNELASAQADAVGVAR269 ○ |
| BCAM2761 | Giant cable pilus (cblA) | 49 | 6 | 95.9 | 0.929 | 80LATAPALKNQTSPGAAEIPLSVK102 ◑ |
| BCAS0104 | A-type flagellar hook-associated protein 2 (HAP2) (fliD2) | 30 | 9 | 96.6 | 0.884 | 52VATLAASQASGNTR65 ● 481MNTNSQYLTRLFGGANSNGTLSK503 ● |
| BCAS0236 # | Trimeric autotransporter adhesin | 6 | 2 | 92.2 | 0.917 | - |
| BCAS0522 | HP | 14 | 2 | 82.2 | 0.471 | - |
| ORF 1 | Product 1 | PSMs 2 | Unique Peptides 2 | Vaxign-ML Score 3 | Surface-Exposed Protein Moieties with Predicted B Cell Linear Epitopes 4 |
|---|---|---|---|---|---|
| BCAL0076 * | Putative lipoprotein | 7 | 2 | 48.2 | 86QEANDMSAQHNGGLSGDEQR105 ◑ |
| BCAL0151 # | Extracellular ligand binding protein | 90 | 15 | 98.9 | 73ITLQLDPQDDAADPRQATQVAQK95 ◑ 119IYSDAGVVQISPSATNPAYTQQGFK143 ◑ 199VMSHDATNDKAVDFR213 ○ 325ANSTDPAKILAAMPATKYTGVIGTTTFDS353 ◑ |
| BCAL0343 | Putative type VI secretion system protein (TssD) | 9 | 4 | 90.9 | 30SWDHSIVQPR39 ● 40SATASTAGGHTMTR53 ◑ |
| BCAL0360 * | HP | 12 | 3 | 81.7 | 48DNAPLDER55 ● 72AANHQVIGTSETYSSVQAR90 ● |
| BCAL0389 # | Thiol:disulfide interchange protein (DsbC) | 5 | 2 | 91.9 | 40LGNDAPIK47 ◑ 224RLPGAVSADQLNQALASSK242 ○ |
| BCAL0562 | Flagellin synthesis anti-sigma-28 factor (FglM) | 8 | 4 | 90.9 | 22APSGTAQSSAQAGDAGSTGGDTTVNLSGLSGQLR55 ● |
| BCAL0849 | Metallo peptidase, subfamily M48B | 13 | 2 | 95.9 | 159SAAGAASPGVAALSSSQLGDITEK182 ● |
| BCAL1105 * | HP | 10 | 2 | 90.4 | 37DAMGHDAMAK46 ◑ |
| BCAL1390 * | Glucanase | 3 | 2 | 90.9 | 267ADPLAAPLLAK277 ● 365FGADGTLDTR374 ● |
| BCAL1848 * | HP | 6 | 2 | 87.0 | 98APQALVVTTRSAGSGGYVGAQAYVTTSR125 ◑ |
| BCAL1849 * | HP | 13 | 4 | 90.9 | 72GGGTGQLEYTVK83 ◑ 153GVANVQLSFQAAAPK167 ◑ |
| BCAL1938 * | Cysteine peptidase, family C40 | 3 | 2 | 90.9 | 346TSTADDPIAR355 ● |
| BCAL1961 * | HP | 51 | 11 | 91.4 | 58LDPNTLAPNGDPILVIAAR76 ◑ 82VAAAIATTPNVDLEK96 ○ 82VAAAIATTPNVDLEKEDK99 ◑ 174GNHASTVTLLLDQGADPQ VK193 ○ 194NQLGITALEFAK205 ◑ 223IGASTPADAQK233 ◑ |
| BCAL2229 * | HP | 36 | 11 | 90.9 | 292VGIIDLASRKLVQTIAVGR310 ◑ |
| BCAL2476 * | HP | 11 | 4 | 80.8 | 72KFIIDDNLK80 ◑ |
| BCAL3149 # | HP | 10 | 4 | 77.0 | 122NVHALQQGGATVTEGEEAVGGR143 ◑ |
| BCAL3311 | BcnA | 35 | 8 | 94.4 | 64AAQGSAQMTIDVASFDLGDKMYNDQVAGK92 ◑ 157SAFNVGTGEWKDTSIVADEVQIK179 ◑ |
| BCAL3394 * | Putative exported ribonuclease | 9 | 4 | 84.7 | - |
| BCAL3427 * | Histone H1-like protein (HctB) | 16 | 2 | 90.8 | - |
| BCAM0900 * | HP | 31 | 8 | 82.8 | - |
| BCAM1242 * | HP | 12 | 5 | 90.9 | - |
| BCAM1576 | Phosphoesterase family protein | 29 | 14 | 93.7 | 145ITDAQGKPLPNGVITR160 ○ 256VEGDDPAGTR265 ◑ 268LADDSPASALDGPPK282 ◑ |
| BCAM1761 * | Putative lipoprotein | 14 | 2 | 84.9 | 52LSGTEQSQHNGVTDIAVGSNSYFVTLTPSGNGSVIK87 ○ 91GSGSEPAEEAMR102 ◑ |
| BCAM1876 * | HP | 15 | 4 | 89.7 | - |
| BCAM1920 * | HP | 7 | 3 | 90.9 | 13ATLSSDSGSIR23 ◑ |
| BCAM1921 * | Putative phage membrane protein | 13 | 2 | 90.9 | 330LGGQVSNDVVYAR342 ◑ |
| BCAM1933 * | Putative cyclase | 28 | 5 | 88.7 | 33DLAAEEANRQLVLTFYDR50 ◑ 138IVEHWDVIQPVPETSANR155 ◑ |
| BCAM2181 * | HP | 4 | 2 | 85.9 | 150VEALLADESTAAR162 ◑ |
| BCAM2603 * | HP | 15 | 4 | 56.0 | 19YDGLTALNAYDEDGR33 ○ 37YAITEGPYAGAK48 ◑ 73ATVVHIDDFAAGTSR87 ◑ |
| BCAM2686 * | HP | 8 | 3 | 90.9 | - |
| BCAS0151 * | HP | 14 | 3 | 90.9 | - |
| BCAS0750 | HP | 28 | 3 | 90.9 | - |
| ORF 1 | Product 1 | MW (kDa) 1 | PSMs 2 | Unique Peptides 2 | Vaxign-ML Score 3 | Surface-Exposed Protein Moieties with Predicted B Cell Linear Epitopes 4 |
|---|---|---|---|---|---|---|
| BCAL1985 | Putative exported isomerase | 28.6 | 26 | 6 | 90.9 | 77EGIPNRPDVK86 ◑ 235AQIAQQLVQQKLQAFEEGLR254 ◑ |
| BCAL2645 | Putative OmpA family membrane protein | 21.5 | 22 | 3 | 90.9 | 79LAPSAAQTGTQVTEQPDGSLK99 ● 178LSAQGMGASNPIADNATEAGR198 ◑ |
| BCAM1931 | Putative porin | 37.5 | 62 | 10 | 92.5 | 44SLWSMGSGIDQSR56 ◑ 61GSEDLGGGLK70 ◑ 199LGAAYSQANLGDGTNANGATNIAAQGR225 ◑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seixas, A.M.M.; Silva, C.; Marques, J.M.M.; Mateus, P.; Rodríguez-Ortega, M.J.; Feliciano, J.R.; Leitão, J.H.; Sousa, S.A. Surface-Exposed Protein Moieties of Burkholderia cenocepacia J2315 in Microaerophilic and Aerobic Conditions. Vaccines 2024, 12, 398. https://doi.org/10.3390/vaccines12040398
Seixas AMM, Silva C, Marques JMM, Mateus P, Rodríguez-Ortega MJ, Feliciano JR, Leitão JH, Sousa SA. Surface-Exposed Protein Moieties of Burkholderia cenocepacia J2315 in Microaerophilic and Aerobic Conditions. Vaccines. 2024; 12(4):398. https://doi.org/10.3390/vaccines12040398
Chicago/Turabian StyleSeixas, António M. M., Carolina Silva, Joana M. M. Marques, Patrícia Mateus, Manuel J. Rodríguez-Ortega, Joana R. Feliciano, Jorge H. Leitão, and Sílvia A. Sousa. 2024. "Surface-Exposed Protein Moieties of Burkholderia cenocepacia J2315 in Microaerophilic and Aerobic Conditions" Vaccines 12, no. 4: 398. https://doi.org/10.3390/vaccines12040398
APA StyleSeixas, A. M. M., Silva, C., Marques, J. M. M., Mateus, P., Rodríguez-Ortega, M. J., Feliciano, J. R., Leitão, J. H., & Sousa, S. A. (2024). Surface-Exposed Protein Moieties of Burkholderia cenocepacia J2315 in Microaerophilic and Aerobic Conditions. Vaccines, 12(4), 398. https://doi.org/10.3390/vaccines12040398








