Immunoinformatics Design and In Vivo Immunogenicity Evaluation of a Conserved CTL Multi-Epitope Vaccine Targeting HPV16 E5, E6, and E7 Proteins
Abstract
:1. Introduction
2. Materials and Methods
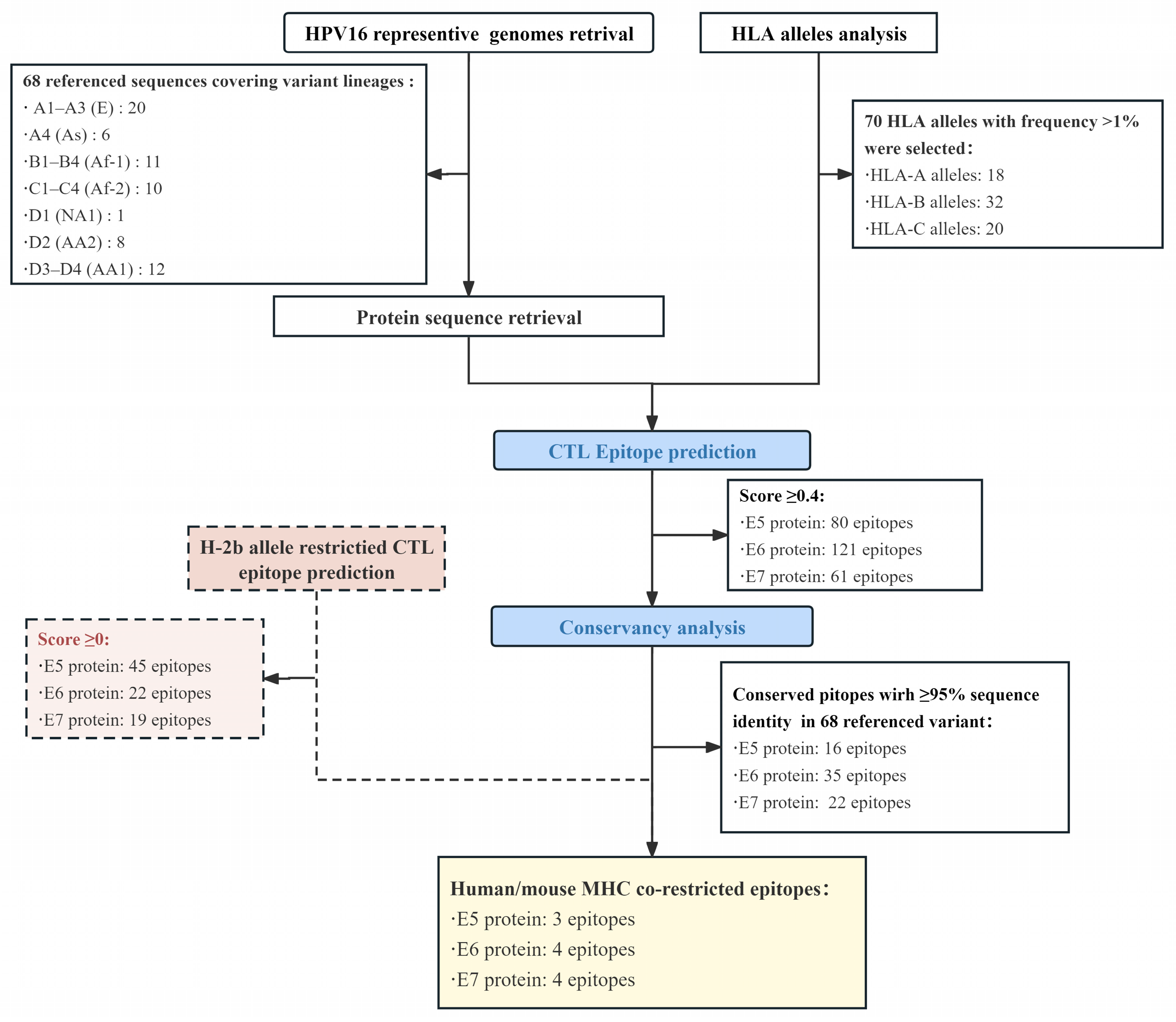
2.1. Sequence Retrieval of the HPV16 Genome and E5, E6, and E7 Proteins
2.2. Immunoinformatics Prediction
2.3. Conserved CTL Epitope Identification and Population Coverage Analysis
2.4. Immunogenicity, Antigenicity and Toxicity Evaluation of Candidate Epitopes
2.5. Preparation of the Mixed-Peptide Vaccine
2.6. Construction of Recombinant Protein Vaccines
2.7. Physiochemical Property Evaluation
2.8. Cell Line and Mice
2.9. IFN-γ ELISpot Assay
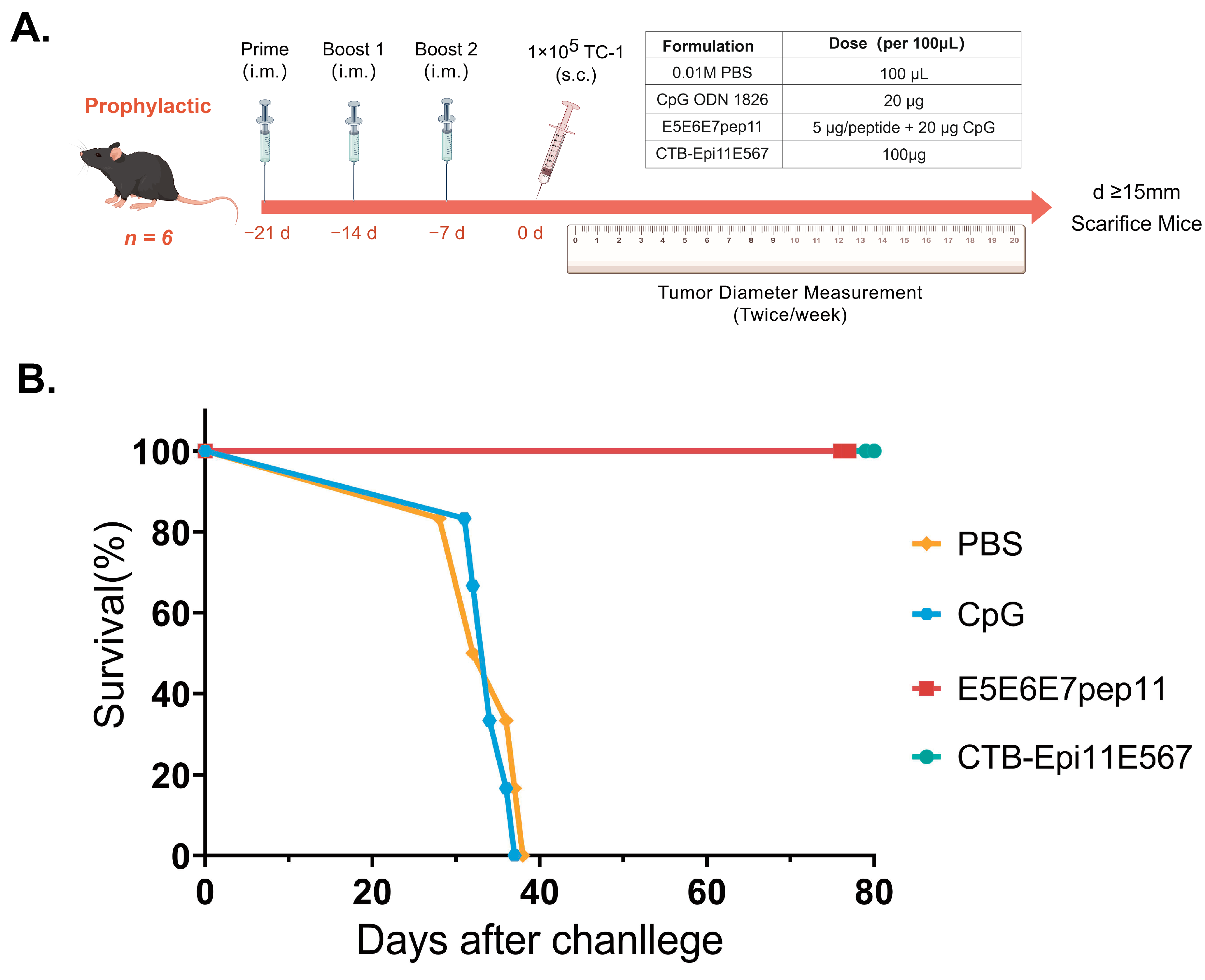
2.10. In Vivo Tumor Prevention Studies
2.11. In Vivo Tumor Treatment Study
2.12. Statistical Analysis
3. Results
3.1. Epitope Prediction and Selection
3.2. Characterization of CTL Epitopes for Vaccine Development
3.3. Evaluation of Antigen-Specific IFN-γ Response
3.4. Construction and Expression of Recombinant Multi-Epitope Protein
3.5. Prophylactic Efficacy of the Multi-Epitope Vaccine
3.6. Therapeutic Efficacy of the Multi-Epitope Vaccine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deshmukh, A.A.; Damgacioglu, H.; Georges, D.; Sonawane, K.; Ferlay, J.; Bray, F.; Clifford, G.M. Global burden of HPV-attributable squamous cell carcinoma of the anus in 2020, according to sex and HIV status: A worldwide analysis. Int. J. Cancer 2022, 152, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- de Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Fan, T.; Zhang, M.; Yang, J.; Zhu, Z.; Cao, W.; Dong, C. Therapeutic cancer vaccines: Advancements, challenges, and prospects. Signal Transduct. Target Ther. 2023, 8, 450. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Ma, J.; Zhang, H.; Shen, J.; Chen, J.; Hong, J.; Xu, Y.; Qian, C. Prophylactic and Therapeutic HPV Vaccines: Current Scenario and Perspectives. Front. Cell Infect. Microbiol. 2022, 12, 909223. [Google Scholar] [CrossRef] [PubMed]
- Coleman, N.; Birley, H.D.; Renton, A.M.; Hanna, N.F.; Ryait, B.K.; Byrne, M.; Taylor-Robinson, D.; Stanley, M.A. Immunological events in regressing genital warts. Am. J. Clin. Pathol. 1994, 102, 768–774. [Google Scholar] [CrossRef]
- Stanley, M. Immune responses to human papillomavirus. Vaccine 2006, 24 (Suppl. 1), S16–S22. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Maraj, B.; Tran, N.P.; Knoff, J.; Chen, A.; Alvarez, R.D.; Hung, C.F.; Wu, T.C. Emerging human papillomavirus vaccines. Expert. Opin. Emerg. Drugs 2012, 17, 469–492. [Google Scholar] [CrossRef]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2019, 10, 3116. [Google Scholar] [CrossRef]
- Gutierrez-Xicotencatl, L.; Pedroza-Saavedra, A.; Chihu-Amparan, L.; Salazar-Piña, A.; Maldonado-Gama, M.; Esquivel-Guadarrama, F. Cellular Functions of HPV16 E5 Oncoprotein during Oncogenic Transformation. Mol. Cancer Res. 2021, 19, 167–179. [Google Scholar] [CrossRef]
- Kim, M.K.; Kim, H.S.; Kim, S.H.; Oh, J.M.; Han, J.Y.; Lim, J.M.; Juhnn, Y.S.; Song, Y.S. Human papillomavirus type 16 E5 oncoprotein as a new target for cervical cancer treatment. Biochem. Pharmacol. 2010, 80, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Ilahi, N.E.; Bhatti, A. Impact of HPV E5 on viral life cycle via EGFR signaling. Microb. Pathog. 2020, 139, 103923. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Cullen, M.; Boland, J.F.; Chen, Z.; Wentzensen, N.; Zhang, X.; Yu, K.; Yang, Q.; Mitchell, J.; et al. HPV16 Sublineage Associations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016, 108, djw100. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Tenet, V.; Georges, D.; Alemany, L.; Pavón, M.A.; Chen, Z.; Yeager, M.; Cullen, M.; Boland, J.F.; Bass, S.; et al. Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res. 2019, 7, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Londesborough, P.; Ho, L.; Terry, G.; Cuzick, J.; Wheeler, C.; Singer, A. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int. J. Cancer 1996, 69, 364–368. [Google Scholar] [CrossRef]
- Dai, S.; Li, C.; Yan, Z.; Zhou, Z.; Wang, X.; Wang, J.; Sun, L.; Shi, L.; Yao, Y. Association of Human Papillomavirus Type 16 Long Control Region Variations with Cervical Cancer in a Han Chinese Population. Int. J. Med. Sci. 2020, 17, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yan, Z.; Dai, S.; Li, C.; Yang, L.; Liu, S.; Zhang, X.; Shi, L.; Yao, Y. Human Papillomavirus Type 16 E1 Mutations Associated with Cervical Cancer in a Han Chinese Population. Int. J. Med. Sci. 2019, 16, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yang, H.; Yang, L.; Yao, Y.; Dai, S.; Shi, L.; Li, C.; Yang, L.; Yan, Z.; Yao, Y. Human papillomavirus type 16 E6 and E7 gene variations associated with cervical cancer in a Han Chinese population. Infect. Genet. Evol. 2019, 73, 13–20. [Google Scholar] [CrossRef]
- De Groot, A.S.; Moise, L.; Terry, F.; Gutierrez, A.H.; Hindocha, P.; Richard, G.; Hoft, D.F.; Ross, T.M.; Noe, A.R.; Takahashi, Y.; et al. Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools. Front. Immunol. 2020, 11, 442. [Google Scholar] [CrossRef]
- Smith, B.; Chen, Z.; Reimers, L.; van Doorslaer, K.; Schiffman, M.; Desalle, R.; Herrero, R.; Yu, K.; Wacholder, S.; Wang, T.; et al. Sequence imputation of HPV16 genomes for genetic association studies. PLoS ONE 2011, 6, e21375. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharmacol. 2016, 120, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Soria-Guerra, R.E.; Nieto-Gomez, R.; Govea-Alonso, D.O.; Rosales-Mendoza, S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J. Biomed. Inform. 2015, 53, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Li, F.; Leier, A.; Marquez-Lago, T.T.; Giam, K.; Croft, N.P.; Akutsu, T.; Smith, A.I.; Li, J.; Rossjohn, J.; et al. A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction. Brief. Bioinform. 2020, 21, 1119–1135. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, P.I. A brief history of HLA. Immunol. Res. 2007, 38, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Rothbard, J.B. Antigen presentation. One size fits all. Curr. Biol. 1994, 4, 653–655. [Google Scholar] [CrossRef] [PubMed]
- Stern, L.J.; Wiley, D.C. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure 1994, 2, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Yadav, I.S.; Hussain, S.; Das, B.C.; Bharadwaj, M. Identification of immunotherapeutic epitope of E5 protein of human papillomavirus-16: An in silico approach. Biologicals 2015, 43, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Lin, C.W.; Tsao, Y.P.; Chen, S.L. Cytotoxic-T-lymphocyte human papillomavirus type 16 E5 peptide with CpG-oligodeoxynucleotide can eliminate tumor growth in C57BL/6 mice. J. Virol. 2004, 78, 1333–1343. [Google Scholar] [CrossRef]
- Liu, D.W.; Tsao, Y.P.; Hsieh, C.H.; Hsieh, J.T.; Kung, J.T.; Chiang, C.L.; Huang, S.J.; Chen, S.L. Induction of CD8 T cells by vaccination with recombinant adenovirus expressing human papillomavirus type 16 E5 gene reduces tumor growth. J. Virol. 2000, 74, 9083–9089. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, D.; Petti, L.M. The E5 proteins. Virology 2013, 445, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Suprynowicz, F.A.; Disbrow, G.L.; Krawczyk, E.; Simic, V.; Lantzky, K.; Schlegel, R. HPV-16 E5 oncoprotein upregulates lipid raft components caveolin-1 and ganglioside GM1 at the plasma membrane of cervical cells. Oncogene 2008, 27, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Maufort, J.P.; Shai, A.; Pitot, H.C.; Lambert, P.F. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res. 2010, 70, 2924–2931. [Google Scholar] [CrossRef]
- Campo, M.S.; Graham, S.V.; Cortese, M.S.; Ashrafi, G.H.; Araibi, E.H.; Dornan, E.S.; Miners, K.; Nunes, C.; Man, S. HPV-16 E5 down-regulates expression of surface HLA class I and reduces recognition by CD8 T cells. Virology 2010, 407, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.L.; Tsao, Y.P.; Liu, D.W.; Huang, S.J.; Lee, W.H.; Chen, S.L. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 2001, 8, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhang, W.; Hu, X.; Wang, W.; Deng, D.; Wang, H.; Wang, C.; Zhou, J.; Wang, S.; Zhang, H.; et al. A novel “priming-boosting” strategy for immune interventions in cervical cancer. Mol. Immunol. 2015, 64, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Blatnik, R.; Mohan, N.; Bonsack, M.; Falkenby, L.G.; Hoppe, S.; Josef, K.; Steinbach, A.; Becker, S.; Nadler, W.M.; Rucevic, M.; et al. A Targeted LC-MS Strategy for Low-Abundant HLA Class-I-Presented Peptide Detection Identifies Novel Human Papillomavirus T-Cell Epitopes. Proteomics 2018, 18, e1700390. [Google Scholar] [CrossRef] [PubMed]
- Riemer, A.B.; Keskin, D.B.; Zhang, G.; Handley, M.; Anderson, K.S.; Brusic, V.; Reinhold, B.; Reinherz, E.L. A conserved E7-derived cytotoxic T lymphocyte epitope expressed on human papillomavirus 16-transformed HLA-A2+ epithelial cancers. J. Biol. Chem. 2010, 285, 29608–29622. [Google Scholar] [CrossRef]
- Jemon, K.; Young, V.; Wilson, M.; McKee, S.; Ward, V.; Baird, M.; Young, S.; Hibma, M. An enhanced heterologous virus-like particle for human papillomavirus type 16 tumour immunotherapy. PLoS ONE 2013, 8, e66866. [Google Scholar] [CrossRef]
- He, X.; Zhou, S.; Quinn, B.; Jahagirdar, D.; Ortega, J.; Abrams, S.I.; Lovell, J.F. HPV-Associated Tumor Eradication by Vaccination with Synthetic Short Peptides and Particle-Forming Liposomes. Small 2021, 17, e2007165. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Zwaveling, S.; Ferreira Mota, S.C.; Nouta, J.; Johnson, M.; Lipford, G.B.; Offringa, R.; van der Burg, S.H.; Melief, C.J. Established human papillomavirus type 16-expressing tumors are effectively eradicated following vaccination with long peptides. J. Immunol. 2002, 169, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Rosalia, R.A.; Quakkelaar, E.D.; Redeker, A.; Khan, S.; Camps, M.; Drijfhout, J.W.; Silva, A.L.; Jiskoot, W.; van Hall, T.; van Veelen, P.A.; et al. Dendritic cells process synthetic long peptides better than whole protein, improving antigen presentation and T-cell activation. Eur. J. Immunol. 2013, 43, 2554–2565. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Monie, A.; Pang, X.; Hung, C.F.; Wu, T.C. Improving therapeutic HPV peptide-based vaccine potency by enhancing CD4+ T help and dendritic cell activation. J. Biomed. Sci. 2010, 17, 88. [Google Scholar] [CrossRef] [PubMed]
- Zom, G.G.; Willems, M.; Khan, S.; van der Sluis, T.C.; Kleinovink, J.W.; Camps, M.G.M.; van der Marel, G.A.; Filippov, D.V.; Melief, C.J.M.; Ossendorp, F. Novel TLR2-binding adjuvant induces enhanced T cell responses and tumor eradication. J. Immunother. Cancer 2018, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Sabbatini, P.; Tsuji, T.; Ferran, L.; Ritter, E.; Sedrak, C.; Tuballes, K.; Jungbluth, A.A.; Ritter, G.; Aghajanian, C.; Bell-McGuinn, K.; et al. Phase I trial of overlapping long peptides from a tumor self-antigen and poly-ICLC shows rapid induction of integrated immune response in ovarian cancer patients. Clin. Cancer Res. 2012, 18, 6497–6508. [Google Scholar] [CrossRef]
- Maynard, S.K.; Marshall, J.D.; MacGill, R.S.; Yu, L.; Cann, J.A.; Cheng, L.I.; McCarthy, M.P.; Cayatte, C.; Robbins, S.H. Vaccination with synthetic long peptide formulated with CpG in an oil-in-water emulsion induces robust E7-specific CD8 T cell responses and TC-1 tumor eradication. BMC Cancer 2019, 19, 540. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, Y.; Hsi, J.; Wang, H.; Tao, R.; Shao, Y. Cholera toxin B subunit acts as a potent systemic adjuvant for HIV-1 DNA vaccination intramuscularly in mice. Hum. Vaccin. Immunother. 2014, 10, 1274–1283. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.M.; Kastenmüller, W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Sanami, S.; Rafieian-Kopaei, M.; Dehkordi, K.A.; Pazoki-Toroudi, H.; Azadegan-Dehkordi, F.; Mobini, G.R.; Alizadeh, M.; Nezhad, M.S.; Ghasemi-Dehnoo, M.; Bagheri, N. In silico design of a multi-epitope vaccine against HPV16/18. BMC Bioinform. 2022, 23, 311. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, A.K.; Kaufmann, A.M.; Riemer, A.B. Identification of promiscuous HPV16-derived T helper cell epitopes for therapeutic HPV vaccine design. Int. J. Cancer 2015, 136, 212–224. [Google Scholar] [CrossRef] [PubMed]



| Protein | Epitope | Sequence | Position | Length | Coverage | Immunogenicity | Antigency | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|---|---|
| E5 | E5p1 | LIRPLLLSV | 28–36 | 9 | 1.95% | −0.12 | 0.33 | Non-Allergen | Non-Toxin |
| E5p2 | TAASAFRCF | 52–60 | 9 | 30.81% | 0.01 | 0.26 | Non-Allergen | Non-Toxin | |
| E5p3 | FVYIPLFLI | 66–73 | 9 | 28.62% | 0.2 | 0.39 | Non-Allergen | Non-Toxin | |
| E6 | E6p4 | EVYDFAFRDL | 41–50 | 10 | 32.66% | 0.34 | 1.46 | Non-Allergen | Non-Toxin |
| E6p5 | YRDGNPYAV | 54–62 | 9 | 44.37% | 0.04 | 0.39 | Allergen | Non-Toxin | |
| E6p6 | KFYSKISEY | 68–76 | 9 | 37.65% | −0.33 | 0.23 | Non-Allergen | Non-Toxin | |
| E6p7 | RHLDKKQRF | 117–125 | 9 | 3.23% | −0.46 | 0.35 | Non-Allergen | Non-Toxin | |
| E7 | E7p8 | TLHEYMLDL | 7–15 | 9 | 39.08% | −0.05 | 0.33 | Non-Allergen | Non-Toxin |
| E7p9 | RAHYNIVTF | 49–57 | 9 | 70.50% | 0.18 | 0.59 | Non-Allergen | Non-Toxin | |
| E7p10 | STHVDIRTL | 71–79 | 9 | 16.98% | 0.27 | 0.58 | Allergen | Non-Toxin | |
| E7p11 | RTLEDLLMGTL | 77–87 | 11 | 17.96% | −0.02 | 0.64 | Allergen | Non-Toxin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, N.; Niu, Z.; Yan, Z.; Liu, W.; Shi, L.; Li, C.; Yao, Y.; Shi, L. Immunoinformatics Design and In Vivo Immunogenicity Evaluation of a Conserved CTL Multi-Epitope Vaccine Targeting HPV16 E5, E6, and E7 Proteins. Vaccines 2024, 12, 392. https://doi.org/10.3390/vaccines12040392
Guo N, Niu Z, Yan Z, Liu W, Shi L, Li C, Yao Y, Shi L. Immunoinformatics Design and In Vivo Immunogenicity Evaluation of a Conserved CTL Multi-Epitope Vaccine Targeting HPV16 E5, E6, and E7 Proteins. Vaccines. 2024; 12(4):392. https://doi.org/10.3390/vaccines12040392
Chicago/Turabian StyleGuo, Ni, Zhixin Niu, Zhiling Yan, Weipeng Liu, Lei Shi, Chuanyin Li, Yufeng Yao, and Li Shi. 2024. "Immunoinformatics Design and In Vivo Immunogenicity Evaluation of a Conserved CTL Multi-Epitope Vaccine Targeting HPV16 E5, E6, and E7 Proteins" Vaccines 12, no. 4: 392. https://doi.org/10.3390/vaccines12040392
APA StyleGuo, N., Niu, Z., Yan, Z., Liu, W., Shi, L., Li, C., Yao, Y., & Shi, L. (2024). Immunoinformatics Design and In Vivo Immunogenicity Evaluation of a Conserved CTL Multi-Epitope Vaccine Targeting HPV16 E5, E6, and E7 Proteins. Vaccines, 12(4), 392. https://doi.org/10.3390/vaccines12040392






