Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector
Abstract
1. Introduction
1.1. Genomic Organization
1.2. Genotypes
1.3. Clinical Signs and Pathological Lessions
2. Factors Associated with Clinical Manifestation of PCV-Associated Disease (PCVAD)
2.1. Virus-Dependent Factors
2.2. Host-Dependent Factors
2.3. Effect of Immunomodulation
2.4. Management Factors
2.5. Vaccine-Related Factors
2.6. Tramission
2.7. Diagnosis
2.8. Life Cycle
2.9. Prevention and Control
2.10. Antiviral Agents
2.11. Improved Managemental Practices
2.12. Control of Copathogens
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chae, C. A review of porcine circovirus 2-associated syndromes and diseases. Vet. J. 2006, 169, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Cino-Ozuna, A.G.; Henry, S.; Hesse, R.; Nietfeld, J.C.; Bai, J.; Scott, H.M.; Rowland, R.R.R. Characterization of a new disease syndrome associated with porcine circovirus type 2 in previously vaccinated herds. J. Clin. Microbiol. 2011, 49, 2012–2016. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A 1974, 226, 153–167. [Google Scholar]
- Tischer, I.; Gelderblom, H.; Vettermann, W.; Koch, M.A. A very small porcine virus with circular single-stranded DNA. Nature 1982, 295, 64–66. [Google Scholar] [CrossRef]
- Harding, J.C.S. Post-Weaning Multisystemic Wasting Syndrome: Preliminary Epidemiology and Clinical Findings. In Proceedings of the Western Canadian Association of Swine Practitioners, Saskatoon, SK, Canada, 22–25 October 1996. [Google Scholar]
- Clark, E.G. Post-Weaning Multisystemic Wasting Syndrome. In Proceedings of the Western Canadian Association of Swine Practitioners Conference, Saskatoon, SK, Canada, 22–25 October 1996. [Google Scholar]
- Allan, G.M.; McNeilly, F.; Kennedy, S.; Daft, B.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Meehan, B.M.; Adair, B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 1998, 10, 3–10. [Google Scholar] [CrossRef]
- Wellenberg, G.J.; Pesch, S.; Berndsen, F.W.; Steverink, P.J.; Hunneman, W.; Van der Vorst, T.J.; Peperkamp, N.H.; Ohlinger, V.F.; Schippers, R.; Van Oirschot, J.T.; et al. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in The Netherlands. Vet. Q. 2010, 22, 167–172. [Google Scholar] [CrossRef]
- Meehan, B.M.; McNeilly, F.; Todd, D.; Kennedy, S.; Jewhurst, V.A.; Ellis, J.A.; Hassard, L.E.; Clark, E.G.; Haines, D.M.; Allan, G.M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 1998, 79, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Allan, G.M.; McNeilly, F.; Meehan, B.M.; Kennedy, S.; Mackie, D.P.; Ellis, J.A.; Clark, E.G.; Espuna, E.; Saubi, N.; Riera, P.; et al. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 1999, 66, 115–123. [Google Scholar] [CrossRef]
- Olvera, A.; Cortey, M.; Segales, J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.Y.; Opaskornkul, K.; Thanawongnuwech, R.; Arshad, S.S.; Hassan, L.; Ooi, P.T. First molecular detection and complete sequence analysis of porcine circovirus type 3 (PCV3) in Peninsular Malaysia. PLoS ONE 2020, 15, e0235832. [Google Scholar] [CrossRef]
- Prinz, C.; Stillfried, M.; Neubert, L.K.; Denner, J. Detection of PCV3 in German wild boars. Virol. J. 2019, 16, 25. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Serra, F.; Esposito, C.; D’Alessio, N.; Ferrara, G.; Cioffi, B.; Anzalone, A.; Pagnini, U.; De Carlo, E.; Fusco, G.; et al. Prevalence of Infection with Porcine Circovirus Types 2 and 3 in the Wild Boar population in the Campania Region (Southern Italy). Animals 2021, 11, 3215. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Lu, S.-S.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; Zhai, Q.; Chen, Q.-L.; Sun, Y.-W.; Xi, Y. Reservoirs of porcine circoviruses: A mini review. Front. Vet. Sci. 2019, 6, 319. [Google Scholar] [CrossRef]
- Franzo, G.; Delwart, E.; Fux, R.; Hause, B.; Su, S.; Zhou, J.Y.; Segales, J. Genotyping porcine circovirus 3 (PCV3) nowadays: Does it make sense? Viruses 2020, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Nunez, J.I.; Segales, J. Current knowledge on porcine circovirus 3 (PCV3): A novel virus with a yet unknown impact on the swine industry. Front. Vet. Sci. 2018, 5, 315. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, M.; Halbur, P.G.; Gill, M.; Toth, T.E.; Meng, X.-J. Genetic characterization of type 2 porcine circovirus (PCV2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay. J. Clin. Microbiol. 2000, 38, 2494–2503. [Google Scholar] [CrossRef] [PubMed]
- Fux, R.; Sockler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018, 15, 25. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Hu, W.-Q.; Li, J.-Y.; Liu, T.-N.; Zhou, J.-Y.; Opriessnig, T.; Xiao, C.-T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Hamel, A.L.; Lin, L.L.; Nayar, G.P. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 1998, 72, 5262–5267. [Google Scholar] [CrossRef]
- Morozov, I.; Sirinarumitr, T.; Sorden, S.D.; Halbur, P.G.; Morgan, M.K.; Yoon, K.J.; Paul, P.S. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 1998, 36, 2535–2541. [Google Scholar] [CrossRef]
- Palinski, R.; Pineyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Mankertz, A.; Hillenbrand, B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 2001, 279, 429–438. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ge, M.; Ren, J.; Xie, Y.-L.; Zhao, D.; Fan, F.-C.; Song, X.-Q.; Li, M.-X.; Xiao, C.-T. Prevalence and Genetic Analysis of Porcine Circovirus 3 in China From 2019 to 2020. Front. Vet. Sci. 2021, 8, 773912. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; You, D.; Wu, F.; Zhu, L.; Sun, X.-G.; Lai, X.-Y.; Ai, Y.-R.; Zhou, Y.-C.; Xu, Z.W. First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021–2022. Front. Microbiol. 2022, 13, 1052533. [Google Scholar] [CrossRef]
- Todd, D.; Bendinelli, M.; Biagini, P.; Hino, S.; Mankertz, A.; Mishiro, S.; Niel, C.; Okamoto, H.; Raidal, S.; Ritchie, B.W.; et al. Circoviridae. In Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses; Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Elsevier Academic Press: London, UK, 2005; pp. 327–334. [Google Scholar]
- Gillespie, J.; Opriessnig, T.; Meng, X.J.; Pelzer, K.; Buechner-Maxwell, V. Porcine circovirus type 2 and porcine circovirus-associated diseases. J. Vet. Intern. Med. 2009, 23, 1151–1163. [Google Scholar] [CrossRef]
- Huang, L.P.; Lu, Y.H.; Wei, Y.W.; Guo, L.J.; Liu, C.M. Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2. BMC Microbiol. 2011, 11, 188. [Google Scholar] [CrossRef]
- Lefebvre, D.J.; Van Doorsselaere, J.; Delputte, P.L.; Nauwynck, H.J. Recombination of two porcine circovirus type 2 strains. Arch. Virol. 2009, 154, 875–879. [Google Scholar] [CrossRef]
- Liu, Q.; Tikoo, S.K.; Babiuk, L.A. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 2001, 285, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, P.; Morozov, I.; Paul, P.S.; Thangthumniyom, N.; Wajjawalku, W.; Meng, X.J. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 2004, 78, 8135–8145. [Google Scholar] [CrossRef]
- Liu, J.; Chen, I.; Kwang, J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 2005, 79, 8262–8274. [Google Scholar] [CrossRef] [PubMed]
- Saikumar, G.; Das, T. Porcine circovirus. In Recent Advances in Animal Virology; Malik, Y.S., Singh, R.K., Yadav, M.P., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 171–195. [Google Scholar]
- Liu, J.; Chen, I.; Du, Q.; Chua, H.; Kwang, J. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 2006, 80, 5065–5073. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, J.; Zhou, N.; Jin, Y.; Wu, J.; Zhoua, J. Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J. Virol. 2013, 87, 1420–1429. [Google Scholar] [CrossRef]
- Lv, Q.; Guo, K.; Zhang, G.; Zhang, Y. The ORF4 protein of porcine circovirus type 2 antagonizes apoptosis by stabilizing the concentration of ferritin heavy chain through physical interaction. J. Gen. Virol. 2016, 97, 1636–1646. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lv, Q.; Guo, K.; Xu, H.; Wang, T.; Zhang, Y. Identification of putative ORF5 protein of porcine circovirus type 2 and functional analysis of GFP-fused ORF5 protein. PLoS ONE 2015, 10, e0134203. [Google Scholar] [CrossRef]
- Lv, J.; Jiang, Y.; Feng, Q.; Fen, Z.; Sun, Y.; Xu, P.; Hou, Y.; Zhang, X.; Fan, Y.; Xu, X.; et al. Porcine Circovirus Type 2 ORF5 Protein Induces Autophagy to Promote Viral Replication via the PERK-eIF2α-ATF4 and mTOR-ERK1/2-AMPK Signaling Pathways in PK-15 Cells. Front. Microbiol. 2020, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J.; Xu, S.; Cai, S.; Ao, C.; Fang, L.; Xiao, S.; Chen, H.; Jiang, H. Identification and functional analysis of the novel ORF6 protein of porcine circovirus type 2 in vitro. Vet. Res. Commun. 2018, 42, 1–10. [Google Scholar] [CrossRef]
- Franzo, G.; Segal’es, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J. Gen. Virol. 2015, 96, 1830–1841. [Google Scholar] [CrossRef] [PubMed]
- Grau-Roma, L.; Crisci, E.; Sibila, M.; Lopez-Soria, S.; Nofrarias, M.; Cortey, M.; Fraile, L.; Olvera, A.; Segales, J. A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. Vet. Microbiol. 2008, 128, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Carman, S.; McEwen, B.; DeLay, J.; Dreumel, T.V.; Lusis, P.; Cai, H.; Fairles, J. Porcine circovirus-2 associated disease in swine in Ontario (2004 to 2005). Can. Vet. J. 2006, 47, 761–762. [Google Scholar] [PubMed]
- Xiao, C.; Harmon, K.M.; Halbur, P.G.; Opriessnig, T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the U.S. during 2014–2016. Vet. Microbiol. 2016, 197, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Qin, Y.; Zeng, Y.; Ouyang, K.; Chen, Y.; Huang, W.; Wei, Z. Genetic analysis of porcine circovirus type 2 (PCV2) strains between 2002 and 2016 reveals PCV2 mutant predominating in porcine population in Guangxi, China. BMC Vet. Res. 2019, 15, 118. [Google Scholar] [CrossRef]
- Thangthamniyom, N.; Sangthong, P.; Poolperm, P.; Thanantong, N.; Boonsoongnern, A.; Hansoongnern, P.; Semkum, P.; Petcharat, N.; Lekcharoensuk, P. Genetic diversity of porcine circovirus type 2 (PCV2) in Thailand during 2009–2015. Vet. Microbiol. 2017, 208, 239–246. [Google Scholar] [CrossRef]
- Kwon, T.; Lee, D.U.; Yoo, S.J.; Je, S.H.; Shin, J.Y.; Lyoo, Y.S. Genotypic diversity of porcine circovirus type 2 (pcv2) and genotype shift to pcv2d in Korean pig population. Virus Res. 2017, 228, 24–29. [Google Scholar] [CrossRef]
- Ramos, N.; Porley, D.; Mirazo, S.; Castro, G.; Cabrera, K.; Lozano, A.; Arbiza, J. Molecular study of porcine circovirus type 2 in wild boars and domestic pigs in uruguay from 2010 to 2014: Predominance of recombinant circulating strains. Gene 2017, 637, 230–238. [Google Scholar] [CrossRef]
- Dupont, K.; Nielsen, E.O.; Baekbo, P.; Larsen, L.E. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet. Microbiol. 2008, 128, 56–64. [Google Scholar] [CrossRef]
- Franzo, D.; Cortey, M.; de Castro, A.M.M.G.; Piovezan, U.; Szabo, M.P.J.; Drigo, M.; Segales, J.; Richtzenhain, L.J. Genetic characterisation of porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Vet. Microbiol. 2015, 178, 158–162. [Google Scholar] [CrossRef]
- Liu, X.; Wang, F.X.; Zhu, H.W.; Sun, N.; Wu, H. Phylogenetic analysis of porcine circovirus type 2 (PCV2) isolates from China with high homology to PCV2c. Arch. Virol. 2016, 161, 1591–1599. [Google Scholar] [CrossRef]
- Harmon, K.M.; Gauger, P.C.; Zhang, J.; Pineyro, P.E.; Dunn, D.D.; Chriswell, A.J. Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc. 2015, 3, e01315-15. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wei, C.; Dai, A.; Lin, Z.; Fan, K.; Fan, J.; Liu, J.; Luo, M.; Yang, X. Detection of PCV2e strains in Southeast China. Peer J. 2018, 6, e4476. [Google Scholar] [CrossRef]
- Davies, B.; Wang, X.; Dvorak, C.M.T.; Marthaler, D.; Murtaugh, M.P. Diagnostic phylogenetics reveals a new porcine circovirus 2 cluster. Virus Res. 2016, 217, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.-C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef]
- Fanelli, A.; Pellegrini, F.; Camero, M.; Catella, C.; Buonavoglia, D.; Fusco, G.; Martella, M.; Lanave, G. Genetic Diversity of Porcine Circovirus Types 2 and 3 in Wild Boar in Italy. Animals 2022, 12, 953. [Google Scholar] [CrossRef]
- Hu, X.; Chen, Z.; Li, Y.; Ding, Z.; Zeng, Q.; Wan, T.; Huansheng, W. Detection of Porcine Circovirus 1/2/3 and Genetic Analysis of Porcine Circovirus 2 in Wild Boar from Jiangxi Province of China. Animals 2022, 12, 2021. [Google Scholar] [CrossRef]
- Rudova, N.; Buttler, J.; Kovalenko, G.; Sushko, M.; Bolotin, V.; Muzykina, L.; Zinenko, O.; Stegniy, B.; Dunaiev, Y.; Sytiuk, M.; et al. Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine. Viruses 2022, 14, 924. [Google Scholar] [CrossRef]
- Song, S.; Park, G.-N.; Choe, S.; Cha, R.M.; Kim, S.-Y.; Hyun, B.-H.; Park, B.-K.; An, D.-J. Genetic diversity of Porcine Circovirus Isolated from Korean Wild Boars. Pathogens 2020, 9, 457. [Google Scholar] [CrossRef]
- Zhang, F.; Yuan, W.; Li, Z.; Zhang, Y.; Zeng, X.; Zhao, M.; Ye, Y.; Ding, Z.; He, H.; Wu, Q.; et al. Porcine circovirus type 3 in pig farms experiencing diarrhea in Jiangxi, China: Prevalence, genome sequence and pathogenicity. Animals 2020, 10, 2324. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Yoo, S.J.; Park, C.-K.; Lyoo, Y.S. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 2017, 207, 178–180. [Google Scholar] [CrossRef]
- Stadejek, T.; Wozniak, A.; Milek, D.; Biernacka, K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound. Emerg. Dis. 2017, 64, 1350–1353. [Google Scholar] [CrossRef]
- Faccini, S.; Barbieri, I.; Gilioli, A.; Sala, G.; Gibelli, L.R.; Moreno, A.; Sacchi, C.; Rosignoli, C.; Franzini, G.; Nigrelli, A. Detection and genetic characterization of porcine circovirus type 3 in Italy. Transbound. Emerg. Dis. 2017, 64, 1661–1664. [Google Scholar] [CrossRef]
- Tochetto, C.; Lima, D.A.; Varela, A.P.M.; Loiko, M.R.; Paim, W.P.; Scheffer, C.M.; Herpich, J.I.; Cerva, C.; Schmitd, C.; Cibulski, S.P.; et al. Full-genome sequence of porcine circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound. Emerg. Dis. 2018, 65, 5–9. [Google Scholar] [CrossRef]
- Bera, B.C.; Choudhary, M.; Anand, T.; Virmani, N.; Sundaram, K.; Choudhary, B.; Tripathi, B.N. Detection and genetic characterization of porcine circovirus 3 (PCV3) in pigs in India. Transbound. Emerg. Dis. 2020, 67, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Berg, M.; Fossum, C.; Wallgren, P.; Blomstrom, A.L. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes 2018, 54, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Fang, B.; Ma, J.; Liu, Y.; Bu, D.; Zhou, P.; Wang, H.; Jia, K.; Zhang, G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound. Emerg. Dis. 2018, 65, 296–303. [Google Scholar] [CrossRef]
- Dhandapani, G.; Yoon, S.-W.; Noh, J.Y.; Jang, S.S.; Han, S.-H.; Jeong, D.G.; Kim, H.K. Detection of Porcine circovirus 3 from captured wild boars in Korea. Vet. Med. Sci. 2021, 7, 1807–1814. [Google Scholar] [CrossRef]
- Saraiva, G.L.; Vidigal, P.M.P.; Fietto, J.L.R.; Bressan, G.C.; Junior, A.S.; de Almeida, M.R. Evolutionary analysis of porcine circovirus 3 (pcv3) indicates an ancient origin from its current strains and a worldwide dispersion. Virus Genes 2018, 54, 376–384. [Google Scholar] [CrossRef]
- Li, G.; He, W.; Zhu, H.; Bi, Y.; Wang, R.; Xing, G.; Zhang, C.; Zhou, J.; Yuen, K.-Y.; Gao, G.F.; et al. Origin, genetic diversity and evolutionary dynamics of novel porcine circovirus 3. Adv. Sci. 2018, 5, 1800275. [Google Scholar] [CrossRef]
- Li, G.; Wang, H.; Wang, S.; Xing, G.; Zhang, C.; Zhang, W.; Liu, J.; Zhang, J.; Su, S.; Zhou, J. Insights into the genetic and host adaptability of emerging porcine circovirus 3. Virulence 2018, 9, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mai, J.; Lei, B.; Zhang, Y.; Yang, Y.; Wang, N. Structure, antigenic properties, and highly efficient assembly of PCV4 capsid protein. Front. Vet. Sci. 2021, 8, 695466. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-G.; Do, H.-Q.; Huynh, T.-M.-L.; Park, Y.-H.; Park, B.-K.; Chung, H.-C. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound. Emerg. Dis. 2022, 69, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.-Y.; Zhang, L.-H.; Zhang, Y.-H.; Cui, J.-T.; Zhao, L.; Zheng, L.-L.; Chen, H.-Y. Phylogenetic analysis of porcine circovirus 4 in Henan Province of China: A retrospective study from 2011 to 2021. Transbound. Emerg. Dis. 2022, 69, 1890–1901. [Google Scholar] [CrossRef]
- Xu, T.; Hou, C.-Y.; Zhang, Y.-H.; Li, H.-X.; Chen, X.-M.; Pan, J.-J.; Chen, H.-Y. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 2022, 808, 145991. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.M.M.G.; Xiao, C.-T. Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef]
- Opriessnig, T.; Meng, X.J.; Halbur, P.G. Porcine circovirus type 2–associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007, 19, 591–615. [Google Scholar] [CrossRef]
- Segalés, J.; Allan, G.M.; Domingo, M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005, 6, 119–142. [Google Scholar] [CrossRef]
- Segales, J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef]
- Harms, P.A.; Halbur, P.G.; Sorden, S.D. Three cases of porcine respiratory disease complex associated with porcine circovirus type 2 infection. J. Swine Health Prod. 2002, 10, 27–30. Available online: https://www.aasv.org/shap/abstracts/abstract.php?v10n1p27 (accessed on 6 July 2023).
- Harding, J.C.S.; Clark, E.G. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). J. Swine Health Prod. 1997, 5, 201–203. Available online: http://www.aasp.org/shap/issues/v5n5/index.html (accessed on 6 July 2023).
- Segales, J.; Domingo, M. Postweaning multisystemic wasting syndrome (PMWS) in pigs: A review. Vet. Q. 2002, 24, 109–124. [Google Scholar] [CrossRef] [PubMed]
- West, K.H.; Bystrom, J.M.; Wojnarowicz, C.; Shantz, N.; Jacobson, M.; Allan, G.M.; Haines, D.M.; Clark, E.G.; Krakowka, S.; McNeilly, F.; et al. Myocarditis and abortion associated with intrauterine infections of sows with porcine circovirus 2. J. Vet. Diagn. Investig. 1999, 11, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Madson, D.M.; Patterson, A.R.; Ramamoorthy, S.; Pal, N.; Meng, X.J.; Opriessnig, T. Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2. Vet. Pathol. 2019, 46, 707–716. [Google Scholar] [CrossRef]
- Pensaert, M.B.; Sánchez Jr, R.E.; Ladekjaer-Mikkelsen, A.S.; Allan, G.M.; Nauwynck, H.J. Viremia and effect of fetal infection with porcine viruses with special reference to porcine circovirus 2 infection. Vet. Microbiol. 2004, 98, 175–183. [Google Scholar] [CrossRef][Green Version]
- Oropeza-Moe, M.; Delgado, A.J.O.; Framstad, T. Porcine circovirus type 2 associated reproductive failure in a specific pathogen free (SPF) piglet producing herd in Norway: A case report. Porcine Health Manag. 2017, 3, 25. [Google Scholar] [CrossRef]
- O’Connor, B.; Gauvreau, H.; West, K.; Bogdan, J.; Ayroud, M.; Clark, E.G.; Konoby, C.; Allan, G.; Ellis, J.A. Multiple porcine circovirus 2-associated abortions and reproductive failure in a multisite swine production unit. Can. Vet. J. 2001, 42, 551–553. [Google Scholar]
- Halbur, P.G. Porcine respiratory disease. In Proceedings of the 15th International Pig Veterinary Society Congress, Birmingham, UK, 5–9 July 1998; Varley, M.A., Done, S., Thomson, J., Eds.; Scientific Committee of the 15th IPVS Congress: Birmingham, UK, 1998; pp. 1–10. [Google Scholar]
- Thacker, E.L. Porcine respiratory disease complex—What is it and why does it remain a problem? Pig J. 2001, 48, 66–70. [Google Scholar]
- Kim, J.; Chung, H.-K.; Chae, C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet. J. 2003, 166, 251–256. [Google Scholar] [CrossRef]
- Tico, G.; Segales, J.; Martinez, J. The blurred border between porcine circovirus type-2 systemic disease and porcine respiratory disease complex. Vet. Microbiol. 2013, 163, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.S.; Pors, S.E.; Jensen, H.E.; Bille-Hansen, V.; Bisgaard, M.; Flachs, E.M.; Nielsen, O.L. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J. Comp. Pathol. 2010, 143, 120–131. [Google Scholar] [CrossRef]
- Kim, J.; Ha, Y.; Jung, K.; Choi, C.; Chae, C. Enteritis associated with porcine circovirus 2 in pigs. Can. J. Vet. Res. 2004, 68, 218–221. [Google Scholar]
- Baro, J.; Segales, J.; Martinez, J. Porcine circovirus type 2 (PCV2) enteric disease: An independent condition or part of the systemic disease? Vet. Microbiol. 2015, 176, 83–87. [Google Scholar] [CrossRef]
- Drolet, R.; Thibault, S.; D’Allaire, S.; Thomson, J.R.; Done, S.H. Porcine dermatitis and nephropathy syndrome (PDNS): An overview of the disease. J. Swine Health Prod. 1999, 7, 283–285. Available online: https://www.aasp.org/shap.html (accessed on 6 July 2023).
- Segales, J.; Rosell, C.; Domingo, M. Porcine dermatitis and nephropathy syndrome. In Trends in Emerging Viral Infections of Swine; Morilla, A., Yoon, K.-J., Zimmerman, J.J., Eds.; Iowa State Press: Iowa, IA, USA, 2002; pp. 313–318. [Google Scholar]
- Lipej, Z.; Segales, J.; Jemersic, L.; Olvera, A.; Roic, B.; Novosel, D.; Mihaljevic, Z.; Manojlovic, L. First description of postweaning multisystemic wasting syndrome (PMWS) in wild boar (Sus scrofa) in Croatia and phylogenetic analysis of partial PCV2 sequences. Acta Vet. Hung. 2007, 55, 389–404. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.; Billinis, C.; Psychas, V.; Birtsas, P.K.; Sofianidis, G.; Leontides, L.; Knowles, N.J.; Spyrou, V. Detection and Genetic Characterization of Porcine Circovirus 2 Isolates from The First Cases of Postweaning Multisystemic and Wasting Syndrome in Wild Boars in Greece. J. Wildl. Dis 2008, 44, 864–870. [Google Scholar] [CrossRef]
- Castro, A.M.M.G.; Castro Jr, F.G.; Budino, F.E.L.; Baldin, C.M.; Silva, S.O.S.; Brandao, P.E.; Richtzenhain, L.J. Detection of Genetic Characterization of Porcine Circovirus 2 (Pcv2) in Brazilian Wildlife Boars. Braz. J. Microbiol. 2012, 43, 1022–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saporiti, V.; Franzo, G.; Sibila, M.; Segales, J. Porcine circovirus 3 (PCV-3) as a causal agent of disease in swine and a proposal of PCV-3 associated disease case definition. Transbound. Emerg. Dis. 2021, 68, 2936–2948. [Google Scholar] [CrossRef]
- Yuzhakov, A.G.; Raev, S.A.; Alekseev, K.P.; Grebennikova, T.V.; Verkhovsky, O.A.; Zaberezhny, A.D.; Aliper, T.I. First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes 2018, 54, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Lumyai, M.; Kesdangsakonwut, S.; Teankum, K.; Jittimanee, S.; Thanawongnuwech, R. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Vet. Microbiol. 2018, 215, 71–76. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Zhou, X.; Zhang, H.; Hause, B.M.; Lin, T.; Liu, R.; Chen, Q.-L.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol. J. 2017, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Kekarainen, T.; Llorens, A.; Correa-Fiz, F.; Segalés, J. Exploratory metagenomic analyses of periweaning failure-to-thrive syndrome-affected pigs. Vet. Rec. 2019, 184, 25. [Google Scholar] [CrossRef] [PubMed]
- Mora-Diaz, J.; Pineyro, P.; Shen, H.; Schwartz, K.; Vannucci, F.; Li, G.; Arruda, B.; Gimenez-Lirola, L. Isolation of PCV3 from perinatal and reproductive cases of PCV3-associated disease and in vivo characterization of PCV3 replication in CD/CD growing pigs. Viruses 2020, 12, 219. [Google Scholar] [CrossRef]
- Shen, H.; Liu, X.; Zhang, P.; Wang, L.; Liu, Y.; Zhang, L.; Liang, P.; Song, C. Genome characterization of a porcine circovirus type 3 in South China. Transbound. Emerg. Dis. 2018, 65, 264–266. [Google Scholar] [CrossRef]
- Chen, G.H.; Mai, K.J.; Zhou, L.; Wu, R.T.; Tang, X.Y.; Wu, J.L.; He, L.L.; Lan, T.; Xie, Q.M.; Sun, Y.; et al. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound. Emerg. Dis. 2017, 64, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Alomar, J.; Saporiti, V.; Perez, M.; Goncalvez, D.; Sibila, M.; Segales, J. Multisystemic lymphoplasmacytic inflammation associated with PCV3 in wasting pigs. Transbound. Emerg. Dis. 2021, 68, 2969–2974. [Google Scholar] [CrossRef]
- De Conti, E.R.; Resende, T.P.; Marshall-Lund, L.; Rovira, A.; Vannucci, F.A. Histological lesions and replication sites of PCV3 in naturally infected pigs. Animals 2021, 11, 1520. [Google Scholar] [CrossRef]
- Arruda, B.; Pineyro, P.; Derscheid, R.; Hause, B.; Byers, E.; Dion, K.; Long, D.; Sievers, C.; Tangen, J.; Williams, T.; et al. PCV3-associated disease in the United States swine herd. Emerg. Microbes Infect. 2019, 8, 684–698. [Google Scholar] [CrossRef]
- Deb, R.; Sonowal, J.; Sengar, G.S.; Pegu, S.R.; Praharaj, M.R.; Malla, W.A.; Singh, I.; Yadav, A.K.; Rajkhowa, S.; Das, P.J.; et al. Porcine Circovirus type 2 infected myocardial tissue transcriptome signature. Gene 2022, 836, 146670. [Google Scholar] [CrossRef]
- Temeeyasen, G.; Lierman, S.; Arruda, B.L.; Main, R.; Vannucci, F.; Gimenez-Lirola, L.G.; Pineyro, P.E. Pathogenicity and immune response against porcine circovirus type 3 infection in caesarean-derived, colostrum-deprived pigs. J. Gen. Virol. 2021, 102, 001502. [Google Scholar] [CrossRef]
- Klaumann, F.; Correa-Fiz, F.; Sibila, M.; Nunez, J.I.; Segales, J. Infection dynamics of porcine circovirus type 3 in longitudinally sampled pigs from four Spanish farms. Vet. Rec. 2019, 184, 619. [Google Scholar] [CrossRef]
- Sun, J.; Wei, L.; Lu, Z.; Mi, S.; Bao, F.; Guo, H.; Tu, C.; Zhu, Y.; Gong, W. Retrospective study of porcine circovirus 3 infection in China. Transbound. Emerg. Dis. 2018, 65, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Shi, J.; Wu, X.; Peng, Z.; Xin, C.; Zhang, L.; Liu, Y.; Gao, M.; Xu, S.; Han, H.; et al. Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3-positive pigs. Transbound. Emerg. Dis. 2018, 65, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, D.; Wang, J.; Zhu, S.; She, R.; Ren, X.; Tian, J.; Quan, R.; Hou, L.; Li, Z.; et al. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J. Virol. 2019, 93, e02045-18. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Dias-Alves, A.; Cabezon, O.; Mentaberre, G.; Castillo-Contreras, R.; Lopez-Bejar, M.; Casas-Diaz, E.; Sibila, M.; Correa-Fiz, F.; Segales, J. Porcine circovirus 3 is highly prevalent in serum and tissues and may persistently infect wild boar (Sus scrofa scrofa). Transbound. Emerg. Dis. 2019, 66, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.-B.; Zhao, Y.; Cui, J.-T.; Zheng, H.-H.; Xu, T.; Hou, C.-Y.; Wang, Z.-Y.; Li, X.-S.; Zheng, L.-L.; Chen, H.-Y. Molecular detection and phylogenetic analysis of Porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 2021, 68, 276–282. [Google Scholar] [CrossRef]
- Zhang, D.; Bai, C.; Ge, K.; Li, Y.; Gao, W.; Jiang, S.; Wang, Y. Establishment of an SYBR Green-based real-time PCR assay for porcine circovirus type 4 detection. J. Virol. Methods 2020, 285, 113963. [Google Scholar] [CrossRef]
- Afolabi, K.O.; Iweriebor, B.C.; Okoh, A.I.; Obi, L.C. Global status of porcine circovirus type 2 and its associated diseases in Sub-Saharan Africa. Adv. Virol. 2017, 2017, 6807964. [Google Scholar] [CrossRef]
- Sun, J.; Huang, L.; Wei, Y.; Wang, Y.; Chen, D.; Du, W.; Wu, H.; Liu, C. Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch. Virol. 2015, 160, 1339–1344. [Google Scholar] [CrossRef]
- Yi, J.; Liu, C. Molecular characterization of porcine circovirus 2 isolated from diseased pigs co-infected with porcine reproductive and respiratory syndrome virus. Virol. J. 2010, 7, 286. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, R.; She, R.; Mao, J.; Zhao, Y.; Du, F.; Liu, C.; Liu, J.; Cheng, M.; Zhu, R.; et al. Fatal disease associated with swine hepatitis e virus and porcine circovirus 2 co-infection in four weaned pigs in China. BMC Vet. Res. 2015, 11, 77. [Google Scholar] [CrossRef]
- Dorr, P.M.; Baker, R.B.; Almond, G.W.; Wayne, S.R.; Gebreyes, W.A. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. J. Am. Vet. Med. Assoc. 2007, 230, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Dewey, C.E.; Johnston, W.T.; Gould, L.; Whiting, T.L. Postweaning mortality in Manitoba swine. Can. J. Vet. Res. 2006, 70, 161–167. [Google Scholar] [PubMed]
- Watanabe, T.T.N.; Zlotowski, P.; de Oliveira, L.G.; Rolim, V.M.; Gomes, M.J.P.; Snel, G.; Driemeier, D. Rectal stenosis in pigs associated with Salmonella typhimurium and porcine circovirus 2 (PCV2) infection. Pesq. Vet. Bras. 2011, 31, 511–515. [Google Scholar] [CrossRef]
- Pallares, F.J.; Halbur, P.G.; Opriessnig, T.; Sorden, S.D.; Villar, D.; Janke, B.H.; Yaeger, M.J.; Larson, D.J.; Schwartz, K.J.; Yoon, K.J.; et al. Porcine circovirus type 2 (PCV-2) coinfections in US field cases of postweaning multisystemic wasting syndrome (PMWS). J. Vet. Diagn. Investig. 2002, 14, 515–519. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Chen, S.-N.; Wei, Z.-Z.; Zhang, J.-W.; Huang, L.; Lin, T.; Yue, C.; Ran, D.-L.; Yuan, S.-S.; Wei, W.-K.; et al. Co-existence of multiple strains of porcine circovirus type 2 in the same pig from China. Virol. J. 2011, 8, 517. [Google Scholar] [CrossRef]
- Hesse, R.; Sheahan, M.A.; Rowland, R.R.R. Evidence for recombination between PCV2a and PCV2b in the field. Virus Res. 2008, 132, 201–207. [Google Scholar] [CrossRef]
- Li, X.; Qiao, M.; Sun, M.; Tian, K. A Duplex Real-Time PCR Assay for the Simultaneous Detection of Porcine Circovirus 2 and Circovirus 3. Virol. Sin. 2018, 33, 181–186. [Google Scholar] [CrossRef]
- Guo, Z.; Ruan, H.; Qiao, S.; Deng, R.; Zhang, G. Co-infection status of porcine circoviruses (PCV2 and PCV3) and porcine epidemic diarrhea virus (PEDV) in pigs with watery diarrhea in Henan province, central China. Microb. Pathog. 2020, 142, 104047. [Google Scholar] [CrossRef]
- Chae, J.-S.; Choi, K.-S. Genetic diversity of porcine circovirus type 2 from pigs in Republic of Korea. Res. Vet. Sci. 2010, 88, 333–338. [Google Scholar] [CrossRef]
- Opriessnig, T.; Ramamoorthy, S.; Madson, D.M.; Patterson, A.R.; Pal, N.; Carman, S.; Meng, X.-J.; Halbur, P.G. Differences in virulence among porcine circovirus type 2 isolates are unrelated to cluster type 2a or 2b and prior infection provides heterologous protection. J. Gen. Virol. 2008, 89, 2482–2491. [Google Scholar] [CrossRef]
- Trible, B.R.; Rowland, R.R.R. Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis. Virus Res. 2012, 164, 68–77. [Google Scholar] [CrossRef]
- Guo, L.; Fu, Y.; Wang, Y.; Lu, Y.; Wei, Y.; Tang, Q.; Fan, P.; Liu, J.; Zhang, L.; Zhang, F.; et al. A porcine circovirus type 2 (PCV2) mutant with 234 amino acids in capsid protein showed more virulence in vivo, compared with classical PCV2a/b strain. PLoS ONE 2012, 7, e41463. [Google Scholar] [CrossRef]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P.G.; Matzinger, S.R.; Meng, X.-J. Mutant USA strain of porcine circovirus type 2 (mPCV2) exhibits similar virulence to the classical PCV2a and PCV2b strains in caesarean-derived, colostrum deprived pigs. J. Gen. Virol. 2014, 95, 2495–2503. [Google Scholar] [CrossRef]
- Cho, H.; Kang, I.; Oh, T.; Yang, S.; Park, K.H.; Min, K.-D.; Ham, H.J.; Chae, C. Comparative study of the virulence of 3 major Korean porcine circovirus type 2 genotypes (a, b and d). Can. J. Vet. Res. 2020, 84, 235–240. [Google Scholar]
- Suh, J.; Oh, T.; Park, K.; Yang, S.; Cho, H.; Chae, C. A comparison of virulence of three porcine circovirus type 2 (PCV2) genotypes (a, b, and d) in pigs singularly inoculated with PCV2 and dually inoculated with PCV2 and porcine reproductive and respiratory syndrome virus. Pathogens 2021, 10, 891. [Google Scholar] [CrossRef]
- Oh, T.; Suh, J.; Park, K.H.; Yang, S.; Cho, H.; Chae, C. A comparison of pathogenicity and virulence of three porcine circovirus type 2 (PCV2) genotypes (a, b, and d) in pigs singularly inoculated with PCV2 and dually inoculated with Mycoplasma hyopneumoniae and PCV2. Pathogens 2021, 10, 979. [Google Scholar] [CrossRef]
- Wei, R.; Xie, J.; Theuns, S.; Nauwynck, H.J. Changes on the viral capsid surface during the evolution of porcine circovirus type 2 (PCV2) from 2009 till 2018 may lead to a better receptor binding. Virus Evol. 2019, 5, vez026. [Google Scholar] [CrossRef]
- Fenaux, M.; Opriessnig, T.; Halbur, P.G.; Elvinger, F.; Meng, X.J. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J. Virol. 2004, 78, 13440–13446. [Google Scholar] [CrossRef]
- Saha, D.; Lefebvre, D.J.; Ooms, K.; Huang, L.; Delputte, P.L.; Doorsselaere, J.V.; Nauwynck, H.J. Single amino acid mutations in the capsid switch the neutralization phenotype of porcine circovirus 2. J. Gen. Virol. 2012, 93, 1548–1555. [Google Scholar] [CrossRef]
- Cheung, A.K.; Lager, K.M.; Kohutyuk, O.I.; Vincent, A.L.; Henry, S.C.; Baker, R.B.; Rowland, R.R.; Dunham, A.G. Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. Arch. Virol. 2007, 152, 1035–1044. [Google Scholar] [CrossRef]
- Cortey, M.; Pileri, E.; Sibila, M.; Pujols, J.; Balasch, M.; Plana, J.; Segales, J. Genotypic shift of porcine circovirus type 2 from PCV-2a to PCV-2b in Spain from 1985 to 2008. Vet. J. 2011, 187, 363–368. [Google Scholar] [CrossRef]
- Opriessnig, T.; Fenaux, M.; Thomas, P.; Hoogland, M.J.; Rothschild, M.F.; Meng, X.J.; Halbur, P.G. Evidence of breed-dependent differences in susceptibility to porcine circovirus type-2 associated disease and lesions. Vet. Pathol. 2006, 43, 281–293. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Wang, P.; Wang, L.; Sun, Y.; Liu, G.; Zhang, P.; Kang, L.; Jiang, S.; Jiang, Y. RNA-seq analysis reveals genes underlying different disease responses to porcine circovirus type 2 in pigs. PLoS ONE 2016, 11, e0155502. [Google Scholar] [CrossRef]
- Liu, G.; Wang, Y.; Jiang, S.; Sui, M.; Wang, C.; Kang, L.; Sun, Y.; Jiang, Y. Suppression of lymphocyte apoptosis in spleen by CXCL13 after porcine circovirus type 2 infection and regulatory mechanism of CXCL13 expression in pigs. Vet. Res. 2019, 50, 17. [Google Scholar] [CrossRef]
- McKeown, N.E.; Opriessnig, T.; Thomas, P.; Guenette, D.K.; Elvinger, F.; Fenaux, M.; Halbur, P.G.; Meng, X.J. Effects of porcine circovirus type 2 (PCV2) maternal antibodies on experimental infection of piglets with PCV2. Clin. Diagn. Lab. Immunol. 2005, 12, 1347–1351. [Google Scholar] [CrossRef]
- Shen, H.-G.; Loiacono, C.M.; Halbur, P.G.; Opriessnig, T. Age-dependent susceptibility to porcine circovirus type 2 infections is likely associated with declining levels of maternal antibodies. J. Swine Health Prod. 2012, 20, 17–24. Available online: http://www.aasv.org/shap.html (accessed on 6 July 2023).
- Vincent, I.E.; Carrasco, C.P.; Guzylack-Piriou, L.; Herrmann, B.; McNeilly, F.; Allan, G.M.; Summerfield, A.; McCullough, K.C. Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology 2005, 115, 388–398. [Google Scholar] [CrossRef]
- Kekarainen, T.; Montoya, M.; Mateu, E.; Segales, J. Porcine circovirus type 2-induced interleukin-10 modulates recall antigen responses. J. Gen. Virol. 2008, 89, 760–765. [Google Scholar] [CrossRef]
- Rose, N.; Opriessnig, T.; Grasland, B.; Jestin, A. Epidemiology and transmission of porcine circovirus type 2 (PCV2). Virus Res. 2012, 164, 78–89. [Google Scholar] [CrossRef]
- Grau-Roma, L.; Fraile, L.; Segales, J. Recent advances in the epidemiology, diagnosis and control of diseases by porcine circovirus type 2. Vet. J. 2011, 187, 23–32. [Google Scholar] [CrossRef]
- Patterson, R.; Nevel, A.; Diaz, A.V.; Martineau, H.M.; Demmers, T.; Browne, C.; Mavrommatis, B.; Werling, D. Exposure to environmental stressors result in increased viral load and further reduction of production parameters in pigs experimentally infected with PCV2b. Vet. Microbiol. 2015, 177, 261–269. [Google Scholar] [CrossRef]
- Krakowka, S.; Ellis, J.; McNeilly, F.; Waldner, C.; Rings, D.M.; Allan, G. Mycoplasma hyopneumoniae bacterins and porcine circovirus type 2 (PCV2) infection: Induction of postweaning multisystemic wasting syndrome (PMWS) in the gnotobiotic swine model of PCV2-associated disease. Can. Vet. J. 2007, 48, 716–724. [Google Scholar]
- Ha, Y.; Lee, Y.H.; Ahn, K.-K.; Kim, B.; Chae, C. Reproduction of postweaning multisystemic wasting syndrome in pigs by prenatal porcine circovirus 2 infection and postnatal porcine parvovirus infection or immunostimulation. Vet. Pathol. 2008, 45, 842–848. [Google Scholar] [CrossRef]
- Opriessnig, T.; Halbur, P.G.; Yu, S.; Thacker, E.L.; Fenaux, M.; Meng, X.J. Effects of the timing of the administration of Mycoplasma hyopneumoniae bacterin on the development of lesions associated with porcine circovirus type 2. Vet. Rec. 2006, 158, 149–154. [Google Scholar] [CrossRef]
- Resendes, A.; Segales, J.; Balasch, M.; Calsamiglia, M.; Sibila, M.; Ellerbrok, H.; Mateu, E.; Plana-Duran, J.; Mankertz, A.; Domingo, M. Lack of an effect of a commercial vaccine adjuvant on the development of postweaning multisystemic wasting syndrome (PMWS) in porcine circovirus type 2 (PCV2) experimentally infected conventional pigs. Vet. Res. 2004, 35, 83–90. [Google Scholar] [CrossRef][Green Version]
- Hoogland, M.J.; Opriessnig, T.; Halbur, P.G. Effects of adjuvants on porcine circovirus type-2 associated lesions. J. Swine Health Prod. 2006, 14, 133–139. [Google Scholar]
- Allan, G.M.; McNeilly, F.; Kennedy, S.; Meehan, B.M.; Ellis, J.A.; Krakowka, S. Immunostimulation, PCV-2 and PMWS. Vet. Rec. 2000, 147, 170–171. [Google Scholar] [PubMed]
- Kyriakis, S.C.; Saoulidis, K.; Lekkas, S.; Miliotis, C.C.; Papoutsis, P.A.; Kennedy, S. The effects of immuno-modulation on the clinical and pathological expression of postweaning multisystemic wasting syndrome. J. Comp. Pathol. 2002, 126, 38–46. [Google Scholar] [CrossRef]
- Grasland, B.; Loizel, C.; Blanchard, P.; Oger, A.; Nignol, A.-C.; Bigarre, L.; Morvan, H.; Cariolet, R.; Jestin, A. Reproduction of PMWS in immunostimulated SPF piglets transfected with infectious cloned genomic DNA of type 2 porcine circovirus. Vet. Res. 2005, 36, 685–697. [Google Scholar] [CrossRef]
- Ladekjaer-Mikkelsen, A.S.; Nielsen, J.; Stadejek, T.; Storgaard, T.; Krakowka, S.; Ellis, J.; McNeilly, F.; Allan, G.; Botner, A. Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week old piglets experimentally infected with porcine circovirus type 2 (PCV2). Vet. Microbiol. 2002, 89, 97–114. [Google Scholar] [CrossRef]
- Haruna, J.; Hanna, P.; Hurnik, D.; Ikede, B.; Miller, L.; Yason, C. The role of immunostimulation in the development of postweaning multisystemic wasting syndrome in pigs under field conditions. Can. J. Vet. Res. 2006, 70, 269. [Google Scholar] [PubMed]
- Andraud, M.; Grasland, B.; Durand, B.; Cariolet, R.; Jestin, A.; Madec, F.; Rose, N. Quantification of porcine circovirus type 2 (PCV-2) within- and between-pen transmission in pigs. Vet. Res. 2008, 39, 1. [Google Scholar] [CrossRef]
- Lopez-Lorenzo, G.; Diaz-Cao, J.M.; Prieto, A.; Lopez-Novo, C.; Lopez, C.M.; Diaz, P.; Rodriguez-Vega, V.; Diez-Banos, P.; Fernandez, G. Environmental distribution of porcine circovirus type 2 (PCV2) in swine herds with natural infection. Sci. Rep. 2019, 9, 14816. [Google Scholar] [CrossRef]
- Bolin, S.R.; Stoffregen, W.C.; Nayar, G.P.S.; Hamel, A.L. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J. Vet. Diagn. Investig. 2001, 13, 185–194. [Google Scholar] [CrossRef]
- Albina, E.; Truong, C.; Hutet, E.; Blanchard, P.; Cariolet, R.; L’Hospitalier, R.; Mahe, D.; Allee, C.; Morvan, H.; Amenna, N.; et al. An experimental model for post-weaning multisystemic wasting syndrome (PMWS) in growing piglets. J. Comp. Pathol. 2001, 125, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Verreault, D.; Letourneau, V.; Gendron, L.; Masse, D.; Gagnon, C.A.; Duchaine, C. Airborne porcine circovirus in Canadian swine confinement buildings. Vet. Microbiol. 2010, 141, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Dupont, K.; Hjulsager, C.K.; Kristensen, C.S.; Baekbo, P.; Larsen, L.E. Transmission of different variants of PCV2 and viral dynamics in a research facility with pigs mingled from PMWS-affected herds and non-affected herds. Vet. Microbiol. 2009, 139, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Song, D.S.; Kim, S.Y.; Lyoo, K.S.; Park, B.K. Detection of porcine circovirus type 2 in feces of pigs with or without enteric disease by polymerase chain reaction. J. Vet. Diagn. Investig. 2003, 15, 369–373. [Google Scholar] [CrossRef]
- Kristensen, C.S.; Baekbo, P.; Bille-Hansen, V.; Botner, A.; Vigre, H.; Enoe, C.; Larsen, L.E. Induction of porcine post-weaning multisystemic wasting syndrome (PMWS) in pigs from PMWS unaffected herds following mingling with pigs from PMWS-affected herds. Vet. Microbiol. 2009, 138, 244–250. [Google Scholar] [CrossRef]
- Kristensen, C.S.; Hjulsager, C.K.; Vestergaard, K.; Dupont, K.; Bille-Hansen, V.; Enoe, C.; Jorsal, S.E.; Baekbo, P.; Larsen, L.E. Experimental Airborne Transmission of Porcine Postweaning Multisystemic Wasting Syndrome. J. Pathog. 2013, 2013, 534342. [Google Scholar] [CrossRef]
- Segales, J.; Calsamiglia, M.; Olvera, A.; Sibila, M.; Badiella, L.; Domingo, M. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, trachea-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS). Vet. Microbiol. 2005, 111, 223–229. [Google Scholar] [CrossRef]
- Nielsen, G.B.; Nielsen, J.P.; Haugegaard, J.; Leth, S.C.; Larsen, L.E.; Kristensen, C.S.; Pedersen, K.S.; Stege, H.; Hjulsager, C.K.; Houe, H. Comparison of serum pools and oral fluid samples for detection of porcine circovirus type 2 by quantitative real-time PCR in finisher pigs. BMC Porcine Health Manag. 2018, 4, 2. [Google Scholar] [CrossRef]
- Ha, Y.; Ahn, K.K.; Kim, B.; Cho, K.-D.; Lee, B.H.; Oh, Y.-S.; Kim, S.-H.; Chae, C. Evidence of shedding of porcine circovirus type 2 in milk from experimentally infected sows. Res. Vet. Sci. 2009, 86, 108–110. [Google Scholar] [CrossRef]
- Madson, D.M.; Ramamoorthy, S.; Kuster, C.; Pal, N.; Meng, X.-J.; Halbur, P.G.; Opriessnig, T. Characterization of shedding patterns of porcine circovirus types 2a and 2b in experimentally inoculated mature boars. J. Vet. Diagn. Investig. 2008, 20, 725–734. [Google Scholar] [CrossRef]
- Shibata, I.; Okuda, Y.; Kitajima, K.; Asai, T. Shedding of porcine circovirus into colostrum of sows. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 278–280. [Google Scholar] [CrossRef]
- Park, J.-S.; Ha, Y.; Kwon, B.; Cho, K.D.; Lee, B.H.; Chae, C. Detection of porcine circovirus 2 in mammary and other tissues from experimentally infected sows. J. Comp. Pathol. 2009, 140, 208–211. [Google Scholar] [CrossRef]
- Gerber, P.F.; Garrocho, F.M.; Lana, A.M.Q.; Lobato, Z.I.P. Serum antibodies and shedding of infectious porcine circovirus 2 into colostrum and milk of vaccinated and unvaccinated naturally infected sows. Vet. J. 2011, 188, 240–242. [Google Scholar] [CrossRef]
- Gava, D.; Zanella, E.L.; Mores, N.; Ciacci-Zanella, J.R. Transmission of porcine circovirus 2 (PCV2) by semen and viral distribution in different piglet tissues. Pesq. Vet. Bras. 2008, 28, 70–76. [Google Scholar] [CrossRef][Green Version]
- Madson, D.M.; Ramamoorthy, S.; Kuster, C.; Pal, N.; Meng, X.-J.; Halbur, P.G.; Opriessnig, T. Infectivity of porcine circovirus type 2 DNA in semen from experimentally infected boars. Vet. Res. 2009, 40, 10. [Google Scholar] [CrossRef]
- Sarli, G.; Morandi, F.; Panarese, S.; Bacci, B.; Ferrara, D.; Bianco, C.; Fusaro, L.; Bacci, M.L.; Galeati, G.; Dottori, M.; et al. Reproduction in porcine circovirus type 2 (PCV2) seropositive gilts inseminated with PCV2b spiked semen. Acta Vet. Scand. 2012, 54, 51. [Google Scholar] [CrossRef]
- Park, J.-S.; Kim, J.; Ha, Y.; Jung, K.; Choi, C.; Lim, J.-K.; Kim, S.-H.; Chae, C. Birth abnormalities in pregnant sows infected intranasally with porcine circovirus 2. J. Comp. Pathol. 2005, 132, 139–144. [Google Scholar] [CrossRef]
- Farnham, M.W.; Choi, Y.K.; Goyal, S.M.; Joo, H.S. Isolation and characterization of porcine circovirus type-2 from sera and stillborn fetuses. Can. J. Vet. Res. 2003, 67, 108. [Google Scholar]
- Madson, D.M.; Patterson, A.R.; Ramamoorthy, S.; Pal, N.; Meng, X.J.; Opriessnig, T. Effect of natural or vaccine-induced porcine circovirus type 2 (PCV2) immunity on fetal infection after artificial insemination with PCV2 spiked semen. Theriogenology 2009, 72, 747–754. [Google Scholar] [CrossRef]
- Giudici, S.D.; Lo Presti, A.; Bonelli, P.; Angioi, P.P.; Sanna, G.; Zinellu, S.; Balzano, F.; Salis, F.; Ciccozzi, M.; Oggiano, A. Phylogenetic analysis of porcine circovirus type 2 in Sardinia, Italy, shows genotype 2d circulation among domestic pigs and wild boars. Infect. Genet. Evol. 2019, 71, 189–196. [Google Scholar] [CrossRef]
- Franzo, G.; Tinello, S.; Grassi, L.; Tucciarone, C.M.; Legnardi, M.; Cecchinato, M.; Dotto, G.; Mondin, A.; Martini, M.; Pasotto, D.; et al. Free to Circulate: An Update on the Epidemiological Dynamics of Porcine Circovirus 2 (PCV2) in Italy Reveals the Role of Local Spreading, Wild Populations, and Foreign Countries. Pathogens 2022, 9, 221. [Google Scholar] [CrossRef]
- Yang, X.; Hou, L.; Ye, J.; He, Q.; Cao, S. Detection of porcine circovirus type 2 (PCV2) in mosquitoes from pig farms by PCR. Pak. Vet. J. 2012, 32, 134–135. [Google Scholar]
- Blunt, R.; McOrist, S.; McKillen, J.; McNair, I.; Jiang, T.; Mellits, K. House fly vector for porcine circovirus 2b on commercial pig farms. Vet. Microbiol. 2011, 149, 452–455. [Google Scholar] [CrossRef]
- Schwarz, L.; Strauss, A.; Loncaric, I.; Spergser, J.; Auer, A.; Rumenapf, T.; Ladinig, A. The stable fly (Stomoxys calcitrans) as a possible vector transmitting pathogens in Austrian pig farms. Microorganisms 2020, 8, 1476. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Zhou, X.; Lin, T.; Zhang, H.; Wen, X.-H.; Zhou, X.-R.; Jia, C.-L.; Tu, D.; Zhu, X.-L.; Chen, Q.-L.; et al. Reappearance of buffalo-origin-like porcine circovirus type 2 strains in swine herds in southern China. New Microbes New Infect. 2017, 17, 98–100. [Google Scholar] [CrossRef]
- Song, T.; Zhang, S.; Hao, J.; Xin, S.; Hui, W.; Tang, M.; Li, W.; Tian, R.; Liu, X.; Rui, P.; et al. First detection and genetic analysis of fox-origin porcine circovirus type 2. Transbound. Emerg. Dis. 2018, 66, 1–6. [Google Scholar] [CrossRef]
- Wang, G.-S.; Sun, N.; Tian, F.-L.; Wen, Y.-J.; Xu, C.; Li, J.; Chen, Q.; Wang, J.-B. Genetic analysis of porcine circovirus type 2 from dead minks. J. Gen. Virol. 2016, 97, 2316–2322. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Chen, S.-N.; Liu, W.; Li, X.-P.; Deng, S.-F.; Wen, X.-H.; Luo, M.-L.; Lv, D.-H.; Wei, W.-K.; Chen, R.-A. Molecular detection and genome characterization of porcine circovirus type 2 in rats captured on commercial swine farms. Arch. Virol. 2016, 161, 3237–3244. [Google Scholar] [CrossRef]
- Song, T.; Hao, J.; Zhang, R.; Tang, M.; Li, W.; Hui, W.; Fu, Q.; Wang, C.; Xin, S.; Zhang, S.; et al. First detection and phylogenetic analysis of porcine circovirus type 2 in raccoon dogs. BMC Vet. Res. 2019, 15, 107. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, X.; Niu, G.; Ren, L. Recent progress on epidemiology and pathobiology of porcine circovirus 3. Viruses 2021, 13, 1994. [Google Scholar] [CrossRef]
- Grassi, L.; Tagliapietra, V.; Rizzoli, A.; Martini, M.; Drigo, M.; Franzo, G.; Menandro, M.L. Lack of evidence on the susceptibility of ticks and wild rodent species to PCV3 infection. Pathogens 2020, 9, 682. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Vargas-Pinto, M.A.; Mogollon, J.D.; Jaime, J. Field infection of a gilt and its litter demonstrated vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (PCV3). BMC Vet. Res. 2021, 17, 150. [Google Scholar] [CrossRef]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Kesdangsakonwut, S.; Tummaruk, P.; Teankum, K.; Assavacheep, P.; Jittimanee, S.; et al. Porcine circovirus type 3 (PCV3) shedding in sow colostrum. Vet. Microbiol. 2018, 220, 12–17. [Google Scholar] [CrossRef]
- Saporiti, V.; Martorell, S.; Cruz, T.F.; Klaumann, F.; Fiz, F.C.; Balasch, M.; Sibila, M.; Segales, J. Frequency of detection and phylogenetic analysis of porcine circovirus 3 (PCV3) in healthy primiparous and multiparous sows and their mummified fetuses and stillborn. Pathogens 2020, 9, 533. [Google Scholar] [CrossRef]
- Kruger, L.; Langin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.-M.; et al. Transmission of porcine circovirus 3 (PCV3) by xenotransplantation of pig hearts into baboons. Viruses 2019, 11, 650. [Google Scholar] [CrossRef]
- Wang, T.; Chai, W.; Wang, Y.; Liu, W.; Huang, Z.; Chen, L.; Guo, R.; Dong, Y.; Liu, M.; Zheng, Q.; et al. First detection and phylogenetic analysis of porcine circovirus 3 in female donkeys with reproductive disorders. BMC Vet. Res. 2021, 17, 308. [Google Scholar] [CrossRef]
- Kim, J.; Jung, K.; Chae, C. Prevalence of porcine circovirus type 2 in aborted fetuses and stillborn piglets. Vet. Rec. 2004, 155, 489–492. Available online: http://veterinaryrecord.bmj.com/ (accessed on 6 July 2023). [CrossRef]
- Segales, J.; Rosell, C.; Domingo, M. Pathological findings associated with naturally acquired porcine circovirus type 2 associated disease. Vet. Microbiol. 2004, 98, 137–149. [Google Scholar] [CrossRef]
- Baekbo, P.; Kristensen, C.S.; Larsen, L.E. Porcine circovirus diseases: A review of PMWS. Transbound. Emerg. Dis. 2012, 59, 376. [Google Scholar] [CrossRef]
- Caprioli, A.; McNeilly, F.; McNair, I.; Lagan-Tregaskis, P.; Ellis, J.; Krakowka, S.; McKillen, J.; Ostanello, F.; Allan, G. PCR detection of porcine circovirus type 2 (PCV2) DNA in blood, tonsillar and faecal swabs from experimentally infected pigs. Res. Vet. Sci. 2006, 81, 287–292. [Google Scholar] [CrossRef]
- Tan, C.Y.; Lin, C.-N.; Ooi, P.-T. What do we know about porcine circovirus 3 (PCV3) diagnosis so far? Transbound. Emerg. Dis. 2021, 68, 2915–2935. [Google Scholar] [CrossRef]
- Seo, H.W.; Han, K.; Oh, Y.; Kang, I.; Park, C.; Joo, H.E.; Kim, S.-H.; Lee, B.-H.; Chae, C. Evaluation of commercial polyclonal—And monoclonal-antibody-based immunohistochemical tests for 2 genotypes of Porcine circovirus type 2 and comparison with in-situ hybridization assays. Can. J. Vet. Res. 2014, 78, 233–236. [Google Scholar]
- McNeilly, F.; Kennedy, S.; Moffett, D.; Meehan, B.M.; Foster, J.C.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Adair, B.M.; Allan, G.M. A comparison of in situ hybridization and immunohistochemistry for the detection of a new porcine circovirus in formalin-fixed tissues from pigs with post-weaning multisystemic wasting syndrome (PMWS). J. Virol. Methods 1999, 80, 123–128. [Google Scholar] [CrossRef]
- Szczotka, A.; Stadejek, T.; Pejsak, Z. A comparison of immunohistochemistry and in situ hybridization for the detection of porcine circovirus type 2 in pigs. Pol. J. Vet. Sci. 2011, 14, 565–571. [Google Scholar] [CrossRef]
- Kim, J.; Chae, C. A comparison of virus isolation, polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine circovirus 2 and porcine parvovirus in experimentally and naturally coinfected pigs. J. Vet. Diagn. Investig. 2004, 16, 45–50. [Google Scholar] [CrossRef]
- Kim, D.; Ha, Y.; Lee, Y.-H.; Chae, S.; Lee, K.; Han, K.; Kim, J.; Lee, J.-H.; Kim, S.-H.; Hwang, K.-K.; et al. Comparative study of in situ hybridization and immunohistochemistry for the detection of porcine circovirus 2 in formalin-fixed, paraffin-embedded tissues. J. Vet. Med. Sci. 2009, 71, 1001–1004. [Google Scholar] [CrossRef]
- Sorden, S.D.; Harms, P.A.; Nawagitgul, P.; Cavanaugh, D.; Paul, P.S. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J. Vet. Diagn. Investig. 1999, 11, 528–530. [Google Scholar] [CrossRef]
- Li, X.; Bai, Y.; Zhang, H.; Zheng, D.; Wang, T.; Wang, Y.; Deng, J.; Sun, Z.; Tian, K. Production of a monoclonal antibody against Porcine circovirus type 3 cap protein. J. Virol. Methods 2018, 261, 10–13. [Google Scholar] [CrossRef]
- Ramos-Vara, J.A. Technical aspects of immunochemistry. Vet. Pathol. 2005, 42, 405–426. [Google Scholar] [CrossRef]
- Webster, J.D.; Miller, M.A.; DuSold, D.; Ramos-Vara, J. Effects of prolonged formalin fixation on diagnostic immunohistochemistry in domestic animals. J. Histochem. Cytochem. 2009, 57, 753–761. [Google Scholar] [CrossRef]
- Kim, J.; Chae, C. Differentiation of porcine circovirus 1 and 2 in formalin-fixed, paraffin-wax-embedded tissues from pigs with postweaning multisystemic wasting syndrome by in-situ hybridisation. Res. Vet. Sci. 2001, 70, 265–269. [Google Scholar] [CrossRef]
- Ha, Y.; Jung, K.; Choi, C.; Hwang, K.-K.; Chae, C. Synthetic peptide-derived antibody-based immunohistochemistry for the detection of porcine circovirus 2 in pigs with postweaning multisystemic wasting syndrome. J. Comp. Pathol. 2005, 133, 201–204. [Google Scholar] [CrossRef]
- Yang, Z.-Z.; Habib, M.; Shuai, J.-B.; Fang, W.-H. Detection of PCV2 DNA by SYBR Green I-based quantitative PCR. J. Zhejiang Univ. Sci. B 2007, 8, 162–169. [Google Scholar] [CrossRef]
- Zhao, K.; Han, F.; Zou, Y.; Zhu, L.; Li, C.; Xu, Y.; Zhang, C.; Tan, F.; Wang, J.; Tao, S.; et al. Rapid detection of porcine circovirus type 2 using a TaqMan-based real-time PCR. Virol. J. 2010, 7, 374. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Wang, J.; Liu, L.; Pang, X.; Yuan, W. Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J. Virol. Methods 2017, 248, 177–180. [Google Scholar] [CrossRef]
- Chen, G.-H.; Tang, X.-Y.; Sun, Y.; Zhou, L.; Li, D.; Bai, Y.; Mai, K.-J.; Li, Y.-Y.; Wu, Q.-W.; Ma, J.-Y. Development of a SYBR green-based real-time quantitative PCR assay to detect PCV3 in pigs. J. Virol. Methods 2018, 251, 129–132. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 2020, 68, 1615–1624. [Google Scholar] [CrossRef]
- Yang, K.; Jiao, Z.; Zhou, D.; Guo, R.; Duan, Z.; Tian, Y. Development of a multiplex PCR to detect and discriminate porcine circoviruses in clinical samples. BMC Infect. Dis. 2019, 19, 778. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Song, J.K.; Shin, S.; Kim, H. Comparison of multiplex real-time PCR and PCR-reverse blot hybridization assays for the direct and rapid detection of porcine circovirus type 2 genotypes. Front. Vet. Sci. 2020, 7, 200. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, H.; Chen, S.; Yang, M.; Yan, Q.; Wen, C.; Hao, Z.; Yan, Y.; Sun, Y.; Hu, J.; et al. Sensitive detection of Porcine circovirus-2 by droplet digital polymerase chain reaction. J. Vet. Diagn. Investig. 2015, 27, 784–788. [Google Scholar] [CrossRef]
- Liu, Y.; Meng, H.; Shi, L.; Li, L. Sensitive detection of porcine circovirus 3 by droplet digital PCR. J. Vet. Diagn. Investig. 2019, 31, 604–607. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, Y.; Liang, L.; Li, J.; Cui, S. Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in China detected by duplex nanoparticle-assisted PCR. Infect. Genet. Evol. 2018, 60, 1–6. [Google Scholar] [CrossRef]
- Pegu, S.R.; Deb, R.; Das, P.J.; Sengar, G.S.; Yadav, A.K.; Rajkhowa, S.; Paul, S.; Gupta, V.K. Development of multiplex PCR assay for simultaneous detection of African swine fever, porcine circo and porcine parvo viral infection from clinical samples. Anim. Biotechnol. 2022, 28, 1–8. [Google Scholar] [CrossRef]
- Ge, M.; Luo, W.; Jiang, D.; Li, R.; Zhao, W.; Chen, G.; Yang, X. Development and application of a double-antigen sandwich enzyme-linked immunosorbent assay for detection of antibodies to porcine circovirus 2. Clin. Vaccine Immunol. 2012, 19, 1480–1486. [Google Scholar] [CrossRef]
- Yao, L.; Li, C.; Wang, J.; Cheng, Y.; Ghonaim, A.H.; Sun, Q.; Yu, X.; Niu, W.; Fan, S.; He, Q. Development of an indirect immunofluorescence assay for PCV3 antibody detection based on capsid protein. Anim. Dis. 2021, 1, 11. [Google Scholar] [CrossRef]
- Deng, J.; Li, X.; Zheng, D.-D.; Wang, Y.; Chen, L.; Song, H. Establishment and application of an indirect ELISA for porcine circovirus 3. Arch. Virol. 2018, 163, 479–482. [Google Scholar] [CrossRef]
- Zhou, S.; Han, S.; Shi, J.; Wu, J.; Yuan, X.; Cong, X.; Xu, S.; Wu, X.; Li, J.; Wang, J. Loop-mediated isothermal amplification for detection of porcine circovirus type 2. Virol. J. 2011, 8, 497. [Google Scholar] [CrossRef]
- Park, Y.-R.; Kim, H.-R.; Kim, S.-H.; Lee, K.-K.; Lyoo, Y.S.; Yeo, S.-G.; Park, C.-K. Loop-mediated isothermal amplification assay for the rapid and visual detection of novel porcine circovirus 3. J. Virol. Methods 2018, 253, 26–30. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Sun, Y.; Cong, G.; Li, Y.; Zhang, Z. Development of isothermal recombinase polymerase amplification assay for rapid detection of porcine circovirus type 2. BioMed Res. Int. 2017, 2017, 8403642. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Zhang, R.; Han, Q.; Wang, J.; Liu, L.; Li, R.; Yuan, W. Recombinase polymerase amplification assay for rapid detection of porcine circovirus 3. Mol. Cell. Probes 2017, 36, 58–61. [Google Scholar] [CrossRef]
- Petrini, S.; Barocci, S.; Gavaudan, S.; Villa, R.; Briscolini, S.; Sabbatini, M.; Mattozzi, C.; Barchiesi, F.; Salamida, S.; Ferrari, M.; et al. Detection of porcine circovirus type 2 (PCV2) from wild boars in central Italy. Eur. J. Wildl. Res. 2009, 55, 465–469. [Google Scholar] [CrossRef]
- Bi, M.; Li, X.; Zhai, W.; Yin, B.; Tian, K.; Mo, X. Structural insight into the type-specific epitope of porcine circovirus type 3. Biosci. Rep. 2020, 40, BSR20201109. [Google Scholar] [CrossRef]
- Lian, Z.; Liu, J.; Liu, P.; Zhu, Z.; Yao, X.; Yuan, L.; Hu, D.; Jiang, Y.; Chen, C.; Chen, N.; et al. Development and application of an indirect ELISA for the detection of antibody to porcine circovirus 4 in pigs. Transbound. Emerg. Dis. 2021, 68, 2975–2979. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Kim, H.-R.; Park, J.-H.; Kwon, N.-Y.; Kim, J.-M.; Kim, J.-K.; Park, J.; Lee, K.-K.; Kim, S.-H.; Kim, W.-I.; et al. Detection of a novel porcine circovirus 4 in Korean pig herds using a loop-mediated isothermal amplification assay. J. Virol. Methods 2022, 299, 114350. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Y.; Li, C.; Yang, K.; Li, Z.; Shang, W.; Song, X.; Shao, Y.; Qi, K.; Tu, J. Rapid detection of porcine circovirus type 4 via multienzyme isothermal rapid amplification. Front. Vet. Sci. 2022, 9, 949172. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, X.; Yin, D.; Cai, C.; Liu, H.; Yang, Y.; Guo, Z.; Yin, L.; Shen, X.; Dai, Y.; et al. Rapid and Easy-Read Porcine Circovirus Type 4 Detection with CRISPR-Cas13a-Based Lateral Flow Strip. Microorganisms 2023, 11, 354. [Google Scholar] [CrossRef]
- Finsterbusch, T.; Steinfeldt, T.; Doberstein, K.; Rodner, C.; Mankertz, A. Interaction of the replication proteins and the capsid protein of porcine circovirus type 1 and 2 with host proteins. Virology 2009, 386, 122–131. [Google Scholar] [CrossRef]
- Misinzo, G.; Delputte, P.L.; Meerts, P.; Lefebvre, D.J.; Nauwynck, H.J. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J. Virol. 2006, 80, 3487–3494. [Google Scholar] [CrossRef]
- Misinzo, G.; Meerts, P.; Bublot, M.; Mast, J.; Weingartl, H.; Nauwynck, H.J. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J. Gen. Virol. 2005, 86, 2057–2068. [Google Scholar] [CrossRef]
- Wei, R.; Trus, I.; Yang, B.; Huang, L.; Nauwynck, H. Breed differences in PCV2 uptake and disintegration in porcine monocytes. Viruses 2018, 10, 562. [Google Scholar] [CrossRef]
- Misinzo, G.; Delputte, P.L.; Lefebvre, D.J.; Nauwynck, H.J. Porcine circovirus 2 infection of epithelial cells is calthrin-, caveolae- and dynamin-independent, actin and Rho-GTPase-mediated, and enhanced by cholesterol depletion. Virus Res. 2009, 139, 1–9. [Google Scholar] [CrossRef]
- Wei, R.; Renne, N.V.; Nauwynck, H.J. Strain-dependent porcine circovirus type 2 (PCV2) entry and replication in T-lymphoblasts. Viruses 2019, 11, 813. [Google Scholar] [CrossRef]
- Greber, U.F.; Way, M. A superhighway to virus infection. Cell 2006, 124, 741–754. [Google Scholar] [CrossRef]
- Theerawatanasirikul, S.; Phecharat, N.; Prawettongsopon, C.; Chaicumpa, W.; Lekcharoensuk, P. Dynein light chain DYNLL1 subunit facilitates porcine circovirus type 2 intracellular transports along microtubules. Arch. Virol. 2016, 162, 677–686. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, K.; Lin, C.; Zhou, J.; Jin, Y.; Dong, W.; Gu, J.; Zhou, J. Conformational changes and nuclear entry of porcine circovirus without disassembly. J. Virol. 2019, 93, e00824-19. [Google Scholar] [CrossRef]
- Misinzo, G.; Delputte, P.L.; Nauwynck, H.J. Inhibition of endosome-lysosome system acidification enhances porcine circovirus 2 infection of porcine epithelial cells. J. Virol. 2008, 82, 1128–1135. [Google Scholar] [CrossRef]
- Finsterbusch, T.; Mankertz, A. Porcine circoviruses—Small but powerful. Virus Res. 2009, 143, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Vega-Rocha, S.; Byeon, I.J.; Gronenborn, B.; Gronenborn, A.M.; Campos-Olivas, R. Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2. J. Mol. Biol. 2007, 367, 473–487. [Google Scholar] [CrossRef]
- Faurez, F.; Dory, D.; Grasland, B.; Jestin, A. Replication of porcine circoviruses. Virol. J. 2009, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Lin, C.; Wang, H.; Wang, L.; Zhou, N.; Jin, Y.; Liao, M.; Zhou, J. Circovirus transport proceeds via direct interaction of the cytoplasmic dynein IC1 subunit with the viral capsid protein. J. Virol. 2015, 89, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Fossum, C.; Berg, M. Porcine circovirus type 2 replicase binds the capsid protein and an intermediate-filament-like protein. J. Gen. Virol. 2006, 87, 3215–3223. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Du, Q.; Zhu, L.; Chen, N.; Luo, L.; Chen, Q.; Yin, J.; Wu, X.; Tong, D.; Huang, Y. Porcine DNAJB6 promotes PCV2 replication via enhancing the formation of autophagy in host cells. Vet. Res. 2020, 51, 61. [Google Scholar] [CrossRef]
- Zhou, J.; Dai, Y.; Lin, C.; Zhang, Y.; Feng, Z.; Dong, W.; Jin, Y.; Yan, Y.; Zhou, J.; Gu, J. Nucleolar protein NPM1 is essential for circovirus replication by binding to viral capsid. Virulence 2020, 11, 1379–1393. [Google Scholar] [CrossRef]
- Liu, J.; Bai, J.; Zhang, L.; Jiang, Z.; Wang, X.; Li, Y.; Jiang, P. Hsp70 positively regulates porcine circovirus type 2 replication in vitro. Virology 2013, 447, 52–62. [Google Scholar] [CrossRef]
- Hou, Q.; Shaohua, H.; Chen, Q.; Jia, H.; Xin, T.; Jiang, Y.; Guo, X.; Zhu, H. Nuclear localization signal regulates Porcine Circovirus tye 2 Capsid protein nuclear export through phosphorylation. Virus Res. 2017, 246, 12–22. [Google Scholar] [CrossRef]
- Zhu, B.; Xu, F.; Li, J.; Shuai, J.; Li, X.; Fang, W. Porcine circovirus type 2 explores the autophagic machinery for replication in PK-15 cells. Virus Res. 2012, 163, 476–485. [Google Scholar] [CrossRef]
- Zhu, B.; Zhou, Y.; Xu, F.; Shuai, J.; Li, X.; Fang, W. Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J. Virol. 2012, 86, 12003–12012. [Google Scholar] [CrossRef]
- Chen, X.; Ren, F.; Hesketh, J.; Shi, X.; Li, J.; Gan, F.; Huang, K. Reactive oxygen species regulate the replication of porcine circovirus type 2 via NF-κB pathway. Virology 2012, 426, 66–72. [Google Scholar] [CrossRef][Green Version]
- Zhai, N.; Liu, K.; Li, H.; Liu, Z.; Wang, H.; Korolchuk, V.; Carroll, B.; Pan, C.; Gan, F.; Huang, K.; et al. PCV2 replication promoted by oxidative stress is dependent on the regulation of autophagy on apoptosis. Vet. Res. 2019, 50, 19. [Google Scholar] [CrossRef] [PubMed]
- Ha, Z.; Xie, C.-Z.; Li, J.-F.; Wen, S.-B.; Zhang, K.-L.; Nan, F.-L.; Zhang, H.; Guo, Y.-C.; Wang, W.; Lu, H.-J.; et al. Molecular detection and genomic characterization of porcine circovirus 3 in pigs from Northeast China. BMC Vet. Res. 2018, 14, 321. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.; Chae, C. First isolation and genetic characterization of porcine circovirus type 3 using primary porcine kidney cells. Vet. Microbiol. 2020, 241, 108576. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Hou, L.; Wei, L.; Quan, R.; Zhou, B.; Jiang, H.; Wang, J.; Zhu, S.; Song, J.; Wang, D.; et al. Porcine circovirus type 3 enters into PK15 cells through calthrin-and dynamin-2-mediated endocytosis in a Rab5/Rab7 and pH-dependent fashion. Front. Microbiol. 2021, 12, 259. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Hou, L.; Wang, D.; Wei, L.; Zhu, S.; Wang, J.; Quan, R.; Jiang, H.; Shi, R.; Liu, J. Nucleolar phosphoprotein NPM1 interacts with porcine circovirus type 3 cap protein and facilitates viral replication. Front. Microbiol. 2021, 12, 1297. [Google Scholar] [CrossRef]
- Zhou, J.; Qiu, Y.; Zhu, N.; Zhou, L.; Dai, B.; Feng, X.; Hou, L.; Liu, J. The Nucleolar Localization Signal of Porcine Circovirus Type 4 Capsid Protein is Essential for Interaction with Serine-48 Residue of Nucleolar Phosphoprotein Nucleophosmin-1. Front. Microbiol. 2021, 12, 751382. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Qiu, Y.; Wang, Y.; Yang, X.; Liu, C.; Shi, Y.; Feng, X.; Hou, L.; Liu, J. Contribution of DEAD-Box RNA Helicase 21 to the Nucleolar Localization of Porcine Circovirus Type 4 Capsid protein. Front. Microbiol. 2022, 13, 802740. [Google Scholar] [CrossRef]
- Wozniak, A.; Milek, D.; Matyba, P.; Stadejek, T. Real-time PCR detection patterns of porcine circovirus type 2 (PCV2) in Polish farms with different statuses of vaccination against PCV2. Viruses 2019, 11, 1135. [Google Scholar] [CrossRef] [PubMed]
- Madec, F.; Rose, N.; Grasland, B.; Cariolet, R.; Jestin, A. Post-weaning multisystemic wasting syndrome and other PCV2-related problems in pigs: A 12-year experience. Transbound. Emerg. Dis. 2008, 55, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Kolyvushko, O.; Rakibuzzaman, A.G.M.; Pillatzki, A.; Webb, B.; Ramamoorthy, S. Efficacy of a commercial PCV2a vaccine with a tow-dose regimen against PCV2d. Vet. Sci. 2019, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Segales, J. Porcine circovirus 2 genotypes, immunity and vaccines: Multiple genotypes but one single serotype. Pathogens 2020, 9, 1049. [Google Scholar] [CrossRef] [PubMed]
- Pejsak, Z.; Podgόrska, K.; Truszczynski, M.; Karbowiak, P.; Stadejek, T. Efficacy of different protocols of vaccination against porcine circovirus type 2 (PCV2) in a farm affected by postweaning multisystemic wasting syndrome (PMWS). Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, e1–e5. [Google Scholar] [CrossRef]
- Segales, J. Best practice and future challenges for vaccination against porcine circovirus type 2. Expert Rev. Vaccines 2014, 14, 473–487. [Google Scholar] [CrossRef]
- Kixmöller, M.; Ritzmann, M.; Eddicks, M.; Saalmuller, A.; Elbers, K.; Fachinger, V. Reduction of PMWS-associated clinical signs and co-infections by vaccination against PCV2. Vaccine 2008, 26, 3443–3451. [Google Scholar] [CrossRef]
- Park, K.H.; Oh, T.; Yang, S.; Cho, H.; Kang, I.; Chae, C. Evaluation of a porcine circovirus type 2a (PCV2a) vaccine efficacy against experimental PCV2a, PCV2b, and PCV2d challenge. Vet. Microbiol. 2019, 231, 87–92. [Google Scholar] [CrossRef]
- Fort, M.; Sibila, M.; Pérez-Martin, E.; Nofrarias, M.; Mateu, E.; Segales, J. One dose of a porcine circovirus 2 (PCV2) sub-unit vaccine administered to 3-week-old conventional piglets elicits cell-mediated immunity and significantly reduces PCV2 viremia in an experimental model. Vaccine 2009, 27, 4031–4037. [Google Scholar] [CrossRef]
- Figueras-Gourgues, S.; Fraile, L.; Segales, J.; Hernandez-Caravaca, I.; Lopez-Ubeda, R.; Garcia-Vazquez, F.A.; Gomez-Duran, O.; Grosse-Liesner, B. Effect of Porcine circovirus 2 (PCV2) maternally derived antibodies on performance and PCV-2 viremia in vaccinated piglets under field conditions. Porc. Health Manag. 2019, 5, 21. [Google Scholar] [CrossRef]
- Opriessnig, T.; Gerber, P.F.; Xiao, C.-T.; Halbur, P.G.; Matzinger, S.R.; Meng, X.-J. Commercial PCV2a-based vaccines are effective in protecting naturally PCV2b-infected finisher pigs against experimental challenge with a 2012 mutant PCV2. Vaccine 2014, 32, 4342–4348. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Xiao, C.-T.; Halbur, P.G.; Gerber, P.F.; Matzinger, S.R.; Meng, X.-J. A commercial porcine circovirus (PCV) type 2a-based vaccine reduces PCV2d viremia and shedding and prevents PCV2d transmission to naïve pigs under experimental conditions. Vaccine 2017, 35, 248–254. [Google Scholar] [CrossRef]
- Opriessnig, T.; O’Neill, K.; Gerber, P.F.; de Castro, A.M.M.G.; Lirola, L.G.G.; Beach, N.M.; Zhou, L.; Meng, X.-J.; Wang, C.; Halbur, P.G. A PCV2 vaccine based on genotype 2b is more effective than a 2a-based vaccine to protect against PCV2b or combined PCV2a/2b viremia in pigs with concurrent PCV2, PRRSV and PPV infection. Vaccine 2013, 31, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-J. Porcine circovirus type 2 (PCV2): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2013, 1, 43–64. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Hahn, T.-W. Evaluation of novel recombinant porcine circovirus type 2d (PCV2d) vaccine in pigs naturally infected with PCV2d. Vaccine 2021, 39, 529–535. [Google Scholar] [CrossRef]
- Huan, C.; Fan, M.; Cheng, Q.; Wang, X.; Gao, Q.; Wang, W.; Gao, S.; Liu, X. Evaluation of the efficacy and cross-protective immunity of live-attenuated chimeric PCV1-2b vaccine against PCV2b and PCV2d subtype challenge in pigs. Front. Microbiol. 2018, 9, 455. [Google Scholar] [CrossRef]
- Kang, S.-J.; Bae, S.-M.; Lee, H.-J.; Jeong, Y.-J.; Lee, M.-A.; You, S.-H.; Lee, H.-S.; Hyun, B.-H.; Lee, N.; Cha, S.-H. Porcine circoviruse (PCV) genotype 2d-based virus-like particles (VLPs) induced broad cross-neutralizing antibodies against diverse genotypes and provided protection in dual-challenge infection of a PCV2d virus and a type 1 Porcine reproductive and respiratory syndrome virus (PRRSV). Pathogens 2021, 10, 1145. [Google Scholar] [CrossRef]
- Sylla, S.; Cong, Y.-L.; Sun, Y.-X.; Yang, G.-L.; Ding, X.-M.; Yang, Z.-Q.; Zhou, Y.-L.; Yang, M.; Wang, C.-F.; Ding, Z. Protective immunity conferred by porcine circovirus 2 ORF2-based DNA vaccine in mice. Microbiol. Immunol. 2014, 58, 398–408. [Google Scholar] [CrossRef]
- Hou, Z.; Wang, H.; Feng, Y.; Li, Q.; Li, J. A candidate DNA vaccine encoding a fusion protein of porcine complement C3d-P28 and ORF2 of porcine circovirus type 2 induces cross-protective immunity against PCV2b and PCV2d in pigs. Virol. J. 2019, 16, 57. [Google Scholar] [CrossRef]
- Li, D.; Du, Q.; Wu, B.; Li, J.; Chang, L.; Zhao, X.; Huang, Y.; Tong, D. Immunogenicity of adenovirus vaccines expressing the PCV2 capsid protein in pigs. Vaccine 2017, 35, 4722–4729. [Google Scholar] [CrossRef]
- Tian, D.; Sooryanarain, H.; Matzinger, S.R.; Gauger, P.C.; Karuppannan, A.K.; Elankumaran, S.; Opriessnig, T.; Meng, X.-J. Protective efficacy of a virus-vectored multi-component vaccine against porcine reproductive and respiratory syndrome virus, porcine circovirus type 2 and swine influenza virus. J. Gen. Virol. 2017, 98, 3026–3036. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Yang, W.-C.; Chang, Y.-K.; Wang, C.-Y.; Huang, W.-R.; Li, J.-Y.; Chuang, K.-P.; Wu, H.-Y.; Chang, C.-D.; Nielsen, B.L.; et al. Construction of polycistronic baculovirus surface display vectors to express the PCV2 Cap(d41) protein and analysis of its immunogenicity in mice and swine. Vet. Res. 2020, 51, 112. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-C.; Chen, T.-Y.; Chi, J.-N.; Chien, M.-S.; Huang, C. Efficient expression and purification of porcine circovirus type 2 virus-like particles in Escherichia coli. J. Biotechnol. 2016, 220, 78–85. [Google Scholar] [CrossRef]
- Rakibuzzaman, A.G.M.; Ramamoorthy, S. Comparative immunopathogenesis and biology of recently discovered porcine circoviruses. Transbound. Emerg. Dis. 2021, 68, 2957–2968. [Google Scholar] [CrossRef]
- Plut, J.; Jamnikar-Ciglenecki, U.; Golinar-Oven, I.; Knific, T.; Stukelj, M. A molecular survey and phylogenetic analysis of porcine circovirus type 3 using oral fluid, faeces and serum. BMC Vet. Res. 2020, 16, 281. [Google Scholar] [CrossRef]
- Peswani, A.R.; Narkpuk, J.; Krueger, A.; Bracewell, D.G.; Lekcharoensuk, P.; Haslam, S.M.; Dell, A.; Jaru-Ampornpan, P.; Robinson, C. Novel constructs and 1-step chromatography protocols for the production of Porcine circovirus 2d (PCV2d) and circovirus 3 (PCV3) subunit vaccine candidates. Food Bioprod. Process. 2022, 131, 125–135. [Google Scholar] [CrossRef]
- Tian, K.; Li, X.; Xiao, Y.; Xi, X.; Sun, J.; Zhang, X. Bivalent Vaccine Composition for Preventing and/or Treating Porcine Circovirus Infections and Preparation Method and Use Thereof. U.S. Patent 11,116,836, 14 September 2021. [Google Scholar]
- Tian, K.; Li, X.; Xiao, Y.; Sun, J.; Zhang, X. Porcine Circovirus Type 3 Strain, Vaccine Composition, Method of Making the Same and Use Thereof. U.S. Patent 10,869,919, 22 December 2020. [Google Scholar]
- Feng, H.; Blanco, G.; Segales, J.; Sibila, M. Can Porcine circovirus type 2 (PCV2) infection be eradicated by mass vaccination? Vet. Microbiol. 2014, 172, 92–99. [Google Scholar] [CrossRef]
- Madson, D.M.; Patterson, A.R.; Ramamoorthy, S.; Pal, S.; Meng, X.-J.; Opriessnig, T. Effect of porcine circovirus type 2 (PCV2) vaccination of the dam on PCV2 replication in utero. Clin. Vaccine Immunol. 2009, 16, 830–834. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, H.; Sun, P.; Khan, A.; Guo, J.; Zheng, X.; Sun, Y.; Fan, K.; Yin, W.; Li, H. Matrine exhibits antiviral activity in a PRRSV/PCV2 co-infected mouse model. Phytomedicine 2020, 77, 153289. [Google Scholar] [CrossRef]
- Sun, N.; Yu, T.; Zhao, J.-X.; Sun, Y.-G.; Jiang, J.-B.; Duan, Z.-B.; Wang, W.-K.; Hu, Y.-L.; Lei, H.-M.; Li, H.-Q. Antiviral activities of natural compounds derived from traditional Chinese medicines against porcine circovirus type 2 (PCV2). Biotechnol. Bioproc. E 2015, 20, 180–187. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Jin, E.; He, Q.; Yan, W.; Yang, H.; Gong, S.; Guo, Y.; Fu, S.; Chen, X.; et al. The antiviral activity of arctigenin in traditional Chinese medicine on porcine circovirus type 2. Res. Vet. Sci. 2016, 106, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-L.; Yang, J.; Fu, Y.-F.; Meng, X.-N.; Zhao, W.-D.; Hu, T.-J. Effect of total flavonoids of Spatholobus suberectus Dunn on PCV2 induced oxidative stress in RAW264.7 cells. BMC Complement. Altern. Med. 2017, 17, 244. [Google Scholar] [CrossRef] [PubMed]
- Madec, F.; Eveno, E.; Morvan, P.; Harmon, L.; Blanchard, P.; Cariolet, R.; Amenna, N.; Truong, C.; Mahe, D.; Albina, E.; et al. Post-weaning multisystemic wasting syndrome (PMWS) in pigs in France: Clinical observations from follow-up studies on affected farms. Livest. Prod. Sci. 2000, 63, 223–233. Available online: www.elsevier.com/locate/livprodsci (accessed on 6 July 2023).
- Alexopoulos, C.; Kritas, S.K.; Kyriakis, C.S.; Tzika, E.; Kyriakis, S.C. The Impact of Vaccination of Sows and/or their Litters with an Attenuated PRRSV Vaccine on Reduction of PCV2 Effects. In Proceedings of the 4th International Symposium on Emerging and Re-Emerging Pig Diseases, Rome, Italy, 29 June–2 July 2003; pp. 132–133. [Google Scholar]
- Genzow, M.; Schwartz, K.; Gonzalez, G.; Anderson, G.; Chittick, W. The effect of vaccination against porcine reproductive and respiratory syndrome virus (PRRSV) on the porcine circovirus-2 load in porcine circovirus associated disease (PCVAD) affected pigs. Can. J. Vet. Res. 2009, 73, 87–90. [Google Scholar] [PubMed]
- Niederwerder, M.C.; Bawa, B.; Serao, N.V.L.; Trible, B.R.; Kerrigan, M.A.; Lunney, J.K.; Dekkers, J.C.M.; Rowland, R.R.R. Vaccination with a porcine reproductive and respiratory syndrome (PRRS) modified live virus vaccine followed by challenge with PRRS virus and porcine circovirus type 2 (PCV2) protects against PRRS but enhances PCV2 replication and pathogenesis compared to results for nonvaccinated cochallenged controls. Clin. Vaccine Immunol. 2015, 22, 1244–1254. [Google Scholar] [CrossRef]
- Seo, H.W.; Park, S.-J.; Park, C.; Chae, C. Interaction of porcine circovirus type 2 and Mycoplasma hyopneumoniae vaccines on dually infected pigs. Vaccine 2014, 32, 2480–2486. [Google Scholar] [CrossRef]
- Yang, S.; Ahn, Y.; Oh, T.; Cho, H.; Park, K.H.; Chae, C. Field evaluation of a single-dose bivalent vaccine of porcine circovirus type 2b and Mycoplasma hyopneumoniae. Vet. Med. Sci. 2021, 7, 755–765. [Google Scholar] [CrossRef]
- Yang, S.; Oh, T.; Park, K.H.; Cho, H.; Suh, J.; Chae, C. Experimental efficacy of a trivalent vaccine containing porcine circovirus types 2a/b (PCV2a/b) and Mycoplasma hyopneumoniae against PCV2d and M. hyopneumoniae challenges. Vet. Microbiol. 2021, 258, 109100. [Google Scholar] [CrossRef]
- Um, H.; Yang, S.; Oh, T.; Cho, H.; Park, K.H.; Suh, J. A field efficacy trial of a trivalent vaccine containing porcine circovirus type 2a and 2b, and Mycoplasma hyopneumoniae in three herds. Vet. Med. Sci. 2021, 8, 578–590. [Google Scholar] [CrossRef]
- Oh, T.; Park, K.H.; Yang, S.; Jeong, J.; Kang, I.; Park, C.; Chae, C. Evaluation of the efficacy of a trivalent vaccine mixture against a triple challenge with Mycoplasma hyopneumoniae, PCV2, and PRRSV and the efficacy comparison of the respective monovalent vaccines against a single challenge. BMC Vet. Res. 2019, 15, 342. [Google Scholar] [CrossRef]
- Wang, X.; Lv, C.; Ji, X.; Wang, B.; Qiu, L.; Yang, Z. Ivermectin treatment inhibits the replication of Porcine circovirus 2 (PCV2) in vitro and mitigates the impact of viral infection in piglets. Virus Res. 2019, 263, 80–86. [Google Scholar] [CrossRef] [PubMed]



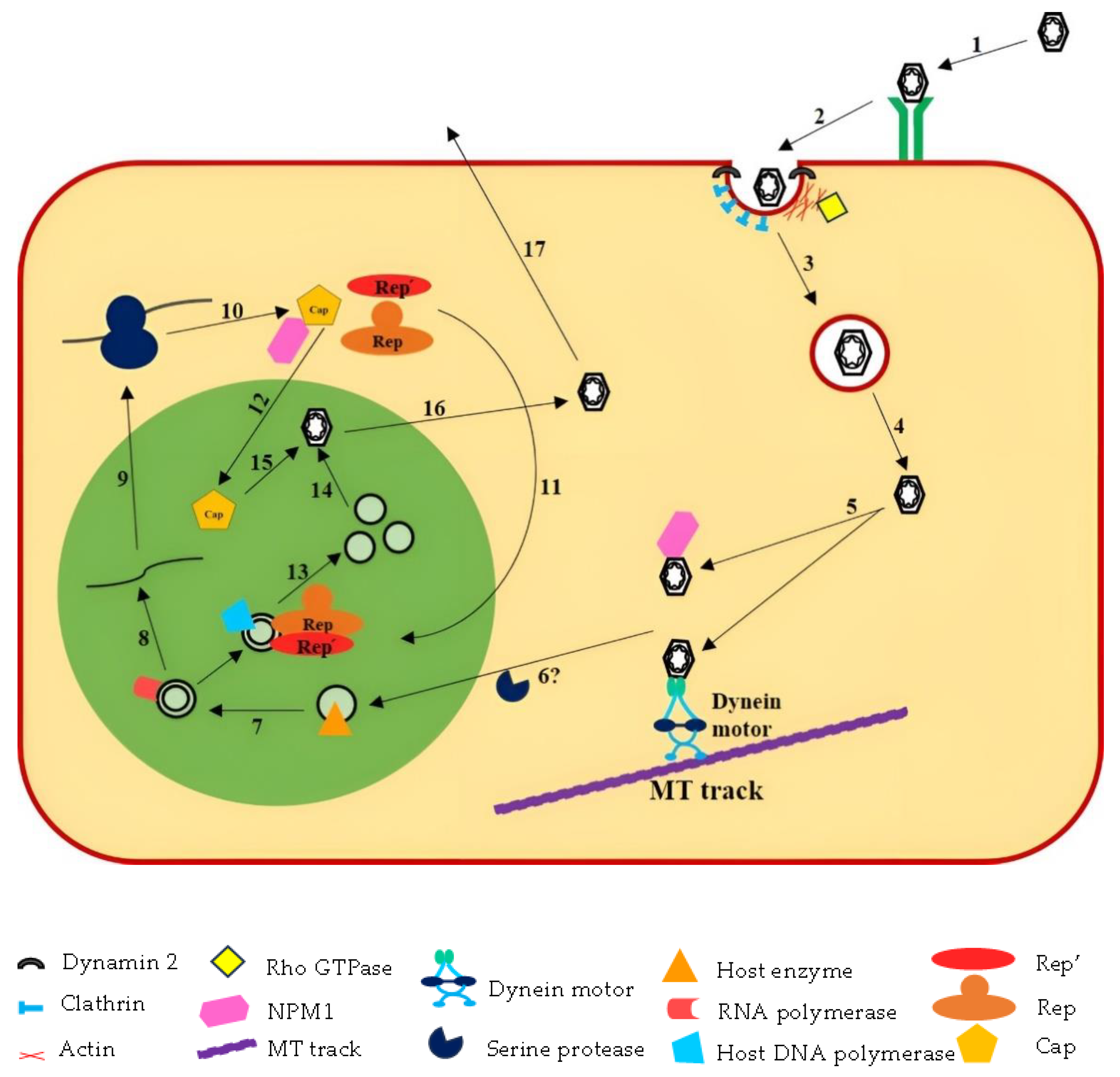
| Porcine Circovirus | Size (nt) | ORF1 | ORF2 | ORF3 | References | |||
|---|---|---|---|---|---|---|---|---|
| Protein | Size (aa) | Protein | Size (aa) | Protein | Size (aa) | |||
| PCV1 | 1758–1760 | Rep | 312 | Cap | 230–233 | NS | 206 | [17,18,21] |
| Rep′ | 168 | |||||||
| PCV2 | 1766–1777 | Rep | 314 | Cap | 233–236 | NS | 104 | [17,18,19,21,25] |
| Rep′ | 297 | |||||||
| PCV3 | 1999–2001 | Rep | 296–297 | Cap | 214 | Unknown | 231 | [12,17,19,23,24] |
| PCV4 | 1770 | Rep | 296 | Cap | 228 | - | - | [20] |
| PCV1–PCV2 | PCV1–PCV3 | PCV1–PCV4 | PCV2–PCV3 | PCV2–PCV4 | PCV3–PCV4 | References | |
|---|---|---|---|---|---|---|---|
| Complete genome (nt) | 68.0–76.0 | 43.5–44.0 | 50.3–51.6 | 42.7–48.0 | 51.5 | 42.9–45.0 | [12,20,21,22,26,27] |
| Replicase (aa) | 86.0 | 45.5–45.9 | 48.1–50.7 | 46.3–48.0 | 16.2–47.2 | 48.4–49.7 | [20,22,24,26,27] |
| Capsid (aa) | 65.0 | 24.0–25.2 | 43.1–44.4 | 25.9–37.0 | 12.7–45.0 | 23.2–24.8 | [20,22,23,24,26,27] |
| Vaccine | Manufacturer | Antigen | Adjuvant | Recommended for | Administration | References |
|---|---|---|---|---|---|---|
| Circovac® | Merial (Duluth, GA, USA) | Inactivated PCV2a (whole virus) | Mineral oil | Females of breeding age | 2 mL IM 2 doses | [124,157] |
| FosteraTM PCV | Pfizer (Leipzig, Germany) | Killed PCV1-2a (chimeric virus) | SL-CD aqueous | Piglets (≥3 weeks of age) | 2 mL IM 1 dose | |
| Ingelvac CircoFLEX® | Boehringer Ingelheim (Ingelheim am Rhein, Germany) | Cap protein of PCV2a (recombinant) | Carbomer | Piglets (>2 weeks of age) | 1 mL IM 1 dose | |
| Circumvent® PCV | Intervet/SP (Merck, Rahway, NJ, USA) | Cap protein of PCV2a (recombinant) | Microsol Diluvac Forte® (MDF) | Piglets (≥3 weeks of age) | 2 mL IM 2 doses | |
| Porcilis® PCV | Schering-Plough (Merck, Kenilworth NJ, USA) | Cap protein of PCV2a (recombinant) | Mineral oil | Piglets (≥3 weeks of age) | 2 mL IM 1/2 dose |
| Class | Type of Antigen | Vector Used | Adjuvant | Route of Administration | Effects under Experimental Conditions | Reference |
|---|---|---|---|---|---|---|
| DNA vaccines | Full-length ORF2 | pEGFP-N1 | Freund’s adjuvant | Intraperitoneal | Immunization of 6-week-old BALB/c mice thrice (15 µg each) at an interval of 14 days provided efficient protection against PCV2 infection through induction of highly specific IgG antibodies and cytokines (IFN-γ and IL-10). Vaccination reduces both viral load and number of microscopic lesions in lymph nodes. | [294] |
| Full-length ORF2 (PCV2d) | pVAX1 | C3d-P28 | Intramuscular | Vaccination of 3-week-old piglets (500 µg each) stimulated both PCV2-specific antibody responses and interferon-γ secreting cells (IFN-γ-SC), reduced viremia and level of genomic DNA and conferred protection against both PCV2b and PCV2d challenge. | [295] | |
| Viral vectored vaccines | Truncated ORF2 | Adenovirus (AdEasyTM) | CD40L, GMCSF | Intramuscular | Vaccination of 4-week-old pigs induced strong humoral and cell-mediated immune responses and provided better protection than commercial inactivated vaccine (PCV2 SH-strain). Viral load was reduced significantly and no obvious gross and microscopic lesions were observed in lungs and lymph nodes. | [296] |
| Full-length ORF2 of PCV2b along with HA Ag of SIV | DS722 (PRRSV) | Not used | Intramuscular | Vaccination of 3-week-old piglets provided good protection against PRRSV but only partial protection against SIV and PCV2b. | [297] | |
| Truncated Cap | Baculovirus (BacDD) | Not used | Intramuscular | Immunization of 9-week-old SPF pigs (80 µg) induced higher levels of neutralizing antibodies and IFN-γ and significantly reduced viral loads of vaccinated pigs as compared with negative control group. | [298] | |
| Virus-like particles (VLPs) | Full-length ORF2 | pET24a (+) | Montanide ISA-201 | Intraperotoneal | Vaccination of SPF mice thrice (30 µg each) with rCap VLPs at 2-week intervals induced strong humoral and cellular immune responses as demonstrated by induction of PCV2-specific neutralizing antibodies and secretion of IFN-γ in splenocytes | [299] |
| Full-length ORF2 (PCV2d) | pOET1 | Not used | Intramuscular | Immunization of 3-week-old piglets induced higher levels of anti-PCV2d IgG and neutralizing antibodies, significantly reduced amount of genomic DNA in blood, saliva, tissues, reduced macroscopic and microscopic lesions in lungs and inguinal lymph nodes and stimulated average daily weight gain in vaccinated group | [293] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maity, H.K.; Samanta, K.; Deb, R.; Gupta, V.K. Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector. Vaccines 2023, 11, 1308. https://doi.org/10.3390/vaccines11081308
Maity HK, Samanta K, Deb R, Gupta VK. Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector. Vaccines. 2023; 11(8):1308. https://doi.org/10.3390/vaccines11081308
Chicago/Turabian StyleMaity, Hemanta Kumar, Kartik Samanta, Rajib Deb, and Vivek Kumar Gupta. 2023. "Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector" Vaccines 11, no. 8: 1308. https://doi.org/10.3390/vaccines11081308
APA StyleMaity, H. K., Samanta, K., Deb, R., & Gupta, V. K. (2023). Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector. Vaccines, 11(8), 1308. https://doi.org/10.3390/vaccines11081308






