Second-Generation Phage Lambda Platform Employing SARS-CoV-2 Fusion Proteins as a Vaccine Candidate
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Production
2.2.1. Cloning, Expression and Purification of the RBD Constructs
2.2.2. Purification of Phage Lambda Proteins
2.3. Biophysical Characterization Studies
2.3.1. Analytical Ultracentrifugation
2.3.2. Circular Dichroism Spectroscopy
2.4. In Vitro Phage Lambda PLP Studies
2.4.1. Particle Decoration
2.4.2. Particle Characterization
2.5. Viruses and In Vivo Murine Studies
2.5.1. Virus and Cell Culture
2.5.2. Animal Study Design
2.5.3. Particle Antigenicity and Immunogenicity Testing
2.5.4. Neutralization Assay
2.5.5. Quantification of SARS-CoV-2 Genomic and Subgenomic RNA
2.6. Graphing and Statistical Analyses
3. Results
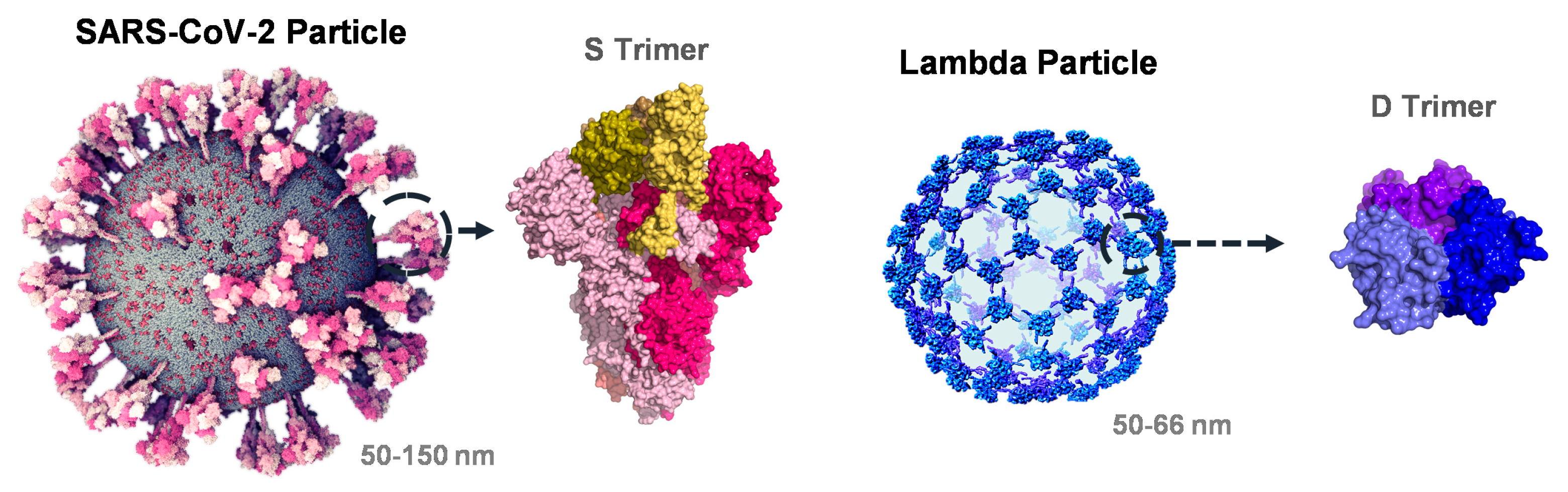
3.1. Interrogation of SARS-CoV-2 and Phage Lambda for RBD Fusion Design
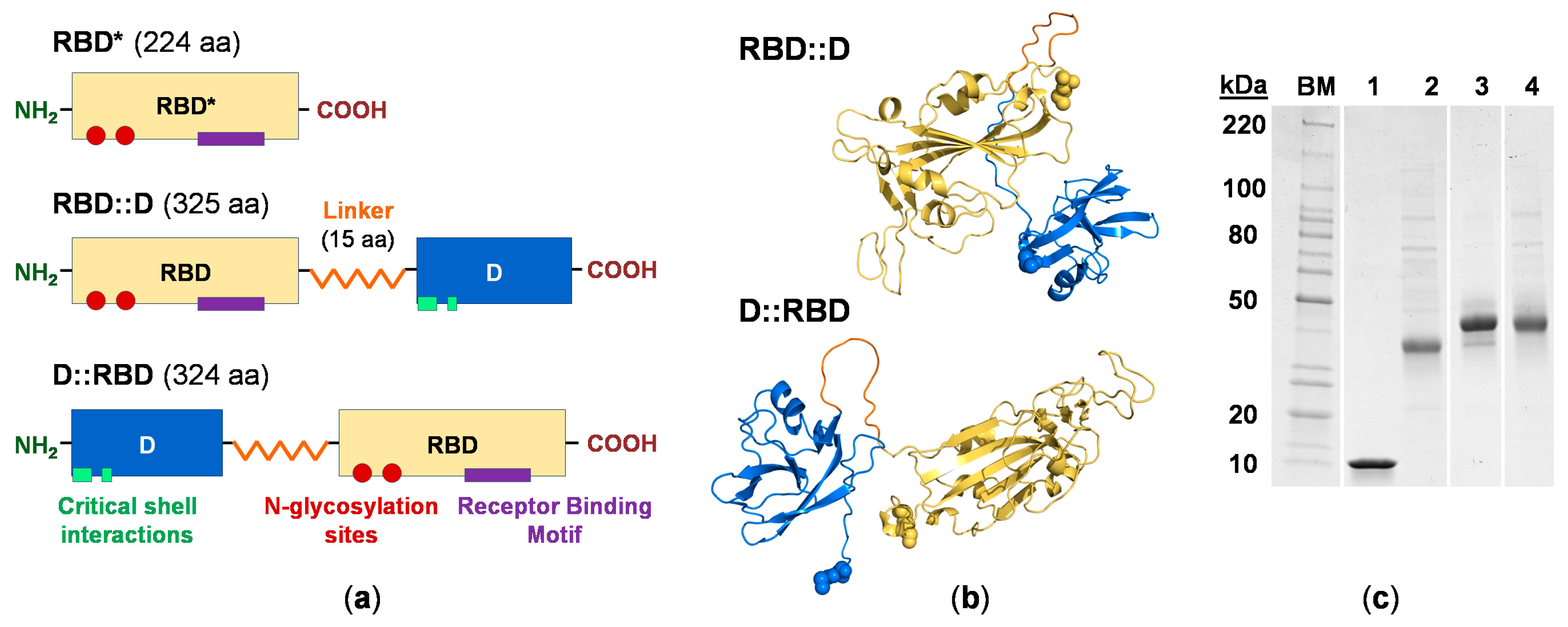
3.2. Engineering the RBD Constructs
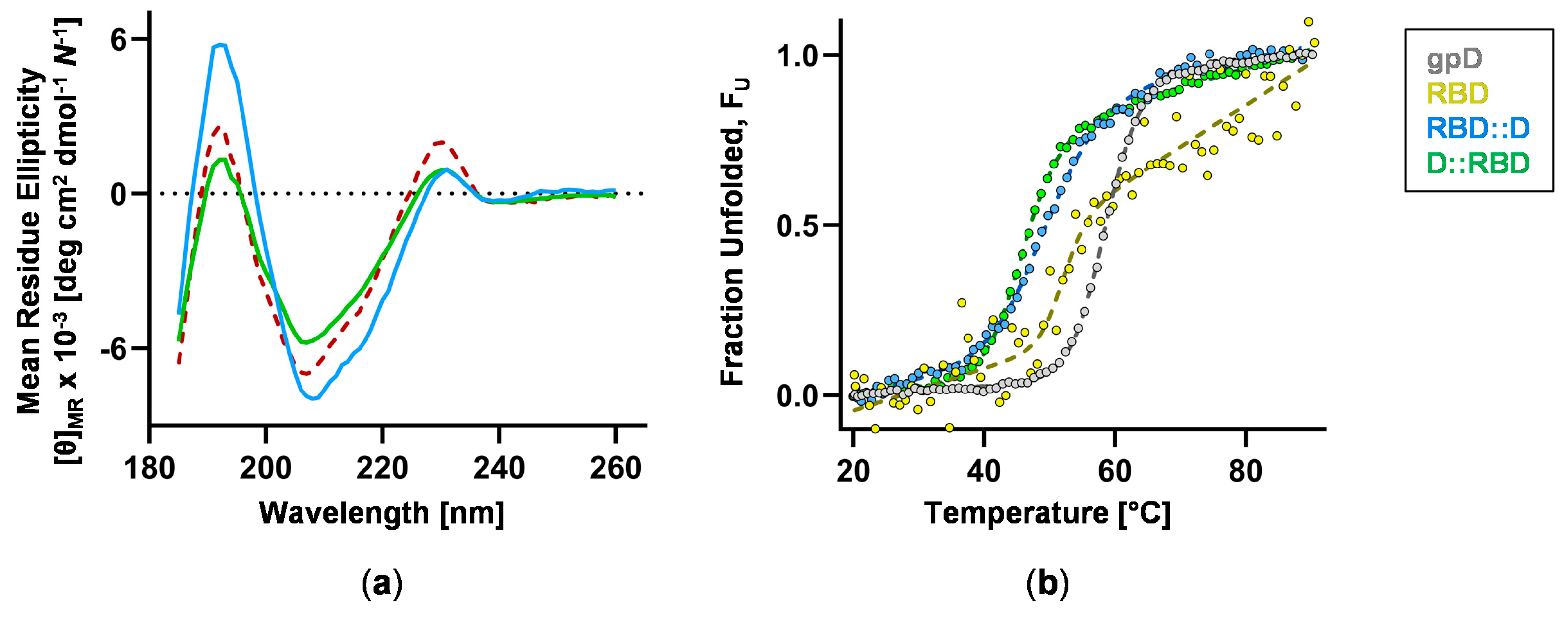
3.3. RBD Construct Characterization
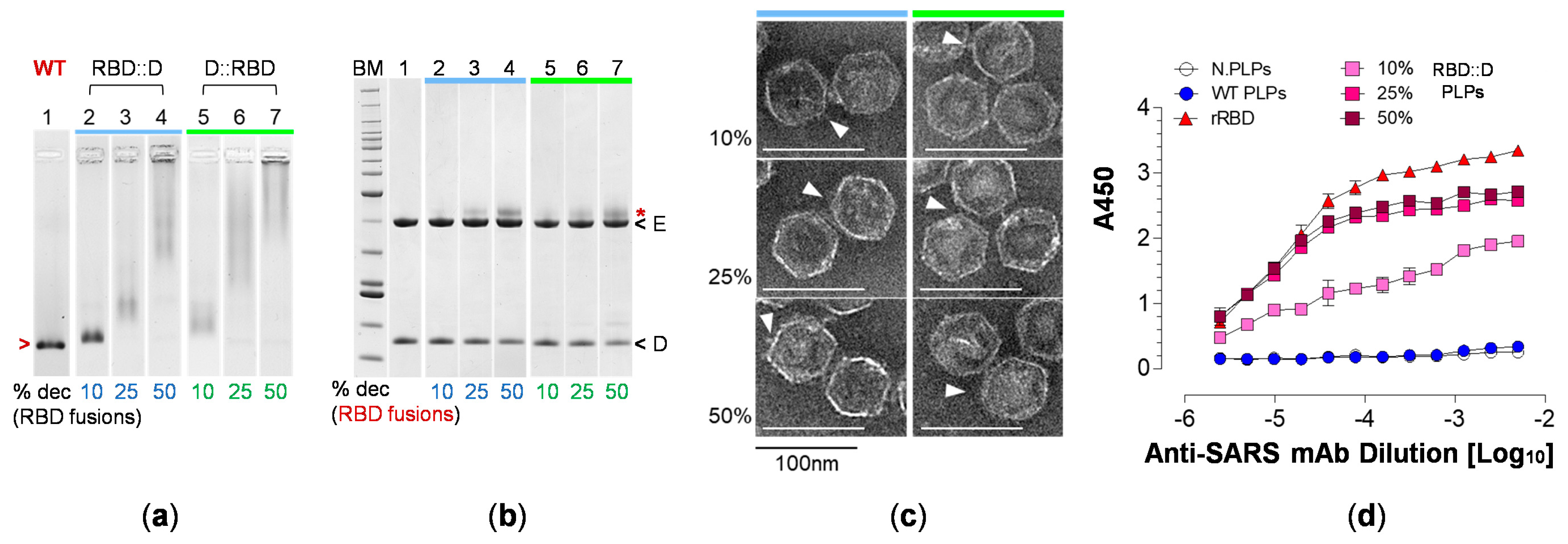
3.4. RBD Fusions Are Efficiently Displayed on PLPs and Remain Antigenic
3.5. H6S-D::RBD PLPs Are Immunogenic
3.6. Vaccination with H6S-D::RBD PLPs Protects Against Virulent SARS-CoV-2 Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Kristensen, D. Opportunities and challenges of developing thermostable vaccines. Expert. Rev. Vaccines 2009, 8, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Rohovie, M.J.; Nagasawa, M.; Swartz, J.R. Virus-like particles: Next-generation nanoparticles for targeted therapeutic delivery. Bioeng. Transl. Med. 2017, 2, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Preston, K.B.; Randolph, T.W. Stability of lyophilized and spray dried vaccine formulations. Adv. Drug Deliv. Rev. 2021, 171, 50–61. [Google Scholar] [CrossRef]
- Smit, M.J.; Sander, A.F.; Ariaans, M.; Fougeroux, C.; Heinzel, C.; Fendel, R.; Esen, M.; Kremsner, P.G.; Ter Heine, R.; Wertheim, H.F.; et al. First-in-human use of a modular capsid virus-like vaccine platform: An open-label, non-randomised, phase 1 clinical trial of the SARS-CoV-2 vaccine ABNCoV2. Lancet Microbe 2023, 4, e140–e148. [Google Scholar] [CrossRef]
- Yong, X.; Liu, J.; Zeng, Y.; Nie, J.; Cui, X.; Wang, T.; Wang, Y.; Chen, Y.; Kang, W.; Yang, Z.; et al. Safety and immunogenicity of a heterologous booster with an RBD virus-like particle vaccine following two- or three-dose inactivated COVID-19 vaccine. Hum. Vaccin. Immunother. 2023, 19, 2267869. [Google Scholar] [CrossRef]
- Hager, K.J.; Perez Marc, G.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F.; et al. Efficacy and Safety of a Recombinant Plant-Based Adjuvanted COVID-19 Vaccine. N. Engl. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef]
- Catalano, C.E. Bacteriophage lambda: The path from biology to theranostic agent. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1517. [Google Scholar] [CrossRef]
- Catalano, C. Bacteriophage Lambda as a Nano Theranostic Platform. In Physical Virology; Springer: Cham, Switzerland, 2023; pp. 307–328. [Google Scholar] [CrossRef]
- Chang, J.R.; Song, E.H.; Nakatani-Webster, E.; Monkkonen, L.; Ratner, D.M.; Catalano, C.E. Phage lambda capsids as tunable display nanoparticles. Biomacromolecules 2014, 15, 4410–4419. [Google Scholar] [CrossRef]
- Catala, A.; Dzieciatkowska, M.; Wang, G.; Gutierrez-Hartmann, A.; Simberg, D.; Hansen, K.C.; D’Alessandro, A.; Catalano, C.E. Targeted Intracellular Delivery of Trastuzumab Using Designer Phage Lambda Nanoparticles Alters Cellular Programs in Human Breast Cancer Cells. ACS Nano 2021, 15, 11789–11805. [Google Scholar] [CrossRef] [PubMed]
- McClary, W.D.; Catala, A.; Zhang, W.; Gamboni, F.; Dzieciatkowska, M.; Sidhu, S.S.; D’Alessandro, A.; Catalano, C.E. A Designer Nanoparticle Platform for Controlled Intracellular Delivery of Bioactive Macromolecules: Inhibition of Ubiquitin-Specific Protease 7 in Breast Cancer Cells. ACS Chem. Biol. 2022, 17, 1853–1865. [Google Scholar] [CrossRef] [PubMed]
- Davenport, B.J.; Catala, A.; Weston, S.M.; Johnson, R.M.; Ardanuy, J.; Hammond, H.L.; Dillen, C.; Frieman, M.B.; Catalano, C.E.; Morrison, T.E. Phage-like particle vaccines are highly immunogenic and protect against pathogenic coronavirus infection and disease. NPJ Vaccines 2022, 7, 57. [Google Scholar] [CrossRef]
- Upadhyay, V.; Lucas, A.; Panja, S.; Miyauchi, R.; Mallela, K.M.G. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021, 297, 101208. [Google Scholar] [CrossRef]
- Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 1967, 6, 1948–1954. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Schuck, P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003, 320, 104–124. [Google Scholar] [CrossRef]
- Micsonai, A.; Bulyaki, E.; Kardos, J. BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. Methods Mol. Biol. 2021, 2199, 175–189. [Google Scholar] [CrossRef]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Leist, S.R.; Dinnon, K.H., 3rd; Schafer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e1012. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Evilevitch, A.; Jeembaeva, M.; Potter, C.S.; Carragher, B.; Johnson, J.E. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure 2008, 16, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Hardenbrook, N.J.; Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 2022, 52, 123–134. [Google Scholar] [CrossRef]
- Turonova, B.; Sikora, M.; Schurmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; von Bulow, S.; Gecht, M.; Bagola, K.; Horner, C.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Hoess, R.H. Bacteriophage lambda as a vehicle for peptide and protein display. Curr. Pharm. Biotechnol. 2002, 3, 23–28. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, R.; Porollo, A.; Meller, J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins 2004, 56, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Zhang, K.; Sanyal, M.; Tang, S.; Weidenbacher, P.A.; Li, S.; Pham, T.D.; Pak, J.E.; Chiu, W.; Kim, P.S. A Single Immunization with Spike-Functionalized Ferritin Vaccines Elicits Neutralizing Antibody Responses against SARS-CoV-2 in Mice. ACS Cent. Sci. 2021, 7, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Keeffe, J.R.; Schiepers, A.; Dross, S.E.; Greaney, A.J.; Rorick, A.V.; Gao, H.; Gnanapragasam, P.N.P.; Fan, C.; West, A.P., Jr.; et al. Mosaic sarbecovirus nanoparticles elicit cross-reactive responses in pre-vaccinated animals. bioRxiv 2024. [Google Scholar] [CrossRef]
- Zhu, J.; Ananthaswamy, N.; Jain, S.; Batra, H.; Tang, W.C.; Lewry, D.A.; Richards, M.L.; David, S.A.; Kilgore, P.B.; Sha, J.; et al. A universal bacteriophage T4 nanoparticle platform to design multiplex SARS-CoV-2 vaccine candidates by CRISPR engineering. Sci. Adv. 2021, 7, eabh1547. [Google Scholar] [CrossRef]
- Li, M.; Guo, P.; Chen, C.; Feng, H.; Zhang, W.; Gu, C.; Wen, G.; Rao, V.B.; Tao, P. Bacteriophage T4 Vaccine Platform for Next-Generation Influenza Vaccine Development. Front. Immunol. 2021, 12, 745625. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Zhang, J.; Xia, N.; Zhao, Q. Escherichia coli-derived virus-like particles in vaccine development. NPJ Vaccines 2017, 2, 3. [Google Scholar] [CrossRef]
- Sharma, J.; Uchida, M.; Miettinen, H.M.; Douglas, T. Modular interior loading and exterior decoration of a virus-like particle. Nanoscale 2017, 9, 10420–10430. [Google Scholar] [CrossRef]
- Hayes, S.; Gamage, L.N.; Hayes, C. Dual expression system for assembling phage lambda display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 2010, 28, 6789–6799. [Google Scholar] [CrossRef] [PubMed]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catala, A.; Davenport, B.J.; Morrison, T.E.; Catalano, C.E. Second-Generation Phage Lambda Platform Employing SARS-CoV-2 Fusion Proteins as a Vaccine Candidate. Vaccines 2024, 12, 1201. https://doi.org/10.3390/vaccines12111201
Catala A, Davenport BJ, Morrison TE, Catalano CE. Second-Generation Phage Lambda Platform Employing SARS-CoV-2 Fusion Proteins as a Vaccine Candidate. Vaccines. 2024; 12(11):1201. https://doi.org/10.3390/vaccines12111201
Chicago/Turabian StyleCatala, Alexis, Bennett J. Davenport, Thomas E. Morrison, and Carlos E. Catalano. 2024. "Second-Generation Phage Lambda Platform Employing SARS-CoV-2 Fusion Proteins as a Vaccine Candidate" Vaccines 12, no. 11: 1201. https://doi.org/10.3390/vaccines12111201
APA StyleCatala, A., Davenport, B. J., Morrison, T. E., & Catalano, C. E. (2024). Second-Generation Phage Lambda Platform Employing SARS-CoV-2 Fusion Proteins as a Vaccine Candidate. Vaccines, 12(11), 1201. https://doi.org/10.3390/vaccines12111201





