Immunogenicity Analysis of Chikungunya Virus DNA Vaccine Based on Mutated Putative N-Linked Glycosylation Sites of the Envelope Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Animals
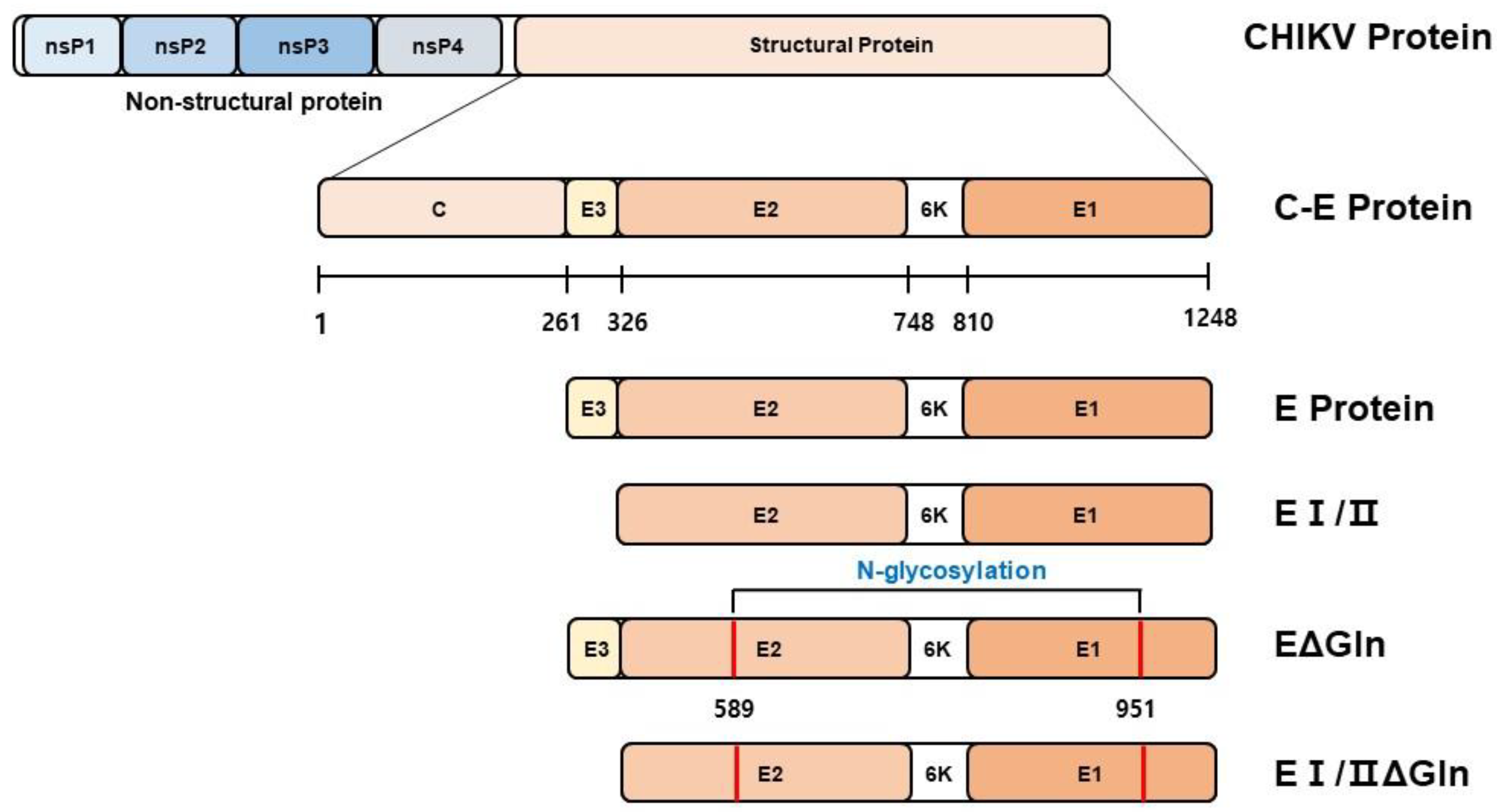
2.2. Construction of DNA-Based CHIKV Vaccine
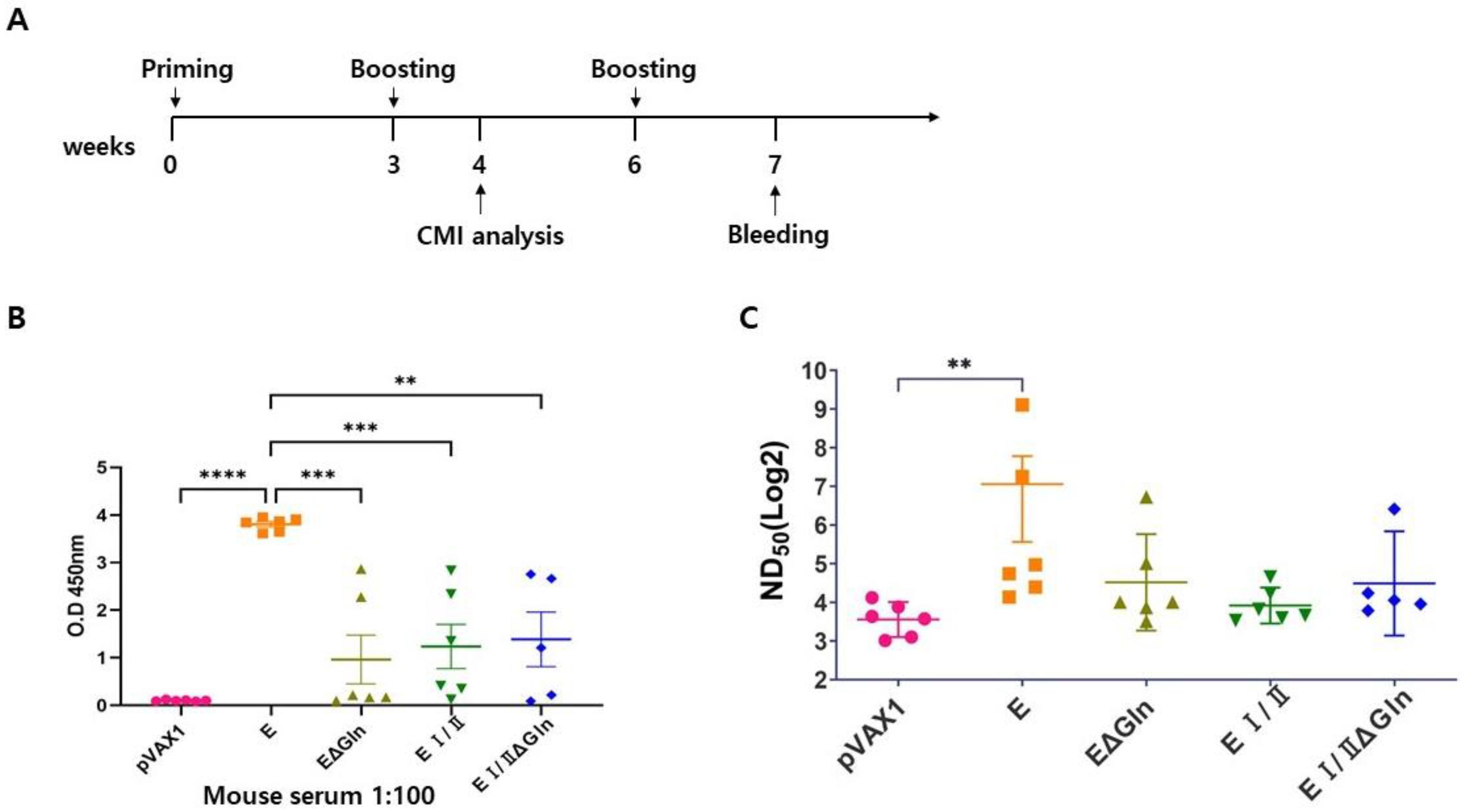
2.3. Immunization of Mice
2.4. IgG Level Measurement by Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. CHIKV Neutralization Assay
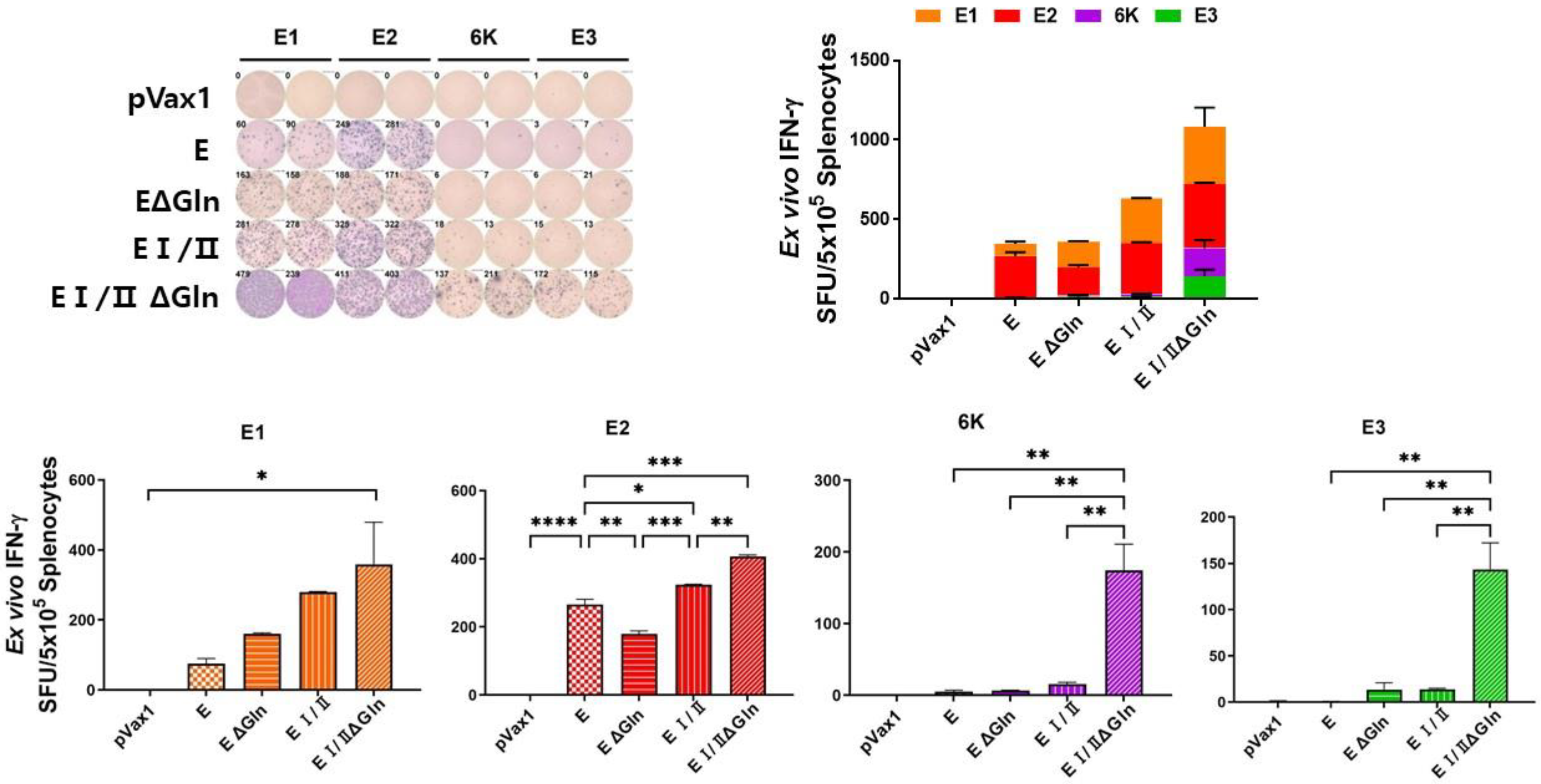
2.6. Cytokine (IFN-γ) Expression Analysis
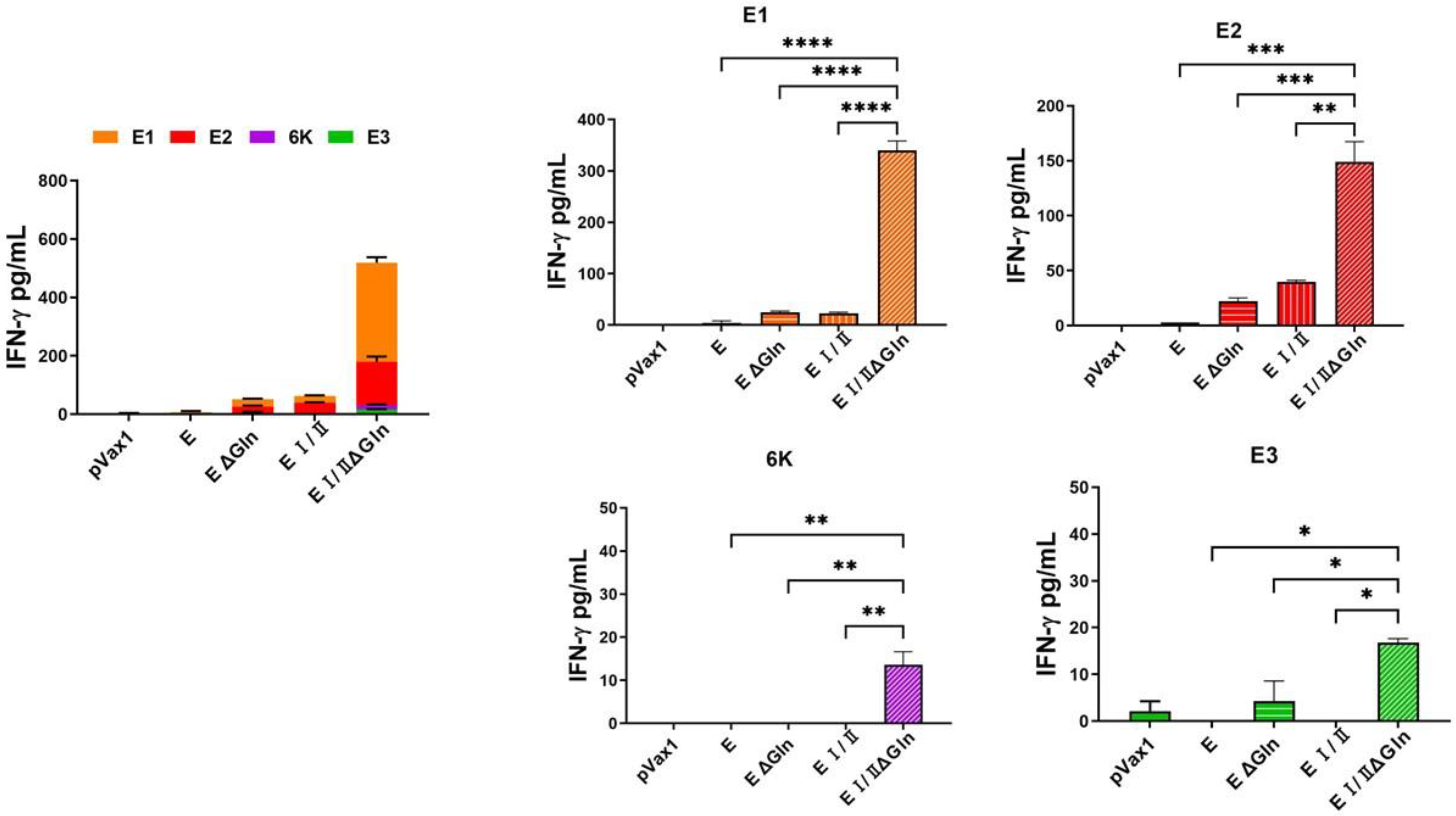
2.7. Multiplex Cytokine Measurement by ELISA
2.8. Statistical Analysis
3. Results
3.1. Designing of DNA Vaccine Candidates against Chikungunya Virus
3.2. Humoral Immune Response Evoked in Mice by the Candidate Vaccines
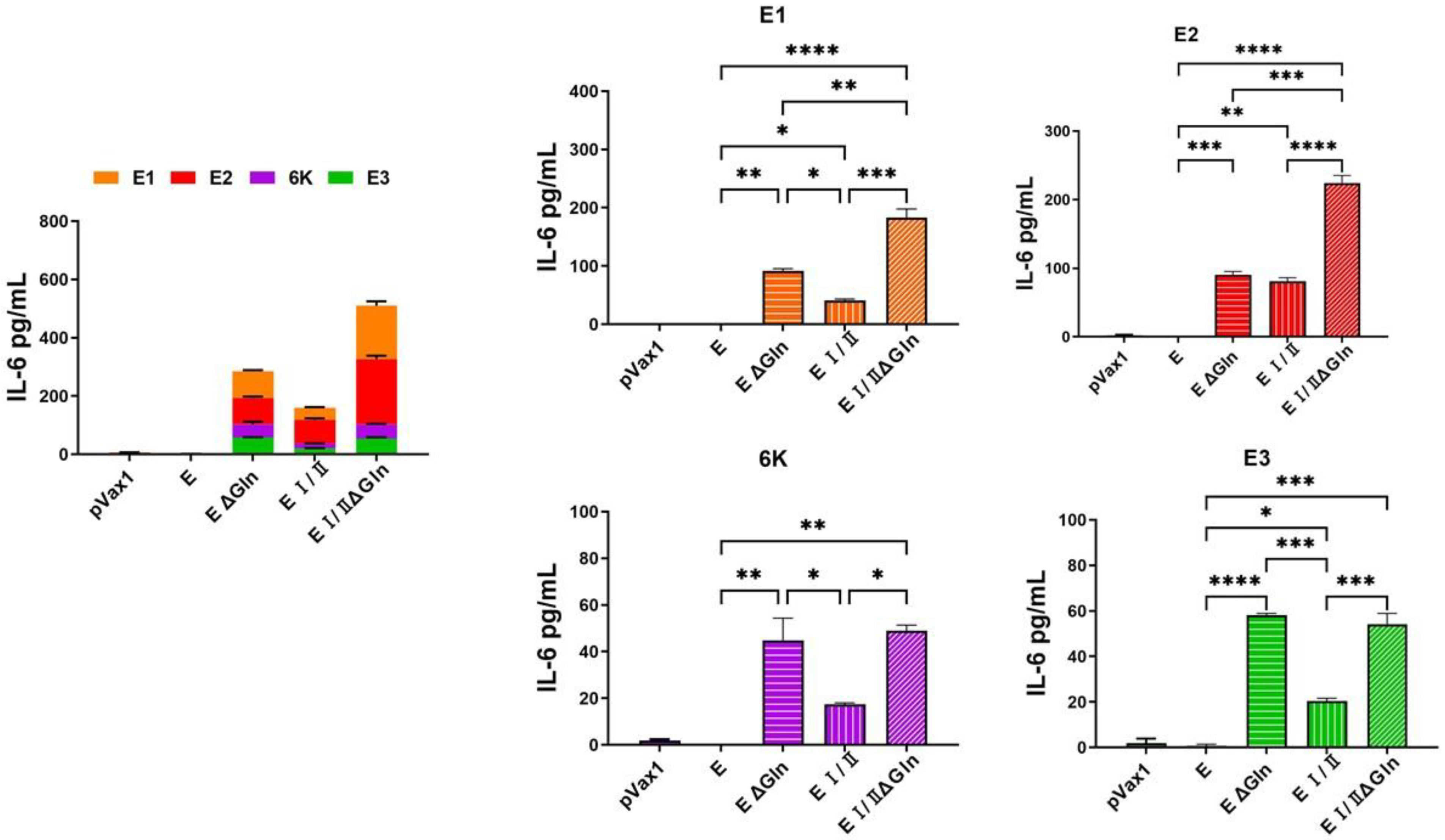
3.3. Cell-Mediated Immune Response Elicited by DNA Vaccines against CHIKV Based on N-Linked Glycan Mutants of the E Protein
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robinson, M.C. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952–1953. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.-C.; Lavenir, R.; Pardigon, N.; Reynes, J.-M.; Pettinelli, F.; et al. Genome Microevolution of Chikungunya Viruses Causing the Indian Ocean Outbreak. PLOS Med. 2006, 3, e263. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.A.; Dermody, T.S. Chikungunya virus: Epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig. 2017, 127, 737–749. [Google Scholar] [CrossRef]
- Leparc-Goffart, I.; Nougairede, A.; Cassadou, S.; Prat, C.; de Lamballerie, X. Chikungunya in the Americas. Lancet 2014, 383, 514. [Google Scholar] [CrossRef]
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.; et al. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef]
- Dupuis-Maguiraga, L.; Noret, M.; Brun, S.; Le Grand, R.; Gras, G.; Roques, P. Chikungunya disease: Infection-associated markers from the acute to the chronic phase of arbo-virus-induced arthralgia. PLoS Neglected Trop. Dis. 2012, 6, e1446. [Google Scholar] [CrossRef] [PubMed]
- Ramphan, S.; Khongwichit, S.; Saisawang, C.; Kovanich, D.; Ketterman, A.J.; Ubol, S.; Auewarakul, P.; Roytrakul, S.; Smith, D.R.; Kuadkitkan, A. Ubiquitin-Conjugating Enzyme E2 L3 is Downregulated by the Chikungunya Virus nsP2 Protease. Proteom.—Clin. Appl. 2018, 12, e1700020. [Google Scholar] [CrossRef] [PubMed]
- Solignat, M.; Gay, B.; Higgs, S.; Briant, L.; Devaux, C. Replication cycle of chikungunya: A re-emerging arbovirus. Virology 2009, 393, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Lecuit, M. Chikungunya Virus and the Global Spread of a Mosquito-Borne Disease. N. Engl. J. Med. 2015, 372, 1231–1239. [Google Scholar] [CrossRef]
- Martín, C.S.-S.; Nanda, S.; Zheng, Y.; Fields, W.; Kielian, M. Cross-Inhibition of Chikungunya Virus Fusion and Infection by Alphavirus E1 Domain III Proteins. J. Virol. 2013, 87, 7680–7687. [Google Scholar] [CrossRef]
- Weger-Lucarelli, J.; Aliota, M.T.; Wlodarchak, N.; Kamlangdee, A.; Swanson, R.; Osorio, J.E. Dissecting the role of E2 protein domains in alphavirus pathogenicity. J. Virol. 2016, 90, 2418–2433. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xiang, Y.; Akahata, W.; Holdaway, H.; Pal, P.; Zhang, X.; Diamond, M.S.; Nabel, G.J.; Rossmann, M.G.; Diseases, I.; et al. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2013, 2, e00435. [Google Scholar] [CrossRef] [PubMed]
- Voss, J.E.; Vaney, M.-C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 2430–2437. [Google Scholar] [CrossRef]
- Stanley, P. Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 2011, 3, a005199. [Google Scholar] [CrossRef]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.L.; Schultz, K.L.W.; Kent, R.J.; Venkatesan, M.; Griffin, D.E. Role of N-Linked Glycosylation for Sindbis Virus Infection and Replication in Vertebrate and Invertebrate Systems. J. Virol. 2009, 83, 5640–5647. [Google Scholar] [CrossRef]
- Chen, L.; Wang, M.; Zhu, D.; Sun, Z.; Ma, J.; Wang, J.; Kong, L.; Wang, S.; Liu, Z.; Wei, L.; et al. Implication for alphavirus host-cell entry and assembly indicated by a 3.5A resolution cryo-EM structure. Nat. Commun. 2018, 9, 5326. [Google Scholar] [CrossRef]
- Ribeiro-Filho, H.V.; Coimbra, L.D.; Cassago, A.; Rocha, R.P.F.; Guerra, J.; de Felicio, R.; Carnieli, C.M.; Leme, L.; Padilha, A.C.; Paes Leme, A.F.; et al. Cryo-EM structure of the mature and infective Mayaro virus at 4.4 A resolution reveals features of arthritogenic alphaviruses. Nat. Commun. 2021, 12, 3038. [Google Scholar] [CrossRef]
- Cohen, B.J.; Audet, S.; Andrews, N.; Beeler, J. Plaque reduction neutralization test for measles antibodies: Description of a standardised laboratory method for use in immunogenicity studies of aerosol vaccination. Vaccine 2007, 26, 59–66. [Google Scholar] [CrossRef]
- Cherian, N.; Bettis, A.; Deol, A.; Kumar, A.; Di Fabio, J.L.; Chaudhari, A.; Yimer, S.; Fahim, R.; Endy, T. Strategic considerations on developing a CHIKV vaccine and ensuring equitable access for countries in need. NPJ Vaccines 2023, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Ly, H. Ixchiq (VLA1553): The first FDA-approved vaccine to prevent disease caused by Chikungunya virus infection. Virulence 2024, 15, 2301573. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Narciso-Abraham, M.; Hadl, S.; McMahon, R.; Toepfer, S.; Fuchs, U.; Hochreiter, R.; Bitzer, A.; Kosulin, K.; Larcher-Senn, J.; et al. Safety and immunogenicity of a single-shot live-attenuated chikungunya vaccine: A double-blind, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2023, 401, 2138–2147. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Liao, H.Y.; Chen, X.; Wang, S.W.; Cheng, C.W.; Shahed-Al-Mahmud, M.; Liu, Y.M.; Mohapatra, A.; Chen, T.H.; Lo, J.M.; et al. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci. Transl. Med. 2022, 14, eabm0899. [Google Scholar] [CrossRef] [PubMed]
- Goyal, K.; Goel, H.; Baranwal, P.; Tewary, A.; Dixit, A.; Pandey, A.K.; Benjamin, M.; Tanwar, P.; Dey, A.; Khan, F.; et al. Immunological Mechanisms of Vaccine-Induced Protection against SARS-CoV-2 in Humans. Immuno 2021, 1, 442–456. [Google Scholar] [CrossRef]
- Dowling, W.; Thompson, E.; Badger, C.; Mellquist, J.L.; Garrison, A.R.; Smith, J.M.; Paragas, J.; Hogan, R.J.; Schmaljohn, C. Influences of Glycosylation on Antigenicity, Immunogenicity, and Protective Efficacy of Ebola Virus GP DNA Vaccines. J. Virol. 2007, 81, 1821–1837. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, P.; Cao, R.; Gu, J. Mutation of putative N-Linked Glycosylation Sites in Japanese encephalitis Virus Premembrane and Envelope proteins enhances humoral immunity in BALB/C mice after DNA vaccination. Virol. J. 2011, 8, 138. [Google Scholar] [CrossRef]
- Ozdilek, A.; Avci, F.Y. Glycosylation as a key parameter in the design of nucleic acid vaccines. Curr. Opin. Struct. Biol. 2022, 73, 102348. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Moon, S.Y.; Kim, S.; Ouh, I.-O.; Lee, Y.; Lim, H. Immunogenicity Analysis of Chikungunya Virus DNA Vaccine Based on Mutated Putative N-Linked Glycosylation Sites of the Envelope Protein. Vaccines 2024, 12, 1097. https://doi.org/10.3390/vaccines12101097
Kim K, Moon SY, Kim S, Ouh I-O, Lee Y, Lim H. Immunogenicity Analysis of Chikungunya Virus DNA Vaccine Based on Mutated Putative N-Linked Glycosylation Sites of the Envelope Protein. Vaccines. 2024; 12(10):1097. https://doi.org/10.3390/vaccines12101097
Chicago/Turabian StyleKim, Kwangwook, Seo Young Moon, Seungyeon Kim, In-Ohk Ouh, Yookyoung Lee, and Heeji Lim. 2024. "Immunogenicity Analysis of Chikungunya Virus DNA Vaccine Based on Mutated Putative N-Linked Glycosylation Sites of the Envelope Protein" Vaccines 12, no. 10: 1097. https://doi.org/10.3390/vaccines12101097
APA StyleKim, K., Moon, S. Y., Kim, S., Ouh, I.-O., Lee, Y., & Lim, H. (2024). Immunogenicity Analysis of Chikungunya Virus DNA Vaccine Based on Mutated Putative N-Linked Glycosylation Sites of the Envelope Protein. Vaccines, 12(10), 1097. https://doi.org/10.3390/vaccines12101097





