Evaluation of the Robustness Verification of Downstream Production Process for Inactivated SARS-CoV-2 Vaccine and Different Chromatography Medium Purification Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions and Virus Production
2.2. Harvest and Inactivation
2.3. Clarification and Concentration
2.4. SEC
2.5. AEC
2.6. Detection of Quality Parameters
2.6.1. Residual DNA Content
2.6.2. Vero Cell HCP
2.6.3. BSA Residual Levels
2.6.4. Total Protein Assay
2.6.5. Antigen Content Detection
2.7. Statistical Methods
3. Results
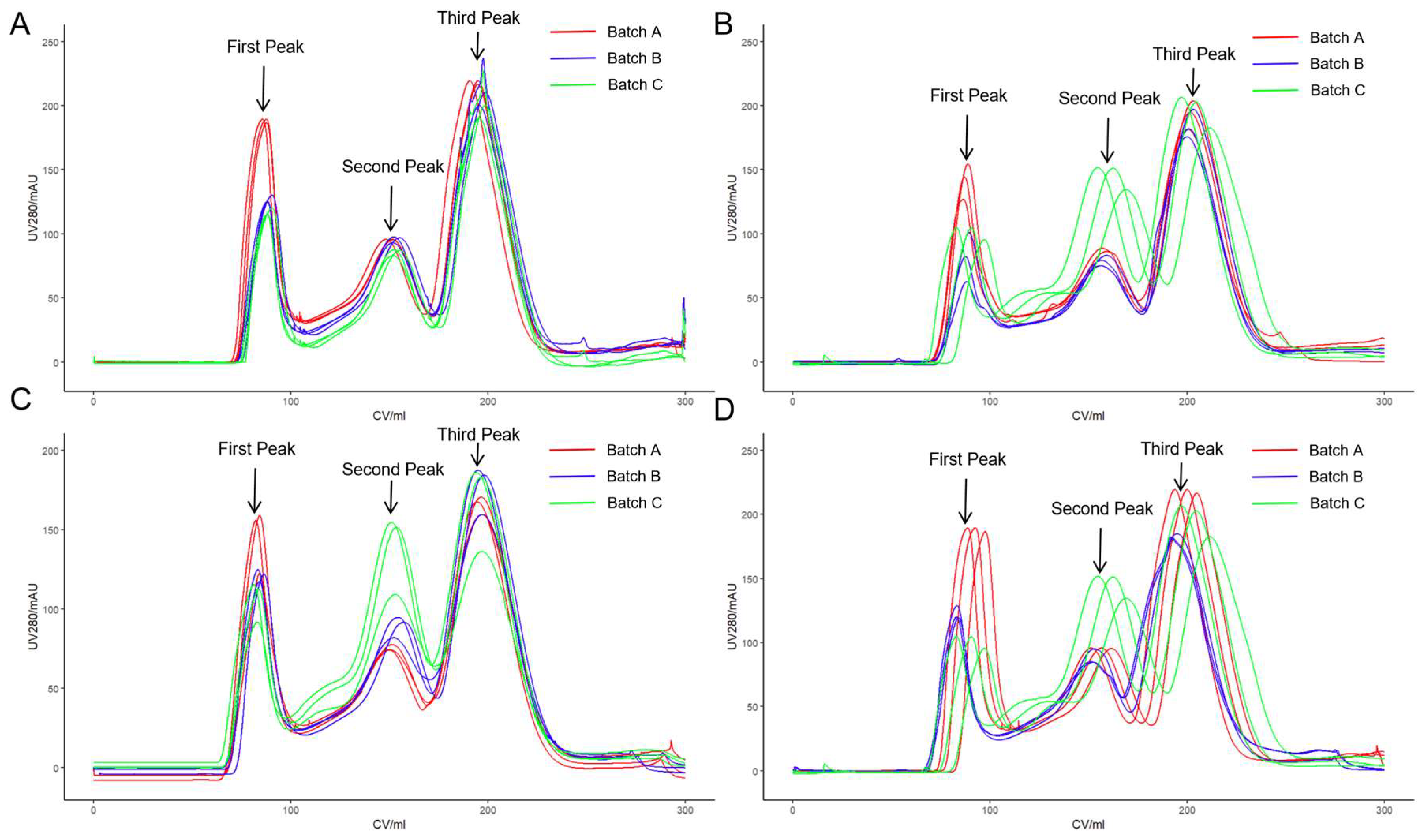
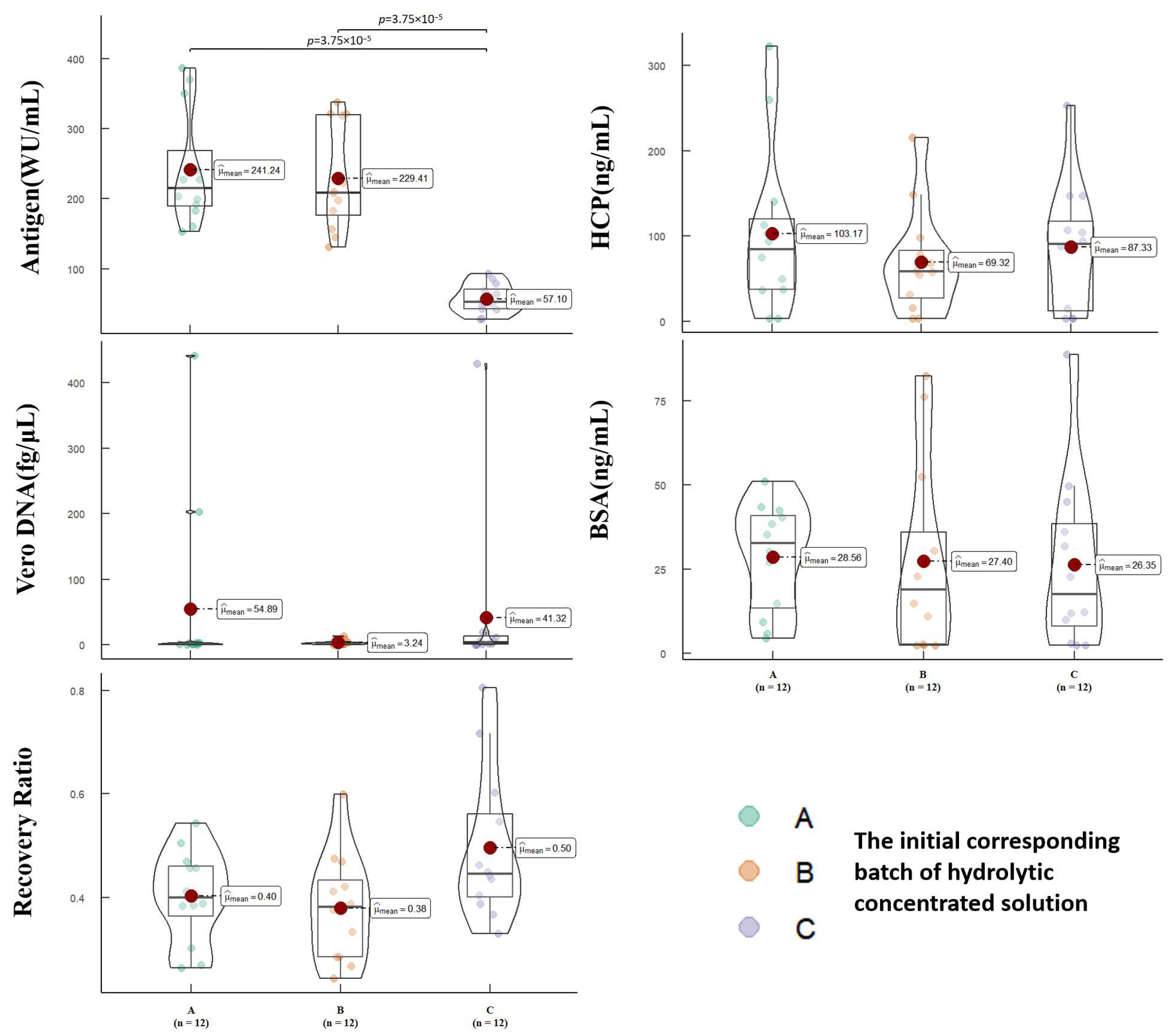
3.1. Virus Purification by SEC
3.2. Analysis of First Peak Areas
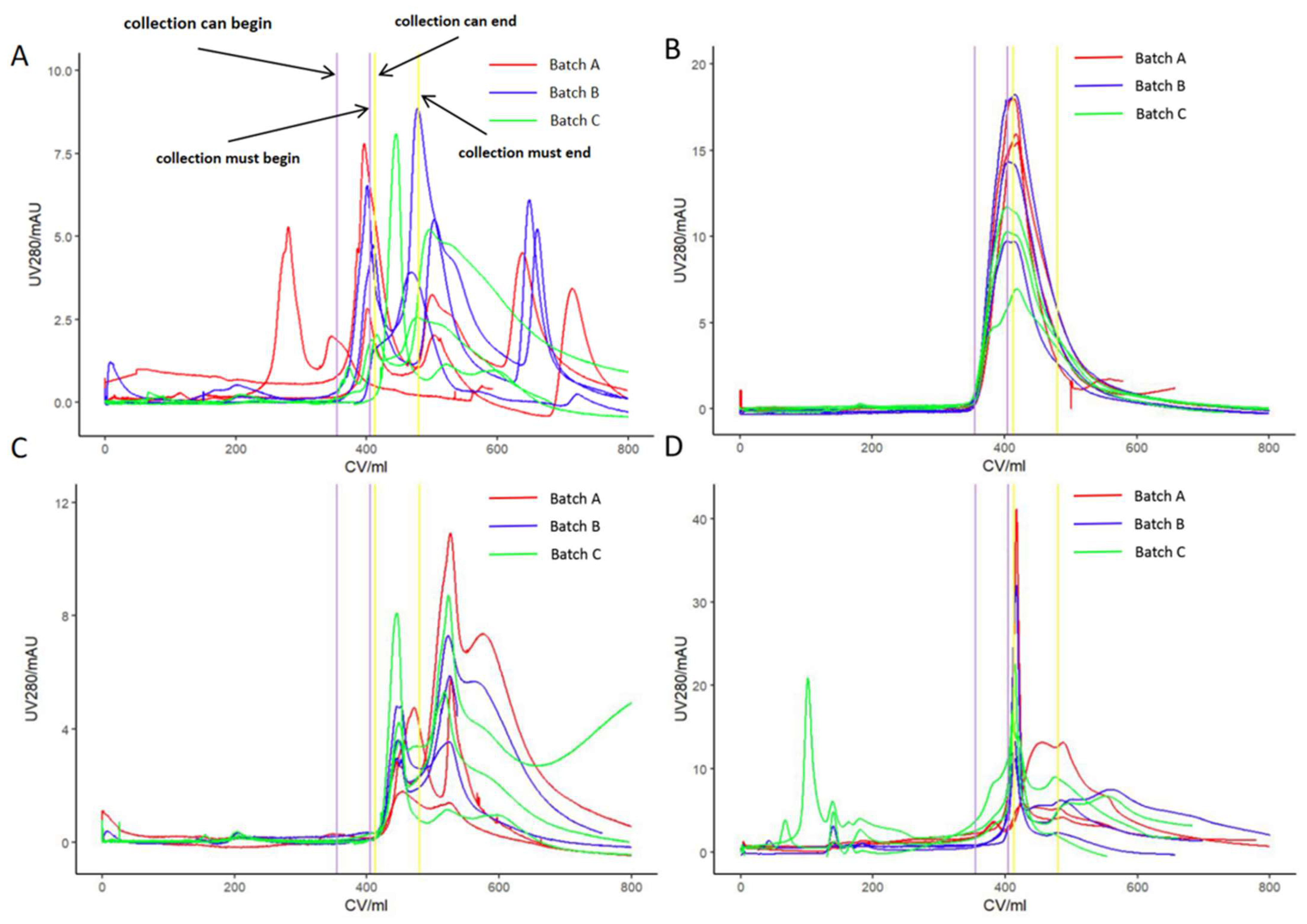
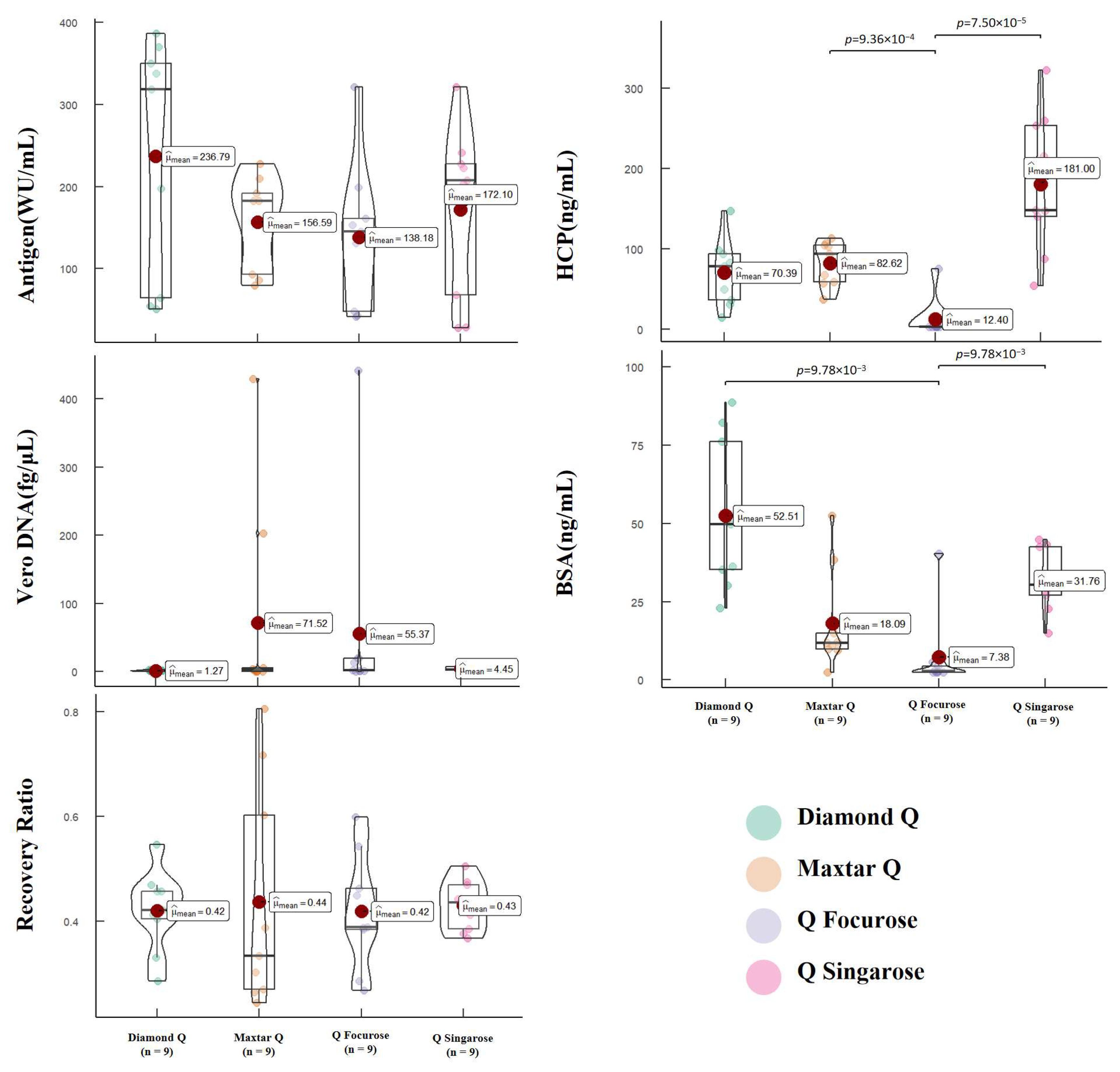
3.3. Virus Purification by Anion Exchange Chromatography
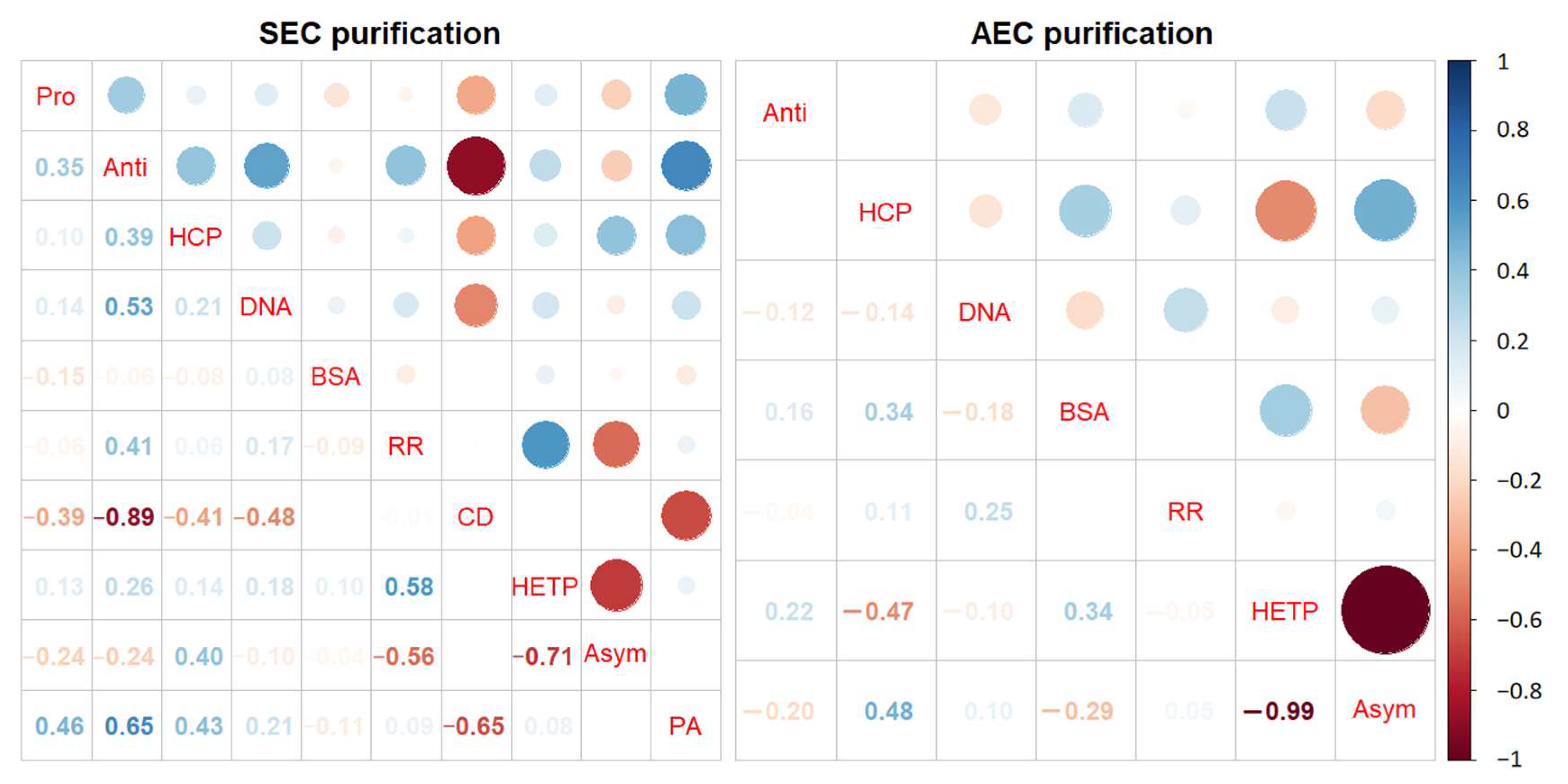
3.4. Correlation Analysis of Relevant Quality Parameters and Process Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273, Erratum in Nature 2020, 588, E6. [Google Scholar] [CrossRef]
- Han, X.; Ye, Q. The variants of SARS-CoV-2 and the challenges of vaccines. J. Med. Virol. 2022, 94, 1366–1372. [Google Scholar] [CrossRef]
- Hawman, D.W.; Meade-White, K.; Archer, J.; Leventhal, S.S.; Wilson, D.; Shaia, C.; Randall, S.; Khandhar, A.P.; Krieger, K.; Hsiang, T.Y.; et al. SARS-CoV2 variant-specific replicating RNA vaccines protect from disease following challenge with heterologous variants of concern. Elife 2022, 11, e75537. [Google Scholar] [CrossRef]
- Andre, F.E.; Booy, R.; Bock, H.L.; Clemens, J.; Datta, S.K.; John, T.J.; Lee, B.W.; Lolekha, S.; Peltola, H.; Ruff, T.A.; et al. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull. World Health Organ. 2008, 86, 140–146. [Google Scholar] [CrossRef]
- Rodrigues, C.M.C.; Plotkin, S.A. Impact of Vaccines; Health, Economic and Social Perspectives. Front. Microbiol. 2020, 11, 1526. [Google Scholar] [CrossRef]
- Nguyen, N.N.; Houhamdi, L.; Hoang, V.T.; Delerce, J.; Delorme, L.; Colson, P.; Brouqui, P.; Fournier, P.E.; Raoult, D.; Gautret, P. SARS-CoV-2 reinfection and COVID-19 severity. Emerg. Microbes Infect. 2022, 11, 894–901. [Google Scholar] [CrossRef]
- Montoy, J.C.C.; Ford, J.; Yu, H.; Gottlieb, M.; Morse, D.; Santangelo, M.; O’Laughlin, K.N.; Schaeffer, K.; Logan, P.; Rising, K.; et al. Innovative Support for Patients with SARS-CoV-2 Infections Registry (INSPIRE) Group. Prevalence of Symptoms ≤ 12 Months after Acute Illness, by COVID-19 Testing Status Among Adults—United States, December 2020–March 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 859–865. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, T.; Mu, Q.; Zhang, R.; Liu, C.; Xu, F.; Liang, L.; Zhao, L.; Zhao, S.; Cai, X.; et al. Immune-Boosting Effect of the COVID-19 Vaccine: Real-World Bidirectional Cohort Study. JMIR Public Health Surveill. 2023, 9, e47272. [Google Scholar] [CrossRef]
- Keulen, D.; Geldhof, G.; Bussy, O.L.; Pabst, M.; Ottens, M. Recent advances to accelerate purification process development: A review with a focus on vaccines. J. Chromatogr. A 2022, 1676, 463195. [Google Scholar] [CrossRef]
- Cappione, A.; Mabuchi, M.; Suhrawardy, S.; Briggs, D.; Nadler, T. The Amicon Pro system—A centrifugal device capable of performing all steps in the protein purification workflow. Postepy. Biochem. 2013, 59, 327–332. [Google Scholar]
- Santry, L.A.; Jacquemart, R.; Vandersluis, M.; Zhao, M.; Domm, J.M.; McAusland, T.M.; Shang, X.; Major, P.M.; Stout, J.G.; Wootton, S.K. Interference chromatography: A novel approach to optimizing chromatographic selectivity and separation performance for virus purification. BMC Biotechnol. 2020, 20, 32. [Google Scholar] [CrossRef]
- Hossienizadeh, S.M.J.; Bagheri, M.; Alizadeh, M.; Rahimi, M.; Azimi, S.M.; Kamalzade, M.; Es-Haghi, A.; Ghassempour, A. Two Dimensional Anion Exchange-Size Exclusion Chromatography Combined with Mathematical Modeling for Downstream Processing of Foot and Mouth Disease Vaccine. J. Chromatogr. A 2021, 1643, 462070. [Google Scholar] [CrossRef]
- Loa, C.C.; Lin, T.L.; Wu, C.C.; Bryan, T.A.; Thacker, H.L.; Hooper, T.; Schrader, D. Purification of turkey coronavirus by Sephacryl size-exclusion chromatography. J. Virol. Methods 2002, 104, 187–194. [Google Scholar] [CrossRef]
- Transfiguracion, J.; Jaalouk, D.E.; Ghani, K.; Galipeau, J.; Kamen, A. Size-exclusion chromatography purification of high-titer vesicular stomatitis virus G glycoprotein-pseudotyped retrovectors for cell and gene therapy applications. Hum. Gene Ther. 2003, 14, 1139–1153. [Google Scholar] [CrossRef]
- Segura, M.M.; Kamen, A.; Trudel, P.; Garnier, A. A novel purification strategy for retrovirus gene therapy vectors using heparin affinity chromatography. Biotechnol. Bioeng. 2005, 90, 391–404. [Google Scholar] [CrossRef]
- Nayak, D.P.; Lehmann, S.; Reichl, U. Downstream processing of MDCK cell-derived equine influenza virus. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2005, 823, 75–81. [Google Scholar] [CrossRef]
- Konz, J.O.; Lee, A.L.; Lewis, J.A.; Sagar, S.L. Development of a purification process for adenovirus: Controlling virus aggregation to improve the clearance of host cell DNA. Biotechnol. Prog. 2005, 21, 466–472. [Google Scholar] [CrossRef]
- Knudsen, H.L.; Fahrner, R.L.; Xu, Y.; Norling, L.A.; Blank, G.S. Membrane ion-exchange chromatography for process-scale antibody purification. J. Chromatogr. A 2001, 907, 145–154. [Google Scholar] [CrossRef]
- Biryukov, J.; Boydston, J.A.; Dunning, R.A.; Yeager, J.J.; Wood, S.; Reese, A.L.; Ferris, A.; Miller, D.; Weaver, W.; Zeitouni, N.E.; et al. Increasing Temperature and Relative Humidity Accelerates Inactivation of SARS-CoV-2 on Surfaces. mSphere 2020, 5, e00441-20. [Google Scholar] [CrossRef]
- Espinoza, D.; Andersson, N.; Nilsson, B. Binary separation control in preparative gradient chromatography using iterative learning control. J. Chromatogr. A 2022, 1673, 463078. [Google Scholar] [CrossRef]
- Xia, S.; Duan, K.; Zhang, Y.; Zeng, X.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, W.; et al. Safety and Immunogenicity of an Inactivated COVID-19 Vaccine, WIBP-CorV, in Healthy Children: Interim Analysis of a Randomized, Double-Blind, Controlled, Phase 1/2 Trial. Front. Immunol. 2022, 13, 898151. [Google Scholar] [CrossRef]
- Gränicher, G.; Coronel, J.; Pralow, A.; Marichal-Gallardo, P.; Wolff, M.; Rapp, E.; Karlas, A.; Sandig, V.; Genzel, Y.; Reichl, U. Efficient influenza A virus production in high cell density using the novel porcine suspension cell line PBG.PK2.1. Vaccine 2019, 37, 7019–7028. [Google Scholar] [CrossRef]
- Tapia, F.; Vázquez-Ramírez, D.; Genzel, Y.; Reichl, U. Bioreactors for high cell density and continuous multi-stage cultivations: Options for process intensification in cell culture-based viral vaccine production. Appl. Microbiol. Biotechnol. 2016, 100, 2121–2132. [Google Scholar] [CrossRef]
- Thomassen, Y.E.; Rubingh, O.; Wijffels, R.H.; van der Pol, L.A.; Bakker, W.A. Improved poliovirus D-antigen yields by application of different Vero cell cultivation methods. Vaccine 2014, 32, 2782–2788. [Google Scholar] [CrossRef]
- Guiochon, G. The limits of the separation power of unidimensional column liquid chromatography. J. Chromatogr. A 2006, 1126, 6–49. [Google Scholar] [CrossRef]
- Bishop, L.D.C.; Misiura, A.; Moringo, N.A.; Landes, C.F. Unraveling peak asymmetry in chromatography through stochastic theory powered Monte Carlo simulations. J. Chromatogr. A 2020, 1625, 461323. [Google Scholar] [CrossRef]
- Pápai, Z.; Pap, T.L. Analysis of peak asymmetry in chromatography. J. Chromatogr. A 2002, 953, 31–38. [Google Scholar] [CrossRef]
- Hubbuch, J.; Linden, T.; Knieps, E.; Ljunglöf, A.; Thömmes, J.; Kula, M.R. Mechanism and kinetics of protein transport in chromatographic media studied by confocal laser scanning microscopy. Part I. The interplay of sorbent structure and fluid phase conditions. J. Chromatogr. A 2003, 1021, 93–104. [Google Scholar] [CrossRef]



| NO. | Strain | Bioreactor Volume | Cell Density at Infection | MOI | Trypsin | Temperature at Infection | Time of Harvest | Total Cell Density at Harvest |
|---|---|---|---|---|---|---|---|---|
| Run#1 | Omicron B.1.1.529 | 1000 L | 3.54 × 106 cells/mL | 0.0001 | 1 μg/mL | 37 °C | 113 hpi | 2.655 × 106 cells/mL |
| Run#2 | Omicron B.1.1.529 | 1000 L | 3.42 × 106 cells/mL | 0.0001 | 1 μg/mL | 37 °C | 115 hpi | 2.565 × 106 cells/mL |
| Run#3 | Omicron B.1.1.529 | 1000 L | 4.6 × 106 cells/mL | 0.0001 | 1 μg/mL | 37 °C | 110 hpi | 3.45 × 106 cells/mL |
| # | Medium | Exclusion Limit | Bed Height (cm) | Volume (mL) | HETP | Asym |
|---|---|---|---|---|---|---|
| 1 | Chromstar 6FF | 10~4000 kDa | 85 | 170.9 | 2976 | 1.17 |
| 2 | Singarose 6FF | 10~4000 kDa | 86 | 172.9 | 2599 | 1.39 |
| 3 | Bestarose 6B | 10~4000 kDa | 85 | 170.9 | 2891 | 0.99 |
| 4 | Focurose 6FF | 10~4000 kDa | 85 | 170.9 | 2647 | 1.20 |
| # | Medium | Exclusion Limit | Bed Height (cm) | Volume (mL) | HETP | Asym |
|---|---|---|---|---|---|---|
| 1 | Maxtar Q | 10~4000 kDa | 25 | 490.87 | 2822 | 1.23 |
| 2 | Q Singarose | 10~4000 kDa | 25 | 490.87 | 2977 | 1.22 |
| 3 | Diamond Q | 10~4000 kDa | 25 | 490.87 | 2608 | 0.8 |
| 4 | Q Focurose | 10~4000 kDa | 25 | 490.87 | 2633 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, J.-H.; Guo, C.-F.; Hao, P.-L.; Meng, S.-L.; Guo, J.; Zhang, D.; Ji, Y.-Q.; Ming, P.-G. Evaluation of the Robustness Verification of Downstream Production Process for Inactivated SARS-CoV-2 Vaccine and Different Chromatography Medium Purification Effects. Vaccines 2024, 12, 56. https://doi.org/10.3390/vaccines12010056
Pang J-H, Guo C-F, Hao P-L, Meng S-L, Guo J, Zhang D, Ji Y-Q, Ming P-G. Evaluation of the Robustness Verification of Downstream Production Process for Inactivated SARS-CoV-2 Vaccine and Different Chromatography Medium Purification Effects. Vaccines. 2024; 12(1):56. https://doi.org/10.3390/vaccines12010056
Chicago/Turabian StylePang, Jia-Hui, Chang-Fu Guo, Peng-Liang Hao, Sheng-Li Meng, Jing Guo, Dou Zhang, Ya-Qi Ji, and Ping-Gang Ming. 2024. "Evaluation of the Robustness Verification of Downstream Production Process for Inactivated SARS-CoV-2 Vaccine and Different Chromatography Medium Purification Effects" Vaccines 12, no. 1: 56. https://doi.org/10.3390/vaccines12010056
APA StylePang, J.-H., Guo, C.-F., Hao, P.-L., Meng, S.-L., Guo, J., Zhang, D., Ji, Y.-Q., & Ming, P.-G. (2024). Evaluation of the Robustness Verification of Downstream Production Process for Inactivated SARS-CoV-2 Vaccine and Different Chromatography Medium Purification Effects. Vaccines, 12(1), 56. https://doi.org/10.3390/vaccines12010056





