Symptomatology and IgG Levels before and after SARS-CoV-2 Omicron Breakthrough Infections in Vaccinated Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Serological Analysis
2.3. Variant Identification
2.4. Data Analysis
3. Results
3.1. Cohort Description
3.2. Time Course IgG Titers, Variant and Disease Symptomatology
3.3. Correlation between IgG Levels and COVID-19 Symptomatology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iheanacho, C.O.; Eze, U.I.H.; Adida, E.A. A systematic review of effectiveness of BNT162b2 mRNA and ChAdOx1 adenoviral vector COVID-19 vaccines in the general population. Bull. Natl. Res. Cent. 2021, 45, 150. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira-Silva, T.; Andrews, J.R.; Boaventura, V.S.; Ranzani, O.T.; Oliveira, V.D.A.; Paixão, E.S.; Júnior, J.B.; Machado, T.M.; Hitchings, M.D.T.; Dorion, M.; et al. Effectiveness of CoronaVac, ChAdOx1 nCoV-19, BNT162b2, and Ad26.COV2.S among individuals with previous SARS-CoV-2 infection in Brazil: A test-negative, case-control study. Lancet Infect. Dis. 2022, 22, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira-Silva, T.; Katikireddi, S.V.; Oliveira, V.d.A.; Flores-Ortiz, R.; Júnior, J.B.; Paixão, E.S.; Robertson, C.; Penna, G.O.; Werneck, G.L.; Barreto, M.L.; et al. Vaccine effectiveness of heterologous CoronaVac plus BNT162b2 in Brazil. Nat. Med. 2022, 28, 838–843. [Google Scholar] [CrossRef]
- Fadlyana, E.; Setiabudi, D.; Kartasasmita, C.B.; Putri, N.D.; Hadinegoro, S.R.; Mulholland, K.; Sofiatin, Y.; Suryadinata, H.; Hartantri, Y.; Sukandar, H.; et al. Immunogenicity and safety in healthy adults of full dose versus half doses of COVID-19 vaccine (ChAdOx1-S or BNT162b2) or full-dose CoronaVac administered as a booster dose after priming with CoronaVac: A randomised, observer-masked, controlled trial in Indonesia. Lancet Infect. Dis. 2023, 23, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111, Erratum in Lancet 2021, 397, 98. [Google Scholar] [CrossRef]
- Kahn, R.; Janusz, C.B.; Castro, M.C.; Matos, A.d.R.; Domingues, C.; Ponmattam, J.; Rey-Benito, G.; Toscano, C.M.; de Oliveira, L.H.; Rearte, A.; et al. The effectiveness of COVID-19 vaccines in Latin America, 2021: A multicenter regional case-control study. Lancet Reg. Health Am. 2023, 20, 100474. [Google Scholar] [CrossRef]
- dos Santos, C.V.B.; Valiati, N.C.M.; de Noronha, T.G.; Porto, V.B.G.; Pacheco, A.G.; Freitas, L.P.; Coelho, F.C.; Gomes, M.F.d.C.; Bastos, L.S.; Cruz, O.G.; et al. The effectiveness of COVID-19 vaccines against severe cases and deaths in Brazil from 2021 to 2022: A registry-based study. Lancet Reg. Health Am. 2023, 20, 100465. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ravichandran, S.; Lee, Y.; Grubbs, G.; Coyle, E.M.; Klenow, L.; Akasaka, O.; Koga, M.; Adachi, E.; Saito, M.; Nakachi, I.; et al. Longitudinal antibody repertoire in “mild” versus “severe” COVID-19 patients reveals immune markers associated with disease severity and resolution. Sci. Adv. 2021, 7, eabf2467. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, J.; Liang, B.-Y.; Fang, Y.-H.; Wang, H.; Yang, X.-L.; Shen, S.; Chen, L.-K.; Li, S.-M.; Lu, S.-H.; Xiang, T.-D.; et al. Occurrence of COVID-19 Symptoms During SARS-CoV-2 Infection Defines Waning of Humoral Immunity. Front. Immunol. 2021, 12, 722027. [Google Scholar] [CrossRef]
- Hajilooi, M.; Keramat, F.; Moazenian, A.; Rastegari-Pouyani, M.; Solgi, G. The quantity and quality of anti-SARS-CoV-2 antibodies show contrariwise association with COVID-19 severity: Lessons learned from IgG avidity. Med. Microbiol. Immunol. 2023, 212, 203–220. [Google Scholar] [CrossRef]
- Paula, N.M.; Conzentino, M.S.; Gonçalves, A.C.A.; da Silva, R.; Weissheimer, K.V.; Kluge, C.H.S.; Marins, P.H.S.A.; Camargo, H.S.C.; Farias, L.R.P.; Sant’ana, T.P.; et al. Population-Based Analysis of the Immunoglobulin G Response to Different COVID-19 Vaccines in Brazil. Vaccines 2023, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Moreira, E.D.; Kitchin, N.; Xu, X.; Dychter, S.S.; Lockhart, S.; Gurtman, A.; Perez, J.L.; Zerbini, C.; Dever, M.E.; Jennings, T.W.; et al. Safety and Efficacy of a Third Dose of BNT162b2 Covid-19 Vaccine. N. Engl. J. Med. 2022, 386, 1910–1921. [Google Scholar] [CrossRef] [PubMed]
- Barin, B.; Kasap, U.; Selçuk, F.; Volkan, E.; Uluçkan, Ö. Comparison of SARS-CoV-2 anti-spike receptor binding domain IgG antibody responses after CoronaVac, BNT162b2, ChAdOx1 COVID-19 vaccines, and a single booster dose: A prospective, longitudinal population-based study. Lancet Microbe 2022, 3, e274–e283. [Google Scholar] [CrossRef] [PubMed]
- Pratesi, F.; Caruso, T.; Testa, D.; Tarpanelli, T.; Gentili, A.; Gioè, D.; Migliorini, P. BNT162b2 mRNA SARS-CoV-2 Vaccine Elicits High Avidity and Neutralizing Antibodies in Healthcare Workers. Vaccines 2021, 9, 672. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Feikin, D.R.; Feikin, D.R.; Higdon, M.M.; Higdon, M.M.; Abu-Raddad, L.J.; Abu-Raddad, L.J.; Andrews, N.; Andrews, N.; Araos, R.; Araos, R.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944, Erratum in Lancet 2023, 401, 644. [Google Scholar] [CrossRef]
- Ssentongo, P.; Ssentongo, A.E.; Voleti, N.; Groff, D.; Sun, A.; Ba, D.M.; Nunez, J.; Parent, L.J.; Chinchilli, V.M.; Paules, C.I. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: A systematic review and meta-analysis. BMC Infect. Dis. 2022, 22, 439. [Google Scholar] [CrossRef]
- Shrotri, M.; Navaratnam, A.M.D.; Nguyen, V.; Byrne, T.; Geismar, C.; Fragaszy, E.; Beale, S.; Fong, W.L.E.; Patel, P.; Kovar, J.; et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet 2021, 398, 385–387. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 385, e84. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, W.; Ma, J.; Wu, S.; Sun, F. Reinfection rates among patients previously infected by SARS-CoV-2: Systematic review and meta-analysis. Chin. Med. J. 2022, 135, 145–152. [Google Scholar] [CrossRef]
- Hansen, C.H.; Michlmayr, D.; Gubbels, S.M.; Mølbak, K.; Ethelberg, S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: A population-level observational study. Lancet 2021, 397, 1204–1212. [Google Scholar] [CrossRef]
- Nordström, P.; Ballin, M.; Nordström, A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: A retrospective, total population cohort study in Sweden. Lancet Infect Dis. 2022, 22, 781–790, Erratum in Lancet Infect Dis. 2022, 22, E159. [Google Scholar] [CrossRef] [PubMed]
- Chemaitelly, H.; Nagelkerke, N.; Ayoub, H.H.; Coyle, P.; Tang, P.; Yassine, H.M.; A Al-Khatib, H.; Smatti, M.K.; Hasan, M.R.; Al-Kanaani, Z.; et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J. Travel. Med. 2022, 29, taac109. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.; Weisblum, Y.; Rutkowska, M.; Poston, D.; DaSilva, J.; Zhang, F.; Bednarski, E.; Cho, A.; Schaefer-Babajew, D.J.; Gaebler, C.; et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 2021, 600, 512–516. [Google Scholar] [CrossRef]
- Wratil, P.R.; Stern, M.; Priller, A.; Willmann, A.; Almanzar, G.; Vogel, E.; Feuerherd, M.; Cheng, C.-C.; Yazici, S.; Christa, C.; et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat. Med. 2022, 28, 496–503. [Google Scholar] [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Altarawneh, H.N.; Chemaitelly, H.; Ayoub, H.H.; Tang, P.; Hasan, M.R.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Coyle, P.; Al-Kanaani, Z.; et al. Effects of Previous Infection and Vaccination on Symptomatic Omicron Infections. N. Engl. J. Med. 2022, 387, 21–34. [Google Scholar] [CrossRef]
- Freire-Neto, F.P.; Teixeira, D.G.; da Cunha, D.C.S.; Morais, I.C.; Tavares, C.P.M.; Gurgel, G.P.; Medeiros, S.D.N.; dos Santos, D.C.; Sales, A.D.O.; Jeronimo, S.M.B. SARS-CoV-2 reinfections with BA.1 (Omicron) variant among fully vaccinated individuals in northeastern Brazil. PLoS Negl. Trop. Dis. 2022, 16, e0010337. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.R.; Ruiz, C.M.R.; Machado, R.R.G.; Magawa, J.Y.; Daher, I.P.; Urbanski, A.H.; Schmitz, G.J.H.; Arcuri, H.A.; Ferreira, M.A.; Sasahara, G.L.; et al. Immunodominant antibody responses directed to SARS-CoV-2 hotspot mutation sites and risk of immune escape. Front. Immunol. 2023, 13, 1010105. [Google Scholar] [CrossRef]
- Rhoden, J.; Hoffmann, A.T.; Stein, J.F.; da Silva, M.S.; Gularte, J.S.; Filippi, M.; Demoliner, M.; Girardi, V.; Spilki, F.R.; Fleck, J.D.; et al. Diversity of Omicron sublineages and clinical characteristics in hospitalized patients in the southernmost state of Brazil. BMC Infect. Dis. 2024, 24, 193. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Immunological memory to SARS-CoV-2 infection and COVID-19 vaccines. Immunol. Rev. 2022, 310, 27–46. [Google Scholar] [CrossRef]
- Goel, R.R.; Apostolidis, S.A.; Painter, M.M.; Mathew, D.; Pattekar, A.; Kuthuru, O.; Gouma, S.; Hicks, P.; Meng, W.; Rosenfeld, A.M.; et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021, 6, eabi6950. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Day, T.; Arino, J.; Colijn, C.; Dushoff, J.; Li, M.; Mechai, S.; Van Domselaar, G.; Wu, J.; Earn, D.J.; et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021, 31, R918–R929. [Google Scholar] [CrossRef] [PubMed]
- Calistri, P.; Amato, L.; Puglia, I.; Cito, F.; Di Giuseppe, A.; Danzetta, M.L.; Morelli, D.; Di Domenico, M.; Caporale, M.; Scialabba, S.; et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021, 105, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhong, J.; Xiong, H.; Li, Y.; Guo, T.; Peng, B.; Fang, C.; Kang, Y.; Tan, J.; Ma, Y. Protective Effect of Inactivated COVID-19 Vaccines against Omicron BA.2 Infection in Guangzhou: A Test-Negative Case-Control Real-World Study. Vaccines 2023, 11, 566. [Google Scholar] [CrossRef]
- Garrett, N.; Tapley, A.; Andriesen, J.; Seocharan, I.; Fisher, L.H.; Bunts, L.; Espy, N.; Wallis, C.L.; Randhawa, A.K.; Ketter, N.; et al. High Rate of Asymptomatic Carriage Associated with Variant Strain Omicron. medRxiv. 2022. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; COVID-19 Genomics UK Consortium; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef]
- Huergo, L.F.; Paula, N.M.; Gonçalves, A.C.A.; Kluge, C.H.S.; Marins, P.H.S.A.; Camargo, H.S.C.; Sant’ana, T.P.; Farias, L.R.P.; Aldrighi, J.D.; Lima, S.; et al. SARS-CoV-2 Seroconversion in Response to Infection and Vaccination: A Time Series Local Study in Brazil. Microbiol. Spectr. 2022, 10, e01026-22. [Google Scholar] [CrossRef]
- Conzentino, M.S.; Gonçalves, A.C.A.; Paula, N.M.; Rego, F.G.M.; Zanette, D.L.; Aoki, M.N.; Nardin, J.M.; Huergo, L.F. A magnetic bead immunoassay to detect high affinity human IgG reactive to SARS-CoV-2 Spike S1 RBD produced in Escherichia coli. Braz. J. Microbiol. 2022, 53, 1263–1269. [Google Scholar] [CrossRef]
- Conzentino, M.S.; Santos, T.P.; Selim, K.A.; Wagner, B.; Alford, J.T.; Deobald, N.; Paula, N.M.; Rego, F.G.; Zanette, D.L.; Aoki, M.N.; et al. Ultra-fast, high throughput and inexpensive detection of SARS-CoV-2 seroconversion using Ni2+ magnetic beads. Anal. Biochem. 2021, 631, 114360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huergo, L.F.; Selim, K.A.; Conzentino, M.S.; Gerhardt, E.C.M.; Santos, A.R.S.; Wagner, B.; Alford, J.T.; Deobald, N.; Pedrosa, F.O.; de Souza, E.M.; et al. Magnetic Bead-Based Immunoassay Allows Rapid, Inexpensive, and Quantitative Detection of Human SARS-CoV-2 Antibodies. ACS Sens. 2021, 3, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Conzentino, M.S.; Forchhammer, K.; Souza, E.M.; Pedrosa, F.O.; Nogueira, M.B.; Raboni, S.M.; Rego, F.G.M.; Zanette, D.L.; Aoki, M.N.; Nardin, J.M.; et al. Antigen production and development of an indirect ELISA based on the nucleocapsid protein to detect human SARS-CoV-2 seroconversion. Braz. J. Microbiol. 2021, 52, 2069–2073. [Google Scholar] [CrossRef] [PubMed]
- Bochnia-Bueno, L.; De Almeida, S.M.; Raboni, S.M.; Adamoski, D.; Amadeu, L.L.M.; Carstensen, S.; Nogueira, M.B. Dynamic of humoral response to SARS-CoV-2 anti-Nucleocapsid and Spike proteins after CoronaVac vaccination. Diagn. Microbiol. Infect. Dis. 2022, 102, 115597. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Wei, J.; Pouwels, K.B.; Stoesser, N.; Matthews, P.C.; Diamond, I.; Studley, R.; Rourke, E.; Cook, D.; Bell, J.I.; Newton, J.N.; et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat. Med. 2022, 28, 1072–1082. [Google Scholar] [CrossRef]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef]
- Yamamoto, S.; Mizoue, T.; Ohmagari, N. Analysis of Previous Infection, Vaccinations, and Anti-SARS-CoV-2 Antibody Titers and Protection Against Infection With the SARS-CoV-2 Omicron BA.5 Variant. JAMA Netw. Open. 2023, 6, e233370. [Google Scholar] [CrossRef]
- Murray, S.M.; Ansari, A.M.; Frater, J.; Klenerman, P.; Dunachie, S.; Barnes, E.; Ogbe, A. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat. Rev. Immunol. 2023, 23, 304–316. [Google Scholar] [CrossRef]
- Yang, Z.-R.; Jiang, Y.-W.; Li, F.-X.; Liu, D.; Lin, T.-F.; Zhao, Z.-Y.; Wei, C.; Jin, Q.-Y.; Li, X.-M.; Jia, Y.-X.; et al. Efficacy of SARS-CoV-2 vaccines and the dose-response relationship with three major antibodies: A systematic review and meta-analysis of randomised controlled trials. Lancet Microbe 2023, 4, e236–e246. [Google Scholar] [CrossRef]
- Buhre, J.S.; Pongracz, T.; Künsting, I.; Lixenfeld, A.S.; Wang, W.; Nouta, J.; Lehrian, S.; Schmelter, F.; Lunding, H.B.; Dühring, L.; et al. mRNA vaccines against SARS-CoV-2 induce comparably low long-term IgG Fc galactosylation and sialylation levels but increasing long-term IgG4 responses compared to an adenovirus-based vaccine. Front. Immunol. 2023, 13, 1020844. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Q.; Vishwanath, S.; Carnell, G.W.; Chan, A.C.Y.; Heeney, J.L. Immune imprinting and next-generation coronavirus vaccines. Nat. Microbiol. 2023, 8, 1971–1985. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Jung, J.; Ko, G.Y.; Lee, J.; Oh, E.-J. Evaluation of Long-Term Adaptive Immune Responses Specific to SARS-CoV-2: Effect of Various Vaccination and Omicron Exposure. Vaccines 2024, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Goguet, E.; Olsen, C.H.; Meyer, W.A., III; Ansari, S.; Powers, J.H., III; Conner, T.L.; Coggins, S.A.; Wang, W.; Wang, R.; Illinik, L.; et al. Immune and behavioral correlates of protection against symptomatic post-vaccination SARS-CoV-2 infection. Front. Immunol. 2024, 15, 1287504. [Google Scholar] [CrossRef] [PubMed]
- Spiteri, G.; D’agostini, M.; Abedini, M.; Ditano, G.; Collatuzzo, G.; Boffetta, P.; Vimercati, L.; Sansone, E.; De Palma, G.; Modenese, A.; et al. Protective role of SARS-CoV-2 anti-S IgG against breakthrough infections among European healthcare workers during pre and post-Omicron surge-ORCHESTRA project. Infection 2024, 52, 1347–1356. [Google Scholar] [CrossRef]
- Jia, X.; Cao, S.; Lee, A.S.; Manohar, M.; Sindher, S.B.; Ahuja, N.; Artandi, M.; Blish, C.A.; Blomkalns, A.L.; Chang, I.; et al. Anti-nucleocapsid antibody levels and pulmonary comorbid conditions are linked to post-COVID-19 syndrome. JCI Insight 2022, 7, e156713. [Google Scholar] [CrossRef]
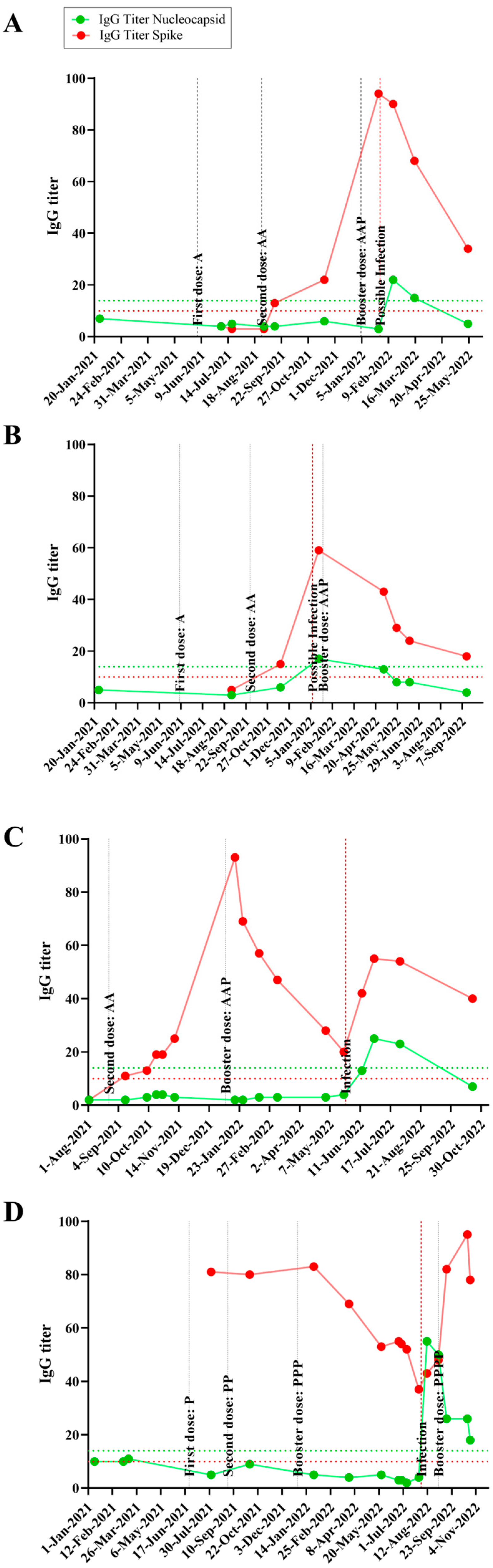
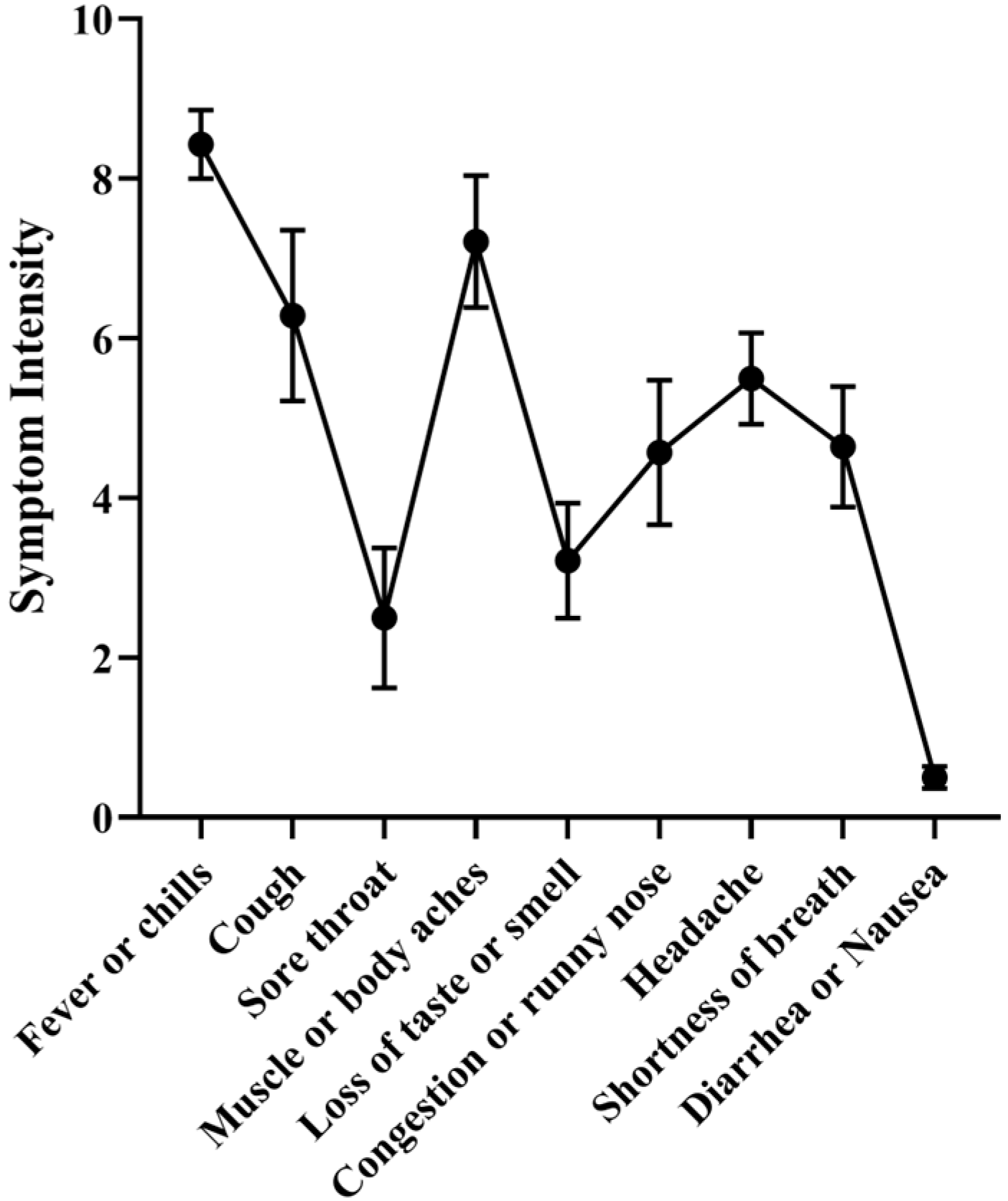
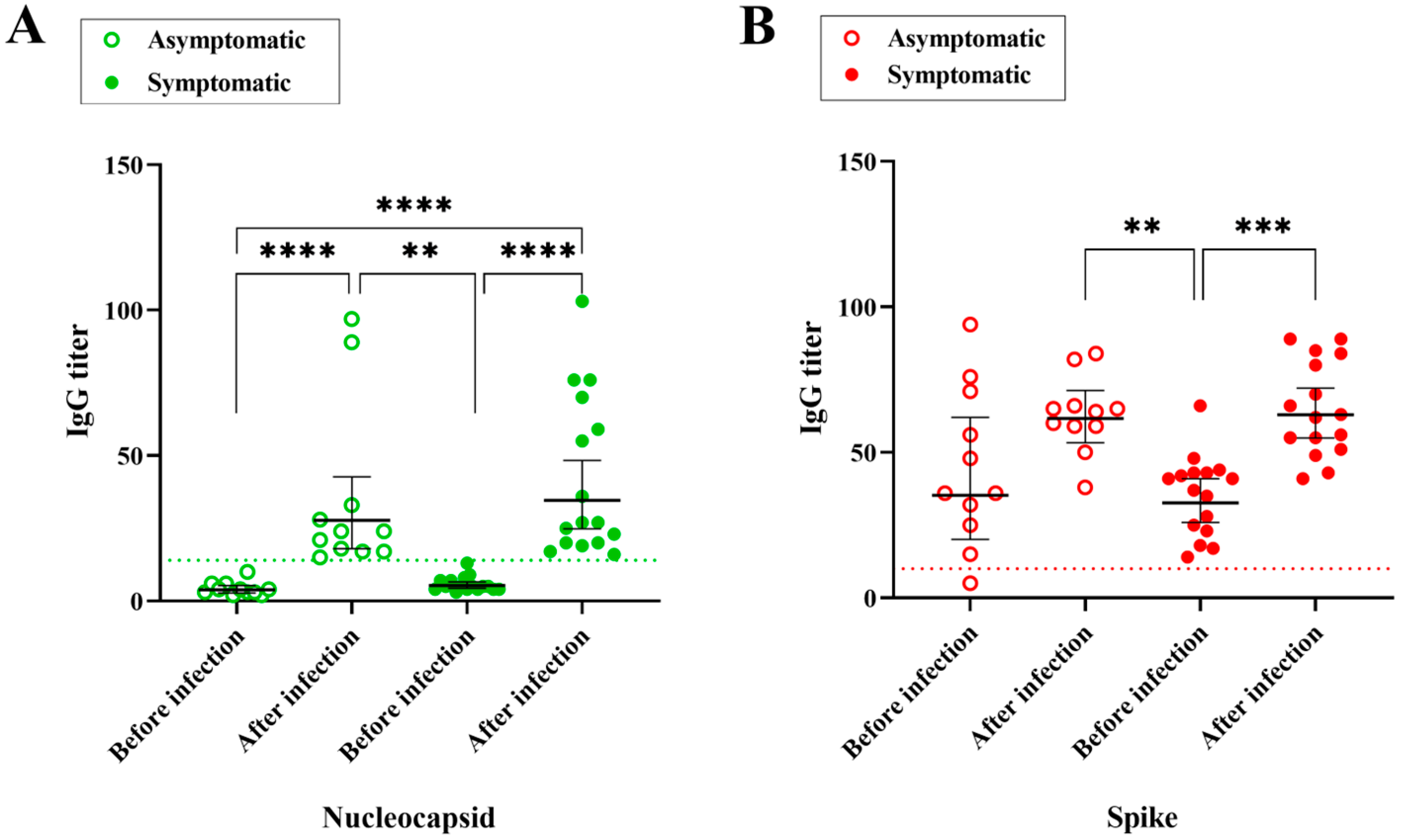
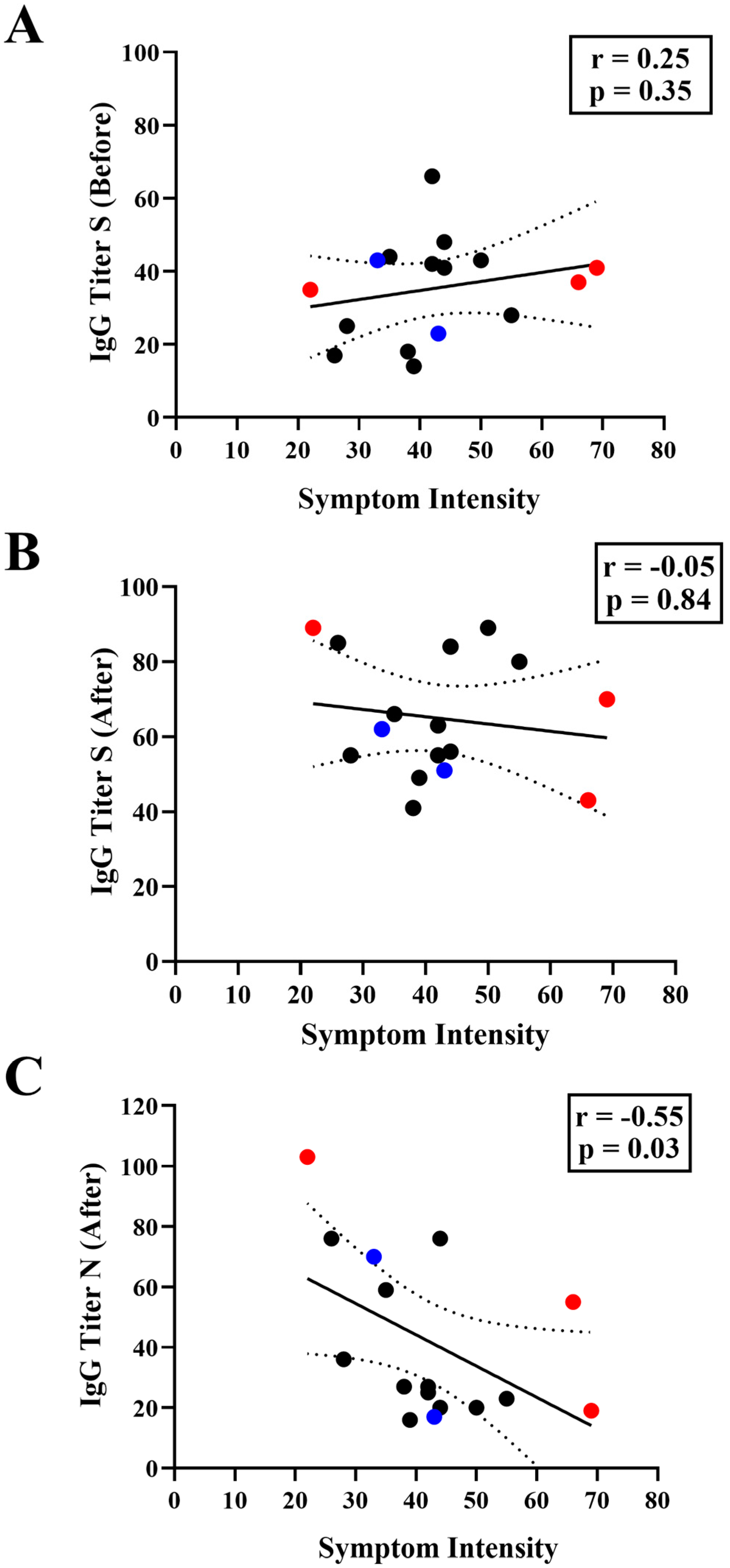
| Individual Code | Age | Gender | Previous Infection | Positive Test | Sum of Symptoms | Vaccination Scheme before Infection | Omicron Sublineage | Anti-N IgG Titer before Infection | Anti-N IgG Titer after Infection | Anti-S IgG Titer before Infection | Anti-S IgG Titer after Infection |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 37 | Female | 0 | PPP | BA.5 | 2 | 15 | 71 | 60 | ||
| AA | 41 | Female | Yes | 35 | AAP | BA.2 | 5 | 59 | 44 | 66 | |
| B | 32 | Female | 0 | AA | BA.5 | 2 | 28 | 5 | 59 | ||
| BB | 19 | Female | 0 | CCP | BA.2 | 6 | 97 | 76 | 50 | ||
| C | 46 | Female | 0 | AAP | BA.2 | 3 | 17 | 36 | 65 | ||
| CC | 38 | Female | Yes | 28 | AAP | BA.5 | 4 | 36 | 25 | 55 | |
| D | 31 | Female | 0 | CCP | BA.5 | 3 | 24 | 32 | 66 | ||
| DD | 41 | Male | 0 | AAP | BA.5 | 4 | 24 | 36 | 38 | ||
| E | 51 | Male | 0 | AAP | BA.5 | 3 | 18 | 94 | 84 | ||
| G | 36 | Male | Yes | 55 | PPP | BA.5 | 3 | 23 | 28 | 80 | |
| GG | 30 | Female | 0 | PPA | BA.5 | 4 | 21 | 56 | 64 | ||
| H | 38 | Male | Yes | 0 | AA | BA.2 | 6 | 17 | 15 | 59 | |
| I | 44 | Female | Yes | 39 | AAPP | BA.5 | 5 | 16 | 14 | 49 | |
| J | 65 | Female | Yes | Yes | 22 | CCPJ | BA.1 | 7 | 103 | 35 | 89 |
| K | 24 | Female | Yes | 33 | PP | BA.1 | 7 | 70 | 43 | 62 | |
| L | 61 | Female | Yes | 44 | AAP | BA.2 | 5 | 20 | 48 | 84 | |
| M | 31 | Male | Yes | 50 | CCP | BA.5 | 8 | 20 | 43 | 89 | |
| N | 44 | Male | Yes | 42 | AAP | BA.2 | 13 | 25 | 42 | 55 | |
| O | 74 | Male | Yes | 26 | CCPJ | BA.1 | 9 | 76 | 17 | 85 | |
| P | 39 | Female | Yes | 44 | CCP | BA.2 | 5 | 76 | 41 | 56 | |
| Q | 39 | Female | Yes | 38 | AAA | BA.2 | 4 | 27 | 18 | 41 | |
| R | 31 | Female | Yes | Yes | 66 | PPPP | BA.5 | 4 | 55 | 37 | 43 |
| S | 39 | Female | Yes | Yes | 69 | AAP | BA.2 | 4 | 19 | 41 | 70 |
| U | 41 | Female | 0 | AAA | BA.5 | 4 | 33 | 25 | 65 | ||
| V | 24 | Female | Yes | 43 | PP | BA.2 | 4 | 17 | 23 | 51 | |
| X | 44 | Male | Yes | 0 | AA | BA.5 | 10 | 89 | 48 | 82 | |
| Y | 48 | Female | Yes | 42 | AAP | BA.2 | 5 | 27 | 66 | 63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paula, N.M.; Joucoski, E.; Baura, V.A.; Souza, E.M.; Pedrosa, F.O.; Gonçalves, A.G.; Huergo, L.F. Symptomatology and IgG Levels before and after SARS-CoV-2 Omicron Breakthrough Infections in Vaccinated Individuals. Vaccines 2024, 12, 1149. https://doi.org/10.3390/vaccines12101149
Paula NM, Joucoski E, Baura VA, Souza EM, Pedrosa FO, Gonçalves AG, Huergo LF. Symptomatology and IgG Levels before and after SARS-CoV-2 Omicron Breakthrough Infections in Vaccinated Individuals. Vaccines. 2024; 12(10):1149. https://doi.org/10.3390/vaccines12101149
Chicago/Turabian StylePaula, Nigella M., Emerson Joucoski, Valter A. Baura, Emanuel M. Souza, Fabio O. Pedrosa, Alan G. Gonçalves, and Luciano F. Huergo. 2024. "Symptomatology and IgG Levels before and after SARS-CoV-2 Omicron Breakthrough Infections in Vaccinated Individuals" Vaccines 12, no. 10: 1149. https://doi.org/10.3390/vaccines12101149







