Deletion of MGF505-2R Gene Activates the cGAS-STING Pathway Leading to Attenuation and Protection against Virulent African Swine Fever Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Antibodies for In Vitro Experiments
2.3. Vectors and Transfection
2.4. Luciferase Assay
2.5. Western Blot
2.6. Co-Immunoprecipitation Assay
2.7. Generation and Isolation of Arm/07-ΔMGF505-2R-GFP Virus by CRISPR/Cas9 Technology
2.8. Viral DNA Extraction for NGS Analysis
2.9. Illumina Sequencing and Data Analysis
2.10. Viral Growth Kinetics
2.11. RT-qPCR Assay
2.12. Experimental Conditions
2.13. Samples Collection and Assessment of Clinical Signs
2.14. ASFV Real-Time qPCR
2.15. ELISA Assay
2.16. Experimental Conditions
2.17. Samples Collection and Assessment of Clinical Signs
2.18. ASFV Real-Time qPCR
2.19. ELISA Assay
2.20. Statistical Analysis
3. Results
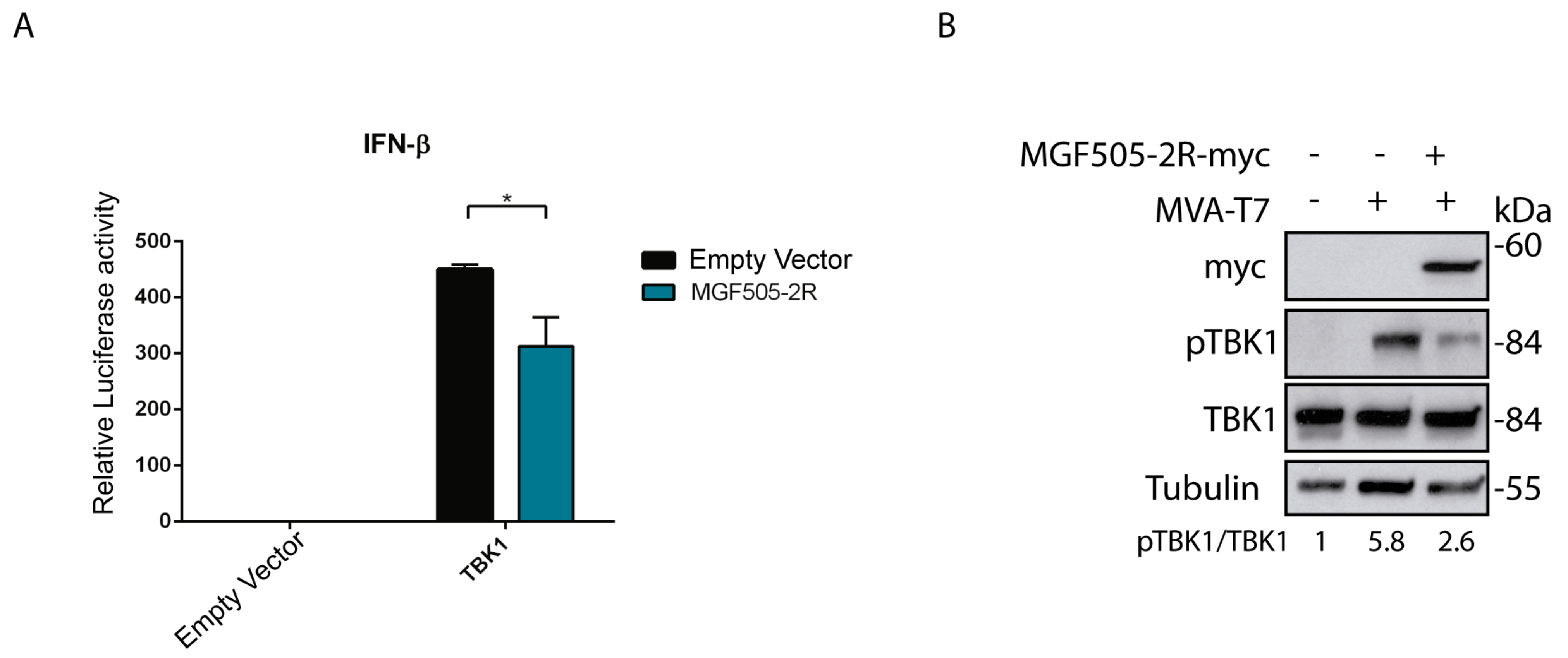
3.1. Ectopic Expression of the ASFV Protein pMGF505-2R Decreases IFN-β Production
3.2. pMGF505-2R Modulates TBK1 Phosphorylation
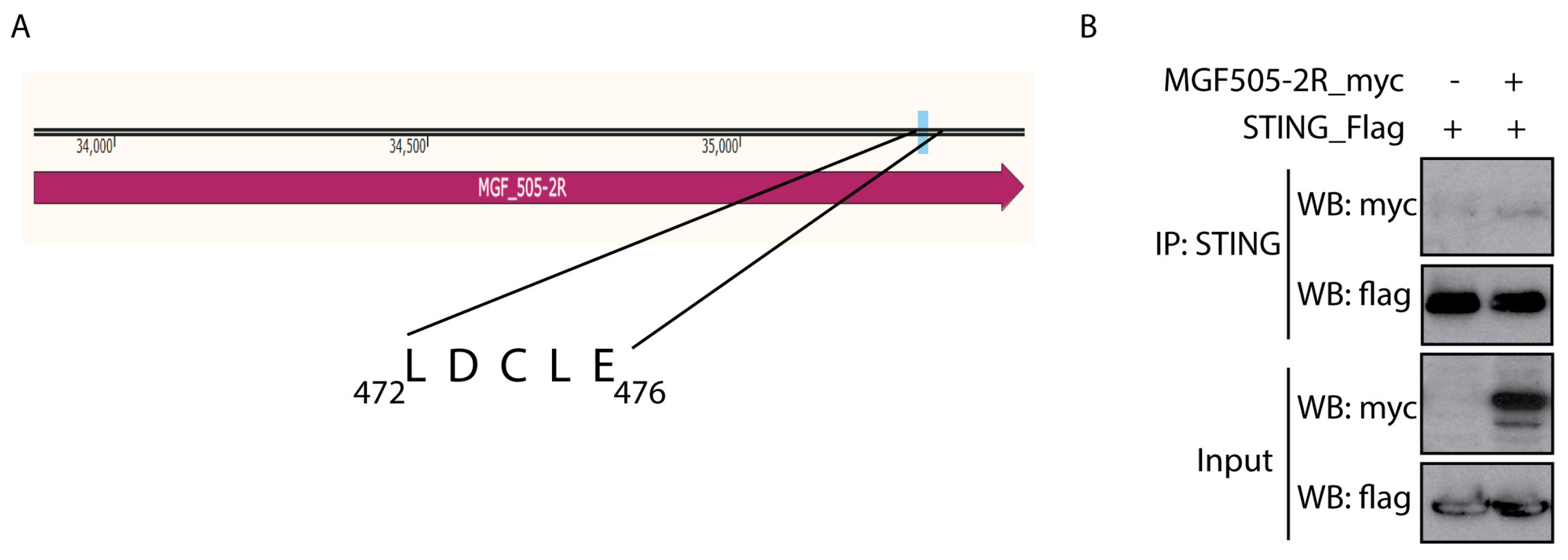
3.3. pMGF505-2R Interacts with STING
3.4. Generation of Recombinant Arm/07-ΔMGF505-2R-GFP Virus
3.5. Growth Kinetic of Recombinant Arm/07-ΔMGF505-2R-GFP Virus
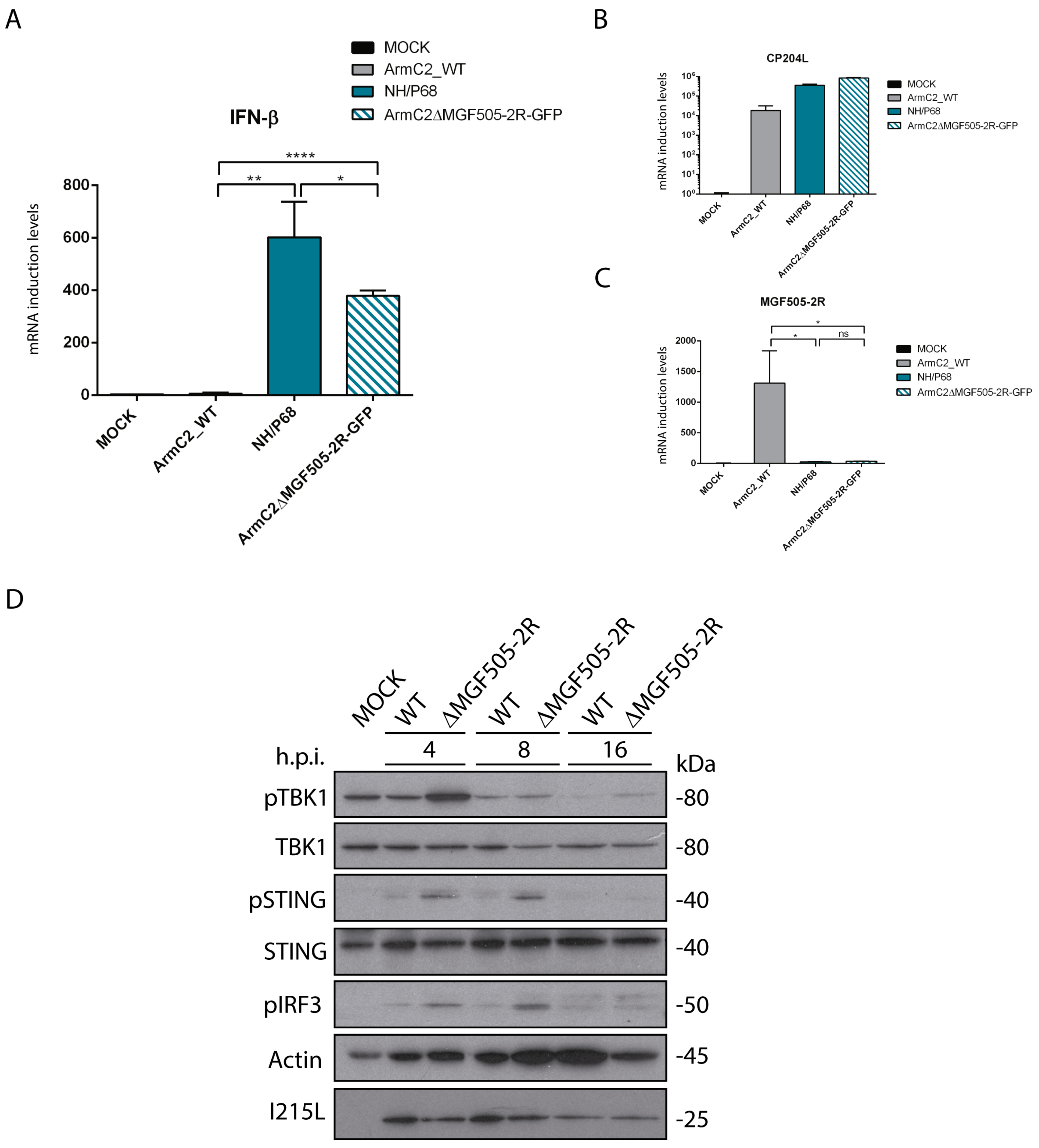
3.6. Arm/07-ΔMGF505-2R-GFP Induces IFN-β Production during Infection
3.7. Infection with Arm/07-ΔMGF505-2R-GFP Does Not Inhibit Phosphorylation of cGAS-STING Pathway Components
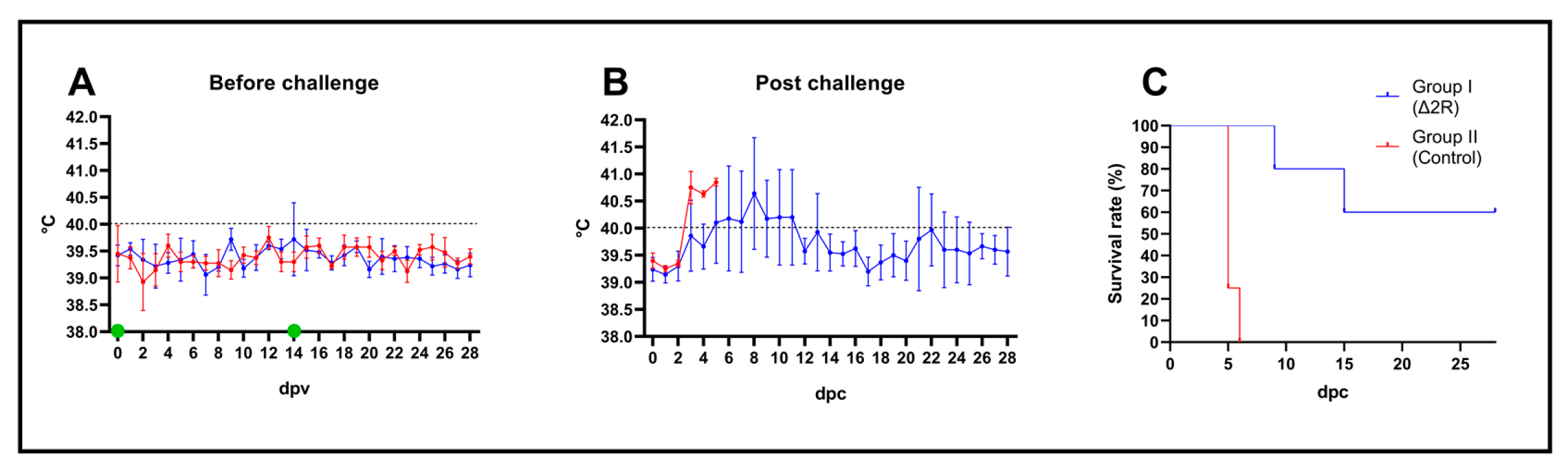
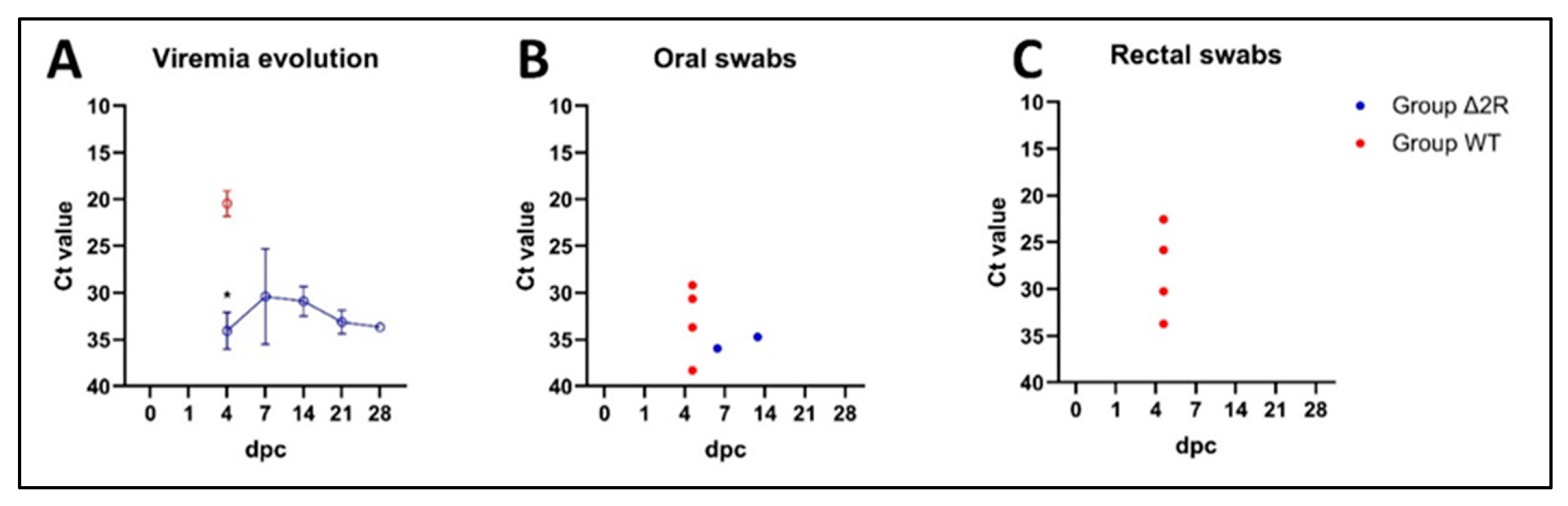
3.8. Arm/07-ΔMGF505-2R-GFP Is Attenuated In Vivo to a Dose of 102 TCID50
3.9. Arm/07-ΔMGF505-2R-GFP Partially Protects against the Circulating Virulent Strain ASFV/Korea/pig/PaJu1/2019
3.10. Activation of Humoral Response after Vaccination with Arm/07-ΔMGF505-2R-GFP
3.11. Arm/07-ΔMGF505-2R-GFP Is Attenuated In Vivo Using a Prime-Boost Protocol at a Dose of 103 TCID50
3.12. Arm/07-ΔMGF505-2R-GFP Partially Protects against the Virulent Strain Arm/07/CBM/c2
3.13. Humoral Response Is Activated after Vaccination with Arm/07-ΔMGF505-2R-GFP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Sauter-Louis, C.; Forth, J.H.; Probst, C.; Staubach, C.; Hlinak, A.; Rudovsky, A.; Holland, D.; Schlieben, P.; Goldner, M.; Schatz, J.; et al. Joining the club: First detection of African swine fever in wild boar in Germany. Transbound. Emerg. Dis. 2021, 68, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Dharmayanti, N.I.; Sendow, I.; Ratnawati, A.; Settypalli, T.B.K.; Saepulloh, M.; Dundon, W.G.; Nuradji, H.; Naletoski, I.; Cattoli, G.; Lamien, C.E. African swine fever in North Sumatra and West Java provinces in 2019 and 2020, Indonesia. Transbound. Emerg. Dis. 2021, 68, 2890–2896. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Park, J.E.; Kim, S.J.; Kim, Y.; Kim, W.; Kim, Y.K.; Jheong, W. Complete genome analysis of the African swine fever virus isolated from a wild boar responsible for the first viral outbreak in Korea, 2019. Front. Vet. Sci. 2022, 9, 1080397. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, D.; Rajukumar, K.; Venkatesh, G.; Singh, F.; Tosh, C.; Kombiah, S.; Dubey, C.K.; Chakravarty, A.; Barman, N.N.; Singh, V.P. Complete genome analysis of African swine fever virus isolated from domestic pigs during the first ASF outbreaks in India. Transbound. Emerg. Dis. 2022, 69, e2020–e2027. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, W.; Moreno, C.; Duran, U.; Henao, N.; Bencosme, M.; Lora, P.; Reyes, R.; Nunez, R.; De Gracia, A.; Perez, A.M. African swine fever in the Dominican Republic. Transbound. Emerg. Dis. 2021, 68, 3018–3019. [Google Scholar] [CrossRef] [PubMed]
- WOAH. African Swine Fever (ASF)—Situation Report 27. Available online: https://www.woah.org/en/document/african-swine-fever-asf-situation-report-27/ (accessed on 4 March 2024).
- Gallardo, C.; Soler, A.; Rodze, I.; Nieto, R.; Cano-Gomez, C.; Fernandez-Pinero, J.; Arias, M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound. Emerg. Dis. 2019, 66, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Leitao, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigario, J.D.; Martins, C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef]
- Garcia-Belmonte, R.; Perez-Nunez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef]
- Dodantenna, N.; Ranathunga, L.; Chathuranga, W.A.G.; Weerawardhana, A.; Cha, J.W.; Subasinghe, A.; Gamage, N.; Haluwana, D.K.; Kim, Y.; Jheong, W.; et al. African Swine Fever Virus EP364R and C129R Target Cyclic GMP-AMP to Inhibit the cGAS-STING Signaling Pathway. J. Virol. 2022, 96, e0102222. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Zheng, X.; Zhu, Y.; Yao, Y.; Li, S.; Xu, Y.; Feng, W.H. African swine fever virus QP383R dampens type I interferon production by promoting cGAS palmitoylation. Front. Immunol. 2023, 14, 1186916. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, W.; Liu, H.; Xue, M.; Dong, S.; Liu, X.; Feng, C.; Cao, S.; Ye, G.; Zhou, Q.; et al. African Swine Fever Virus HLJ/18 CD2v Suppresses Type I IFN Production and IFN-Stimulated Genes Expression through Negatively Regulating cGMP-AMP Synthase-STING and IFN Signaling Pathways. J. Immunol. 2023, 210, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, W.; Li, L.; Li, P.; Ma, Z.; Zhang, J.; Qi, X.; Ren, J.; Ru, Y.; Niu, Q.; et al. African Swine Fever Virus MGF-505-7R Negatively Regulates cGAS-STING-Mediated Signaling Pathway. J. Immunol. 2021, 206, 1844–1857. [Google Scholar] [CrossRef] [PubMed]
- Ranathunga, L.; Dodantenna, N.; Cha, J.W.; Chathuranga, K.; Chathuranga, W.A.G.; Weerawardhana, A.; Subasinghe, A.; Haluwana, D.K.; Gamage, N.; Lee, J.S. African swine fever virus B175L inhibits the type I interferon pathway by targeting STING and 2′3′-cGAMP. J. Virol. 2023, 97, e0079523. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, D.; Zhu, G.; Yang, W.; Ru, Y.; Feng, T.; Qin, X.; Hao, R.; Duan, X.; Liu, X.; et al. Deletion of MGF-110-9L gene from African swine fever virus weakens autophagic degradation of TBK1 as a mechanism for enhancing type I interferon production. FASEB J. 2023, 37, e22934. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yu, S.; Ge, H.; Wang, T.; Li, Y.; Zhou, P.; Pan, L.; Han, Y.; Yang, Y.; Sun, Y.; et al. The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 2022, 96, e0195721. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Xia, N.; Zhang, J.; Cao, Q.; Jiang, S.; Luo, J.; Wang, H.; Chen, N.; Zhang, Q.; Meurens, F.; et al. African Swine Fever Virus Structural Protein p17 Inhibits cGAS-STING Signaling Pathway Through Interacting With STING. Front. Immunol. 2022, 13, 941579. [Google Scholar] [CrossRef] [PubMed]
- Riera, E.; Perez-Nunez, D.; Garcia-Belmonte, R.; Miorin, L.; Garcia-Sastre, A.; Revilla, Y. African Swine Fever Virus Induces STAT1 and STAT2 Degradation to Counteract IFN-I Signaling. Front. Microbiol. 2021, 12, 722952. [Google Scholar] [CrossRef]
- Chen, Q.; Li, L.; Liu, L.; Liu, Z.; Guo, S.; Tan, C.; Chen, H.; Wang, X. African Swine Fever Virus pF778R Attenuates Type I Interferon Response by Impeding STAT1 Nuclear Translocation. Virus Res. 2023, 335, 199190. [Google Scholar] [CrossRef]
- Huang, Z.; Cao, H.; Zeng, F.; Lin, S.; Chen, J.; Luo, Y.; You, J.; Kong, C.; Mai, Z.; Deng, J.; et al. African Swine Fever Virus MGF505-7R Interacts with Interferon Regulatory Factor 9 to Evade the Type I Interferon Signaling Pathway and Promote Viral Replication. J. Virol. 2023, 97, e0197722. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Peng, J.L.; Xu, Z.S.; Xiong, M.G.; Wu, H.N.; Wang, S.Y.; Li, D.; Zhu, G.Q.; Ran, Y.; Wang, Y.Y. African Swine Fever Virus Cysteine Protease pS273R Inhibits Type I Interferon Signaling by Mediating STAT2 Degradation. J. Virol. 2023, 97, e0194222. [Google Scholar] [CrossRef] [PubMed]
- Riera, E.; Garcia-Belmonte, R.; Madrid, R.; Perez-Nunez, D.; Revilla, Y. African swine fever virus ubiquitin-conjugating enzyme pI215L inhibits IFN-I signaling pathway through STAT2 degradation. Front. Microbiol. 2022, 13, 1081035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yang, B.; Shen, C.; Zhang, T.; Hao, Y.; Zhang, D.; Liu, H.; Shi, X.; Li, G.; Yang, J.; et al. MGF360-9L Is a Major Virulence Factor Associated with the African Swine Fever Virus by Antagonizing the JAK/STAT Signaling Pathway. mBio 2022, 13, e0233021. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Sanchez, E.G.; Perez-Nunez, D.; Nogal, M.; de Leon, P.; Carrascosa, A.L.; Nieto, R.; Soler, A.; Arias, M.L.; Revilla, Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine 2018, 36, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Risatti, G.R.; Holinka, L.G.; Krug, P.W.; Carlson, J.; Velazquez-Salinas, L.; Azzinaro, P.A.; Gladue, D.P.; Borca, M.V. Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2017, 91, e01760-16. [Google Scholar] [CrossRef]
- Reis, A.L.; Abrams, C.C.; Goatley, L.C.; Netherton, C.; Chapman, D.G.; Sanchez-Cordon, P.; Dixon, L.K. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 2016, 34, 4698–4705. [Google Scholar] [CrossRef]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Sanchez-Cordon, P.J.; Netherton, C.L.; Chapman, D.A.G.; Dixon, L.K. Deletion of the African Swine Fever Virus Gene DP148R Does Not Reduce Virus Replication in Culture but Reduces Virus Virulence in Pigs and Induces High Levels of Protection against Challenge. J. Virol. 2017, 91, e01428-17. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020, 94, e02017-19. [Google Scholar] [CrossRef]
- Gladue, D.P.; Ramirez-Medina, E.; Vuono, E.; Silva, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Borca, M.V. Deletion of the A137R Gene from the Pandemic Strain of African Swine Fever Virus Attenuates the Strain and Offers Protection against the Virulent Pandemic Virus. J. Virol. 2021, 95, e0113921. [Google Scholar] [CrossRef] [PubMed]
- Perez-Nunez, D.; Sunwoo, S.Y.; Garcia-Belmonte, R.; Kim, C.; Vigara-Astillero, G.; Riera, E.; Kim, D.M.; Jeong, J.; Tark, D.; Ko, Y.S.; et al. Recombinant African Swine Fever Virus Arm/07/CBM/c2 Lacking CD2v and A238L Is Attenuated and Protects Pigs against Virulent Korean Paju Strain. Vaccines 2022, 10, 1992. [Google Scholar] [CrossRef] [PubMed]
- Tran, X.H.; Le, T.T.P.; Nguyen, Q.H.; Do, T.T.; Nguyen, V.D.; Gay, C.G.; Borca, M.V.; Gladue, D.P. African swine fever virus vaccine candidate ASFV-G-DeltaI177L efficiently protects European and native pig breeds against circulating Vietnamese field strain. Transbound. Emerg. Dis. 2022, 69, e497–e504. [Google Scholar] [CrossRef]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef]
- Perez-Nunez, D.; Castillo-Rosa, E.; Vigara-Astillero, G.; Garcia-Belmonte, R.; Gallardo, C.; Revilla, Y. Identification and Isolation of Two Different Subpopulations Within African Swine Fever Virus Arm/07 Stock. Vaccines 2020, 8, 625. [Google Scholar] [CrossRef]
- Monteagudo, P.L.; Lacasta, A.; Lopez, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71DeltaCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91, e01058-17. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Santaren, J.F.; Vinuela, E. Production and titration of African swine fever virus in porcine alveolar macrophages. J. Virol. Methods 1982, 3, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Cotter, C.A.; Earl, P.L.; Wyatt, L.S.; Moss, B. Preparation of Cell Cultures and Vaccinia Virus Stocks. Curr. Protoc. Protein Sci. 2017, 89, 11–18. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- King, K.; Chapman, D.; Argilaguet, J.M.; Fishbourne, E.; Hutet, E.; Cariolet, R.; Hutchings, G.; Oura, C.A.; Netherton, C.L.; Moffat, K.; et al. Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine 2011, 29, 4593–4600. [Google Scholar] [CrossRef]
- Walczak, M.; Zmudzki, J.; Mazur-Panasiuk, N.; Juszkiewicz, M.; Wozniakowski, G. Analysis of the Clinical Course of Experimental Infection with Highly Pathogenic African Swine Fever Strain, Isolated from an Outbreak in Poland. Aspects Related to the Disease Suspicion at the Farm Level. Pathogens 2020, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Afe, A.E.; Shen, Z.J.; Guo, X.; Zhou, R.; Li, K. African Swine Fever Virus Interaction with Host Innate Immune Factors. Viruses 2023, 15, 1220. [Google Scholar] [CrossRef]
- Yang, K.; Xue, Y.; Niu, H.; Shi, C.; Cheng, M.; Wang, J.; Zou, B.; Wang, J.; Niu, T.; Bao, M.; et al. African swine fever virus MGF360-11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production. Vet. Res. 2022, 53, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Xue, Y.; Niu, T.; Li, X.; Cheng, M.; Bao, M.; Zou, B.; Shi, C.; Wang, J.; Yang, W.; et al. African swine fever virus MGF505-7R protein interacted with IRF7and TBK1 to inhibit type I interferon production. Virus Res. 2022, 322, 198931. [Google Scholar] [CrossRef]
- Cui, S.; Wang, Y.; Gao, X.; Xin, T.; Wang, X.; Yu, H.; Chen, S.; Jiang, Y.; Chen, Q.; Jiang, F.; et al. African swine fever virus M1249L protein antagonizes type I interferon production via suppressing phosphorylation of TBK1 and degrading IRF3. Virus Res. 2022, 319, 198872. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, S.; Xin, T.; Wang, X.; Yu, H.; Chen, S.; Jiang, Y.; Gao, X.; Jiang, Y.; Guo, X.; et al. African Swine Fever Virus MGF360-14L Negatively Regulates Type I Interferon Signaling by Targeting IRF3. Front. Cell Infect. Microbiol. 2021, 11, 818969. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Feng, T.; Ma, Z.; Xue, Q.; Wu, P.; Li, P.; Li, S.; Yang, F.; Cao, W.; et al. African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2021, 95, e0082421. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Ye, G.; Xue, M.; Yu, H.; Feng, C.; Zhou, Q.; Liu, X.; Zhang, L.; Jiao, S.; et al. African swine fever virus pE301R negatively regulates cGAS-STING signaling pathway by inhibiting the nuclear translocation of IRF3. Vet. Microbiol. 2022, 274, 109556. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zheng, X.; Li, Y.; Zhu, Y.; Xu, Y.; Yu, Z.; Feng, W.H. African swine fever virus S273R protein antagonizes type I interferon production by interfering with TBK1 and IRF3 interaction. Virol. Sin. 2023, 38, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Kanyema, M.M.; Sun, Y.; Zhao, W.; Lu, Y.; Wang, J.; Li, X.; Shi, C.; Wang, J.; Wang, N.; et al. African Swine Fever Virus L83L Negatively Regulates the cGAS-STING-Mediated IFN-I Pathway by Recruiting Tollip to Promote STING Autophagic Degradation. J. Virol. 2023, 97, e0192322. [Google Scholar] [CrossRef]
- Yang, K.; Huang, Q.; Wang, R.; Zeng, Y.; Cheng, M.; Xue, Y.; Shi, C.; Ye, L.; Yang, W.; Jiang, Y.; et al. African swine fever virus MGF505-11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Vet. Microbiol. 2021, 263, 109265. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Dang, W.; Shi, Z.; Ding, M.; Xu, F.; Li, T.; Feng, T.; Zheng, H.; Xiao, S. Identification of African swine fever virus MGF505-2R as a potent inhibitor of innate immunity in vitro. Virol. Sin. 2023, 38, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, J.; Kang, L.; Huang, L.; Zhou, S.; Hu, L.; Zheng, J.; Li, C.; Zhang, X.; He, X.; et al. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PLoS Pathog. 2021, 17, e1009733. [Google Scholar] [CrossRef]
- Li, D.; Liu, Y.; Qi, X.; Wen, Y.; Li, P.; Ma, Z.; Liu, Y.; Zheng, H.; Liu, Z. African Swine Fever Virus MGF-110-9L-deficient Mutant Has Attenuated Virulence in Pigs. Virol. Sin. 2021, 36, 187–195. [Google Scholar] [CrossRef]

| Position | Type | Location | Mutation | Description |
|---|---|---|---|---|
| 14,005 | InDel | MGF110-11L MGF110-14L | A/AC | Loss of 6 aa N’Term Acquisition of 7 aa C’Term |
| 15,444 | InDel | Intergenic region | A/ACC | |
| 17,403 | InDel | Intergenic region | CG/C | |
| 41,689 | SNP | MGF505-7R | G/A | Gly63/Gln63 |
| 72,293 | SNP | EP424R | T/C | Tyr307/His307 |
| 126,682 | SNP | CP530R | C/T | Ser169/Leu169 |
| Animal | 0 dpv | 3 dpv | 5 dpv | 7 dpv | 10 dpv | 14 dpv | 21 dpv | 0 dpc | 3 dpc | 5 dpc | 7 dpc | 10 dpc | 14 dpc | 21 dpc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #61 | 38.2 | ND | 38.4 | 37.9 | 35.5 | 35.1 | 37.2 | ND | 30.7 | 36.9 | 37.1 | 37.9 | 30.9 | 34.1 |
| #62 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 37.1 | 38.1 | 38.4 | 37.2 | 39.3 |
| #63 | ND | ND | ND | ND | 39.0 | ND | ND | 35.0 | 38.3 | 30.4 | 31.3 | 33.6 | 33.7 | 37.3 |
| #64 | ND | ND | ND | ND | ND | ND | ND | ND | 36.9 | 23.6 | 20.6 | 19.4 | ||
| #65 | ND | ND | ND | 19.8 | 18.2 | |||||||||
| #66 | ND | ND | 20.8 | 17.4 | 17.7 | |||||||||
| #67 | ND | ND | 19.3 | 17.1 | ||||||||||
| #68 | 39.3 | 36.5 | 22.9 |
| Lesion | Arm/07-ΔMGF505-2R: n = 5 | Frequency | Arm/07/CBM/c2: n = 4 | Frequency |
|---|---|---|---|---|
| Splenomegaly | 3 | 60% | 4 | 100% |
| Hyperaemia and/or enlargement of lymph nodes * | 4 | 80% | 4 | 100% |
| Abdomen—exudative fluid | 1 | 20% | 4 | 100% |
| Hyperaemia of tonsil | 2 | 40% | 4 | 100% |
| Petechiae in kidneys | 1 | 20% | 1 | 25% |
| Pleural exudative fluid | 1 | 20% | 2 | 50% |
| Hyperaemia of lungs | 1 | 20% | 2 | 50% |
| Nasal discharge | 1 | 20% | 0 | 0% |
| Pericardial exudative fluid | 2 | 40% | 3 | 75% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sunwoo, S.-Y.; García-Belmonte, R.; Walczak, M.; Vigara-Astillero, G.; Kim, D.-M.; Szymankiewicz, K.; Kochanowski, M.; Liu, L.; Tark, D.; Podgórska, K.; et al. Deletion of MGF505-2R Gene Activates the cGAS-STING Pathway Leading to Attenuation and Protection against Virulent African Swine Fever Virus. Vaccines 2024, 12, 407. https://doi.org/10.3390/vaccines12040407
Sunwoo S-Y, García-Belmonte R, Walczak M, Vigara-Astillero G, Kim D-M, Szymankiewicz K, Kochanowski M, Liu L, Tark D, Podgórska K, et al. Deletion of MGF505-2R Gene Activates the cGAS-STING Pathway Leading to Attenuation and Protection against Virulent African Swine Fever Virus. Vaccines. 2024; 12(4):407. https://doi.org/10.3390/vaccines12040407
Chicago/Turabian StyleSunwoo, Sun-Young, Raquel García-Belmonte, Marek Walczak, Gonzalo Vigara-Astillero, Dae-Min Kim, Krzesimir Szymankiewicz, Maciej Kochanowski, Lihong Liu, Dongseob Tark, Katarzyna Podgórska, and et al. 2024. "Deletion of MGF505-2R Gene Activates the cGAS-STING Pathway Leading to Attenuation and Protection against Virulent African Swine Fever Virus" Vaccines 12, no. 4: 407. https://doi.org/10.3390/vaccines12040407






