Use of Biolayer Interferometry to Identify Dominant Binding Epitopes of Influenza Hemagglutinin Protein of A(H1N1)pdm09 in the Antibody Response to 2010–2011 Influenza Seasonal Vaccine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cloning and Expression of rH1, rHA1 and rHA2
2.2. f-AbBA-2
2.3. Serum Samples
2.4. Influenza Viruses
2.5. Hemagglutination Inhibition (HI) Assay
2.6. Statistical Analysis
3. Results
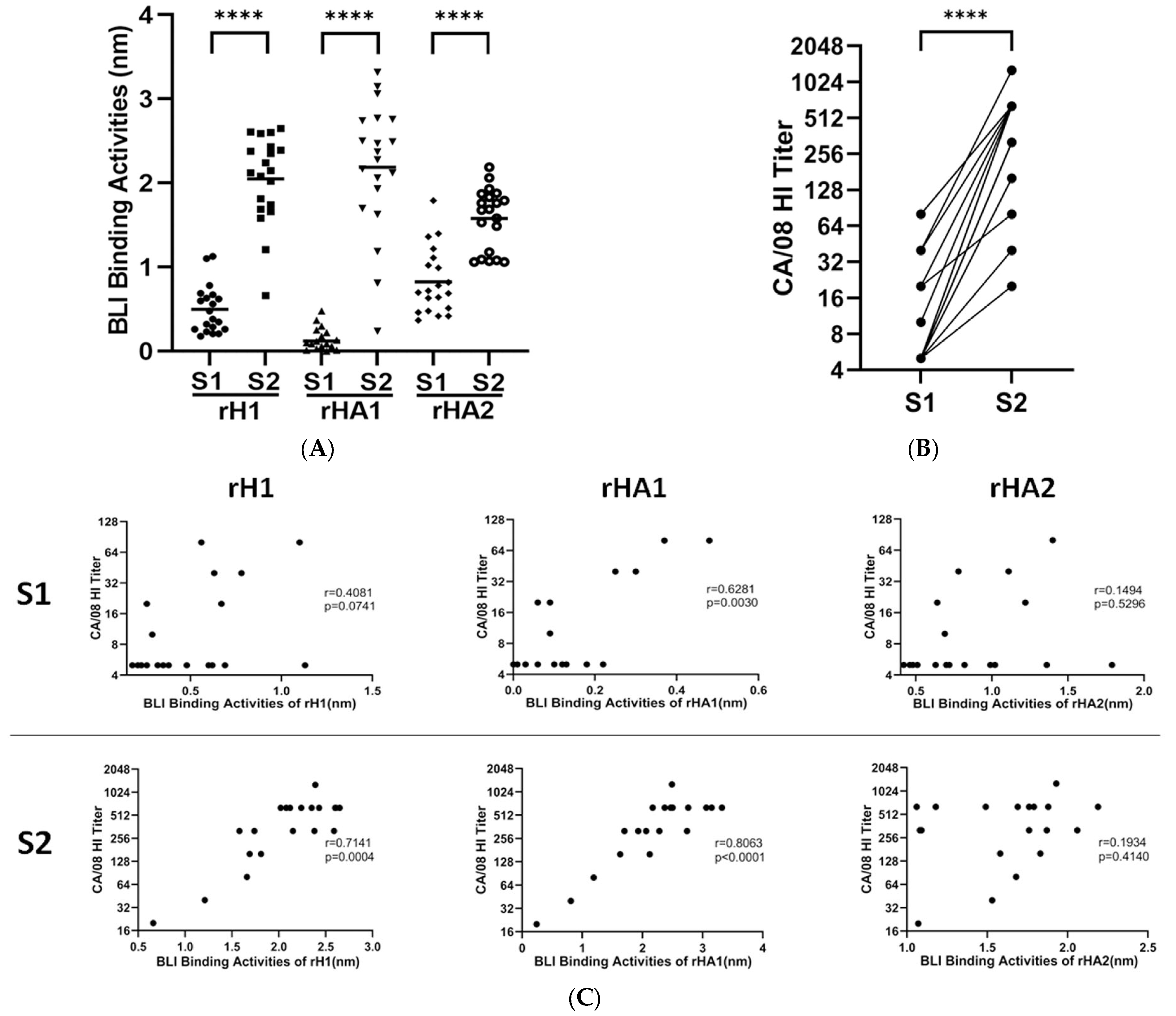
3.1. Assay Development with Human Sera to A(H1N1)pdm09 Vaccine
3.2. Epitope Mapping of Human Sera
3.3. Determination of Dominant Binding Residues Targeted by HI Abs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyon-Plourde, P.; Fakih, I.; Tadount, F.; Fortin, E.; Quach, C. Impact of influenza vaccination on healthcare utilization—A systematic review. Vaccine 2019, 37, 3179–3189. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Palm, A.-K.E.; Utset, H.A.; Huang, M.; Ho, I.Y.; Zheng, N.-Y.; Fitzgerald, T.; Neu, K.E.; Chen, Y.-Q.; Krammer, F.; et al. Monoclonal Antibody Responses after Recombinant Hemagglutinin Vaccine versus Subunit Inactivated Influenza Virus Vaccine: A Comparative Study. J. Virol. 2019, 93, e01150-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, I.A.; Cox, N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 1990, 8, 737–771. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Terajima, M.; Cruz, J.; Co, M.D.T.; Lee, J.-H.; Kaur, K.; Wilson, P.C.; Ennis, F.A. Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J. Virol. 2011, 85, 13463–13467. [Google Scholar] [CrossRef] [Green Version]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 2008, 26 (Suppl. 4), D31–D34. [Google Scholar] [CrossRef]
- Caton, A.J.; Brownlee, G.G.; Yewdell, J.W.; Gerhard, W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 1982, 31, 417–427. [Google Scholar] [CrossRef]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef]
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef]
- Smith, D.J.; Lapedes, A.S.; De Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Koel, B.F.; Burke, D.F.; Bestebroer, T.M.; van der Vliet, S.; Zondag, G.C.M.; Vervaet, G.; Skepner, E.; Lewis, N.S.; Spronken, M.I.J.; Russell, C.A.; et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013, 342, 976–979. [Google Scholar] [CrossRef]
- Huang, K.Y.; Rijal, P.; Schimanski, L.; Powell, T.J.; Lin, T.-Y.; McCauley, J.W.; Daniels, R.S.; Townsend, A.R. Focused antibody response to influenza linked to antigenic drift. J. Clin. Investig. 2015, 125, 2631–2645. [Google Scholar] [CrossRef]
- Li, Y.; Myers, J.L.; Bostick, D.L.; Sullivan, C.B.; Madara, J.; Linderman, S.L.; Liu, Q.; Carter, D.M.; Wrammert, J.; Esposito, S.; et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 2013, 210, 1493–1500. [Google Scholar] [CrossRef]
- Linderman, S.L.; Chambers, B.S.; Zost, S.J.; Parkhouse, K.; Li, Y.; Herrmann, C.; Ellebedy, A.H.; Carter, D.M.; Andrews, S.F.; Zheng, N.-Y.; et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. USA 2014, 111, 15798–15803. [Google Scholar] [CrossRef]
- Chambers, B.S.; Parkhouse, K.; Ross, T.M.; Alby, K.; Hensley, S.E. Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014–2015 Influenza Season. Cell Rep. 2015, 12, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Raymond, D.D.; Stewart, S.M.; Lee, J.; Ferdman, J.; Bajic, G.; Do, K.T.; Ernandes, M.J.; Suphaphiphat, P.; Settembre, E.C.; Dormitzer, P.R.; et al. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat. Med. 2016, 22, 1465–1469. [Google Scholar] [CrossRef]
- Zost, S.J.; Parkhouse, K.; Gumina, M.E.; Kim, K.; Perez, S.D.; Wilson, P.C.; Treanor, J.J.; Sant, A.J.; Cobey, S.; Hensley, S.E. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. USA 2017, 114, 12578–12583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirst, G.K. The Quantitative Determination of Influenza Virus and Antibodies by Means of Red Cell Agglutination. J. Exp. Med. 1942, 75, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Caton, A.J.; Gerhard, W. Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J. Virol. 1986, 57, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Underwood, P.A.; Skehel, J.J.; Wiley, D.C. Receptor-binding characteristics of monoclonal antibody-selected antigenic variants of influenza virus. J. Virol. 1987, 61, 206–208. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bostick, D.L.; Sullivan, C.B.; Myers, J.L.; Griesemer, S.B.; StGeorge, K.; Plotkin, J.B.; Hensley, S.E. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J. Virol. 2013, 87, 9904–9910. [Google Scholar] [CrossRef] [Green Version]
- Hensley, S.E.; Das, S.R.; Bailey, A.L.; Schmidt, L.M.; Hickman, H.D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J.R.; et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 2009, 326, 734–736. [Google Scholar] [CrossRef] [Green Version]
- Medeiros, R.; Escriou, N.; Naffakh, N.; Manuguerra, J.C.; van der Werf, S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology 2001, 289, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Nobusawa, E.; Ishihara, H.; Morishita, T.; Sato, K.; Nakajima, K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): A single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology 2000, 278, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Rowe, T.; Abernathy, R.A.; Hu-Primmer, J.; Thompson, W.W.; Lu, X.; Lim, W.; Fukuda, K.; Cox, N.J.; Katz, J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999, 37, 937–943. [Google Scholar] [CrossRef]
- Lambkin, R.; McLain, L.; Jones, S.E.; Aldridge, S.L.; Dimmock, N.J. Neutralization escape mutants of type A influenza virus are readily selected by antisera from mice immunized with whole virus: A possible mechanism for antigenic drift. J. Gen. Virol. 1994, 75 Pt 12, 3493–3502. [Google Scholar] [CrossRef]
- Sitaras, I.; Kalthoff, D.; Beer, M.; Peeters, B.; de Jong, M.C. Immune escape mutants of Highly Pathogenic Avian Influenza H5N1 selected using polyclonal sera: Identification of key amino acids in the HA protein. PLoS ONE 2014, 9, e84628. [Google Scholar] [CrossRef] [Green Version]
- Schultz-Cherry, S.; Webby, R.J.; Webster, R.G.; Kelso, A.; Barr, I.G.; McCauley, J.W.; Daniels, R.S.; Wang, D.; Shu, Y.; Nobusawa, E.; et al. Influenza Gain-of-Function Experiments: Their Role in Vaccine Virus Recommendation and Pandemic Preparedness. mBio 2014, 5, e02430-14. [Google Scholar] [CrossRef] [Green Version]
- Popova, L.; Smith, K.; West, A.H.; Wilson, P.C.; James, J.A.; Thompson, L.F.; Air, G.M. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE 2012, 7, e41895. [Google Scholar] [CrossRef]
- Dekker, E.L.; Porta, C.; Van Regenmortel, M.H. Limitations of different ELISA procedures for localizing epitopes in viral coat protein subunits. Arch. Virol. 1989, 105, 269–286. [Google Scholar] [CrossRef]
- Gan, S.D.; Patel, K.R. Enzyme immunoassay and enzyme-linked immunosorbent assay. J. Investig. Dermatol. 2013, 133, e12. [Google Scholar] [CrossRef] [Green Version]
- Cabral, T.M.; Berhane, Y.; Schmidt, L.; Tracz, D.M.; Hole, K.; Leith, M.; Corbett, C.R. Development and characterization of neutralizing monoclonal antibodies against the pandemic H1N1 virus (2009). J. Virol. Methods 2012, 183, 25–33. [Google Scholar] [CrossRef]
- Ndifon, W.; Wingreen, N.S.; Levin, S.A. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc. Natl. Acad. Sci. USA 2009, 106, 8701–8706. [Google Scholar] [CrossRef]
- Lubeck, M.; Gerhard, W. Conformational changes at topologically distinct antigenic sites on the influenza A/PR/8/34 virus HA molecule are induced by the binding of monoclonal antibodies. Virology 1982, 118, 1–7. [Google Scholar] [CrossRef]
- Carney, P.J.; Lipatov, A.S.; Monto, A.S.; Donis, R.O.; Stevens, J. Flexible label-free quantitative assay for antibodies to influenza virus hemagglutinins. Clin. Vaccine Immunol. 2010, 17, 1407–1416. [Google Scholar] [CrossRef] [Green Version]
- Abdiche, Y.; Malashock, D.; Pinkerton, A.; Pons, J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008, 377, 209–217. [Google Scholar] [CrossRef]
- Guo, Z.; Wilson, J.R.; York, I.A.; Stevens, J. Biosensor-based epitope mapping of antibodies targeting the hemagglutinin and neuraminidase of influenza A virus. J. Immunol. Methods 2018, 461, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Carney, P.; Stevens, J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010, 2, RRN1152. [Google Scholar] [CrossRef] [PubMed]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.-J.; Kanekiyo, M.; Kong, W.-P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Guo, Z.; Li, Z.-N.; Holiday, C.; Liu, F.; Jefferson, S.; Gross, F.L.; Tzeng, W.-P.; Kumar, A.; York, I.A.; et al. Low quality antibody responses in critically ill patients hospitalized with pandemic influenza A(H1N1)pdm09 virus infection. Sci. Rep. 2022, 12, 14971. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Tzeng, W.-P.; Horner, L.; Kamal, R.P.; Tatum, H.R.; Blanchard, E.G.; Xu, X.; York, I.; Tumpey, T.M.; Katz, J.M.; et al. Influence of Immune Priming and Egg Adaptation in the Vaccine on Antibody Responses to Circulating A(H1N1)pdm09 Viruses After Influenza Vaccination in Adults. J. Infect. Dis. 2018, 218, 1571–1581. [Google Scholar] [CrossRef]
- Li, Z.N.; Liu, F.; Gross, F.L.; Kim, L.; Ferdinands, J.; Carney, P.; Chang, J.; Stevens, J.; Tumpey, T.; Levine, M.Z. Antibody Landscape Analysis following Influenza Vaccination and Natural Infection in Humans with a High-Throughput Multiplex Influenza Antibody Detection Assay. mBio 2021, 12, e02808-20. [Google Scholar] [CrossRef]
- Li, G.M.; Chiu, C.; Wrammert, J.; McCausland, M.; Andrews, S.F.; Zheng, N.-Y.; Lee, J.-H.; Huang, M.; Qu, X.; Edupuganti, S.; et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc. Natl. Acad. Sci. USA 2012, 109, 9047–9052. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Sugawara, K.; Nakauchi, M.; Takahashi, Y.; Onodera, T.; Tsunetsugu-Yokota, Y.; Matsumura, T.; Ato, M.; Kobayashi, K.; Shimotai, Y.; et al. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 influenza virus by using monoclonal antibody escape mutants. J. Virol. 2014, 88, 12364–12373. [Google Scholar] [CrossRef] [Green Version]
- Strengell, M.; Ikonen, N.; Ziegler, T.; Julkunen, I. Minor changes in the hemagglutinin of influenza A(H1N1)2009 virus alter its antigenic properties. PLoS ONE 2011, 6, e25848. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Wang, W.; Zhou, H.; Suguitan, A.L.; Shambaugh, C.; Kim, L.; Zhao, J.; Kemble, G.; Jin, H. Generation of live attenuated novel influenza virus A/California/7/09 (H1N1) vaccines with high yield in embryonated chicken eggs. J. Virol. 2010, 84, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.M.; Rivailler, P.; Hossain, J.; Carney, P.; Balish, A.; Perry, I.; Davis, C.T.; Garten, R.; Shu, B.; Xu, X.; et al. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology 2011, 412, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.T.H.; Behzadi, M.A.; Sun, W.; Freyn, A.W.; Liu, W.-C.; Broecker, F.; Albrecht, R.A.; Bouvier, N.M.; Simon, V.; Nachbagauer, R.; et al. Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J. Clin. Investig. 2018, 128, 4992–4996. [Google Scholar] [CrossRef] [Green Version]


| Mutation | H1 Antigenic Site | Equivalent H3 Antigenic Site | Mutation | H1 Antigenic Site | Equivalent H3 Antigenic Site |
|---|---|---|---|---|---|
| K36E | - a | - | N156D | Sa | B |
| R45G | - | - | K160E | Sa | - |
| G53K | - | E | S162N | Sa | - |
| T72K | Cb | E | K163Q | Sa | - |
| S84N | - | E | D168K | Ca1 | - |
| D97N | - | - | S183P | - | B |
| E115K | - | A | T184K | Sb | B |
| S121K | - | A | S185K | Sb | B |
| N125D | Sa | A | I216T | - | D |
| D127K | - | A | K219E | - | D |
| D127T b | - | A | R221E | Ca2 | D |
| N129D | - | A | D222K | Ca2 | D |
| N129T | - | A | Q223R | - | D/RBS |
| K130E | - | A/RBS | P271K | - | C |
| K130∆ | - | A/RBS | D274K | - | C |
| A139K | Ca2 | A | K283E | - | - |
| K142S | Ca2 | A | S289K | - | - |
| G155K | Sa | B |
| BLI Binding Activities (nm) | Ratio | HI Titer a | Ratio | |||
|---|---|---|---|---|---|---|
| Sample ID | CA/07 rHA1 | Q223R rHA1 | Q223R/WT | MI/45 | Q223R | Q223R/WT |
| #10-S2 | 0.04 (0.02) b | 0.85 (0.03) | 21.25 | 5 | 160 | 32 |
| #14-S2 | 0.22 (0.01) | 1.18 (0.04) | 5.36 | 40 | 640 | 16 |
| #17-S2 | 0.38 (0.02) | 1.02 (0.04) | 2.68 | 20 | 1280 | 64 |
| Sample ID | HI Titer by Virus a | ||||||
|---|---|---|---|---|---|---|---|
| CA/08 | K163Q | 125Gly | 127Gly | K130∆ | MI/45 | Q223R | |
| #1-S2 | 640 | 640 | 640 | 640 | 40 b | 1280 | 1280 |
| #2-S2 | 320 | 640 | 320 | 160 | 1280 | 80 | 640 |
| #3-S2 | 320 | 320 | 320 | 40 | 640 | 320 | 1280 |
| #4-S2 | 160 | 160 | 320 | 80 | 160 | 160 | 640 |
| #5-S2 | 640 | 80 | 320 | 1280 | 1280 | 80 | 320 |
| #6-S2 | 320 | 10 | 40 | 320 | 640 | 5 | 10 |
| #7-S2 | 640 | 1280 | 1280 | 160 | 10 | 1280 | 1280 |
| #8-S2 | 160 | 320 | 320 | 80 | 10 | 320 | 320 |
| #9-S2 | 320 | 320 | 320 | 80 | 5 | 160 | 160 |
| #10-S2 | 20 | 20 | 10 | 10 | 160 | 5 | 160 |
| #11-S2 | 640 | 20 | 80 | 640 | 640 | 20 | 80 |
| #12-S2 | 640 | 320 | 640 | 640 | 640 | 320 | 320 |
| #13-S2 | 320 | 10 | 20 | 320 | 640 | 10 | 40 |
| #14-S2 | 40 | 40 | 80 | 20 | 80 | 40 | 640 |
| #15-S2 | 640 | 160 | 320 | 640 | 640 | 640 | 320 |
| #16-S2 | 640 | 20 | 40 | 640 | 640 | 40 | 160 |
| #17-S2 | 80 | 80 | 80 | 20 | 640 | 20 | 1280 |
| #18-S2 | 1280 | 1280 | 640 | 640 | 40 | 1280 | 1280 |
| #19-S2 | 640 | 10 | 40 | 640 | 1280 | 5 | 5 |
| #20-S2 | 640 | 80 | 160 | 640 | 640 | 40 | 40 |
| Donor Number | Birth Year | Dominant Residues of HA in Bindings by Abs a | |
|---|---|---|---|
| HI | f-AbBA-2 | ||
| #1 | 1991 | K130∆ b | K130E |
| #2 | 1990 | Q223R | K142S, R221E |
| #3 | 1989 | 127Gly, Q223R | K130E, K142S, R221E, Q223R |
| $4 | 1984 | Q223R | K130E, K142S, R221E, Q223R |
| #5 | 1983 | K163Q, Q223R | N125D, S162N, K163Q |
| #6 | 1981 | 125Gly, K163Q | N125D, K163Q |
| #7 | 1981 | 127Gly, K130∆ | K130∆/E |
| #8 | 1981 | K130∆ | K130E |
| #9 | 1981 | 127Gly, K130∆ | K130E |
| #10 | 1980 | Q223R | Q223R |
| #11 | 1979 | 125Gly, K163Q, Q223R | S121K, N125D, 125Gly, K163Q |
| #12 | 1978 | N.D. c | N.D. |
| #13 | 1976 | 125Gly, K163Q, Q223R | N125D, 125Gly, K160E, K163Q |
| #14 | 1974 | Q223R | Q223R |
| #15 | 1971 | K163Q | K163Q |
| #16 | 1970 | 125Gly, K163Q, Q223R | N125D, K163Q |
| #17 | 1970 | 127Gly, Q223R | Q223R |
| #18 | 1969 | K130∆ | K130∆/E |
| #19 | 1969 | 125Gly, K163Q | N125D, K163Q |
| #20 | 1966 | 125Gly, K163Q | N125D, 125Gly, K163Q |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Lu, X.; Carney, P.J.; Chang, J.; Tzeng, W.-p.; York, I.A.; Levine, M.Z.; Stevens, J. Use of Biolayer Interferometry to Identify Dominant Binding Epitopes of Influenza Hemagglutinin Protein of A(H1N1)pdm09 in the Antibody Response to 2010–2011 Influenza Seasonal Vaccine. Vaccines 2023, 11, 1307. https://doi.org/10.3390/vaccines11081307
Guo Z, Lu X, Carney PJ, Chang J, Tzeng W-p, York IA, Levine MZ, Stevens J. Use of Biolayer Interferometry to Identify Dominant Binding Epitopes of Influenza Hemagglutinin Protein of A(H1N1)pdm09 in the Antibody Response to 2010–2011 Influenza Seasonal Vaccine. Vaccines. 2023; 11(8):1307. https://doi.org/10.3390/vaccines11081307
Chicago/Turabian StyleGuo, Zhu, Xiuhua Lu, Paul J. Carney, Jessie Chang, Wen-pin Tzeng, Ian A. York, Min Z. Levine, and James Stevens. 2023. "Use of Biolayer Interferometry to Identify Dominant Binding Epitopes of Influenza Hemagglutinin Protein of A(H1N1)pdm09 in the Antibody Response to 2010–2011 Influenza Seasonal Vaccine" Vaccines 11, no. 8: 1307. https://doi.org/10.3390/vaccines11081307
APA StyleGuo, Z., Lu, X., Carney, P. J., Chang, J., Tzeng, W.-p., York, I. A., Levine, M. Z., & Stevens, J. (2023). Use of Biolayer Interferometry to Identify Dominant Binding Epitopes of Influenza Hemagglutinin Protein of A(H1N1)pdm09 in the Antibody Response to 2010–2011 Influenza Seasonal Vaccine. Vaccines, 11(8), 1307. https://doi.org/10.3390/vaccines11081307







