The Functional Mechanism of BP9 in Promoting B Cell Differentiation and Inducing Antigen Presentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptides, Antigens, and Cells
2.2. Experimental Animals
2.3. Main Reagents
2.4. Main Instruments and Equipment
2.5. Vaccine Preparation and Animal Immunization Protocol
2.6. Serum-Specific Antibody Level Detection
2.7. MTT Assay/Splenic Lymphocyte Activity Assessment
2.8. FCM
2.9. RNA Sequencing of B-Cells
2.10. HD11 Cell Treatment and Detection
2.11. Sample Preparation and miRNA Sequencing
2.12. Verification of DEGs in HD11 Cells with BP9 Treatment
2.13. Data Statistics and Analysis
3. Results
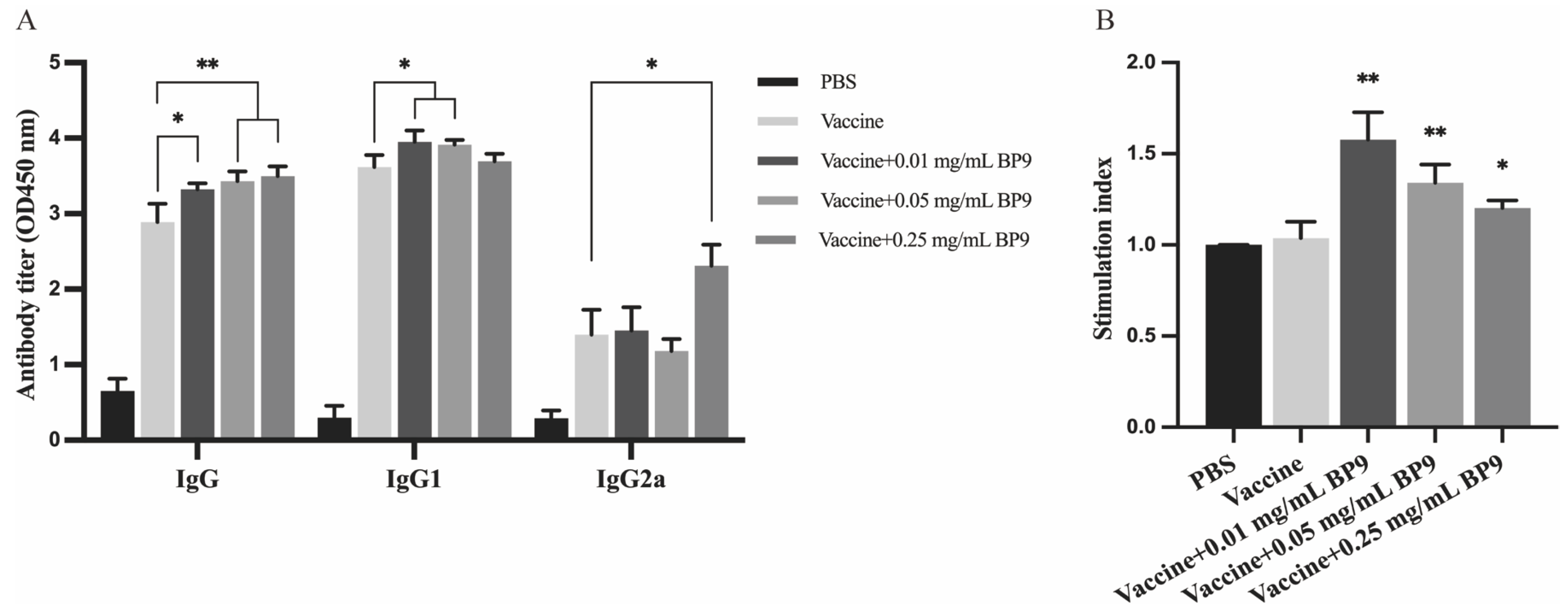
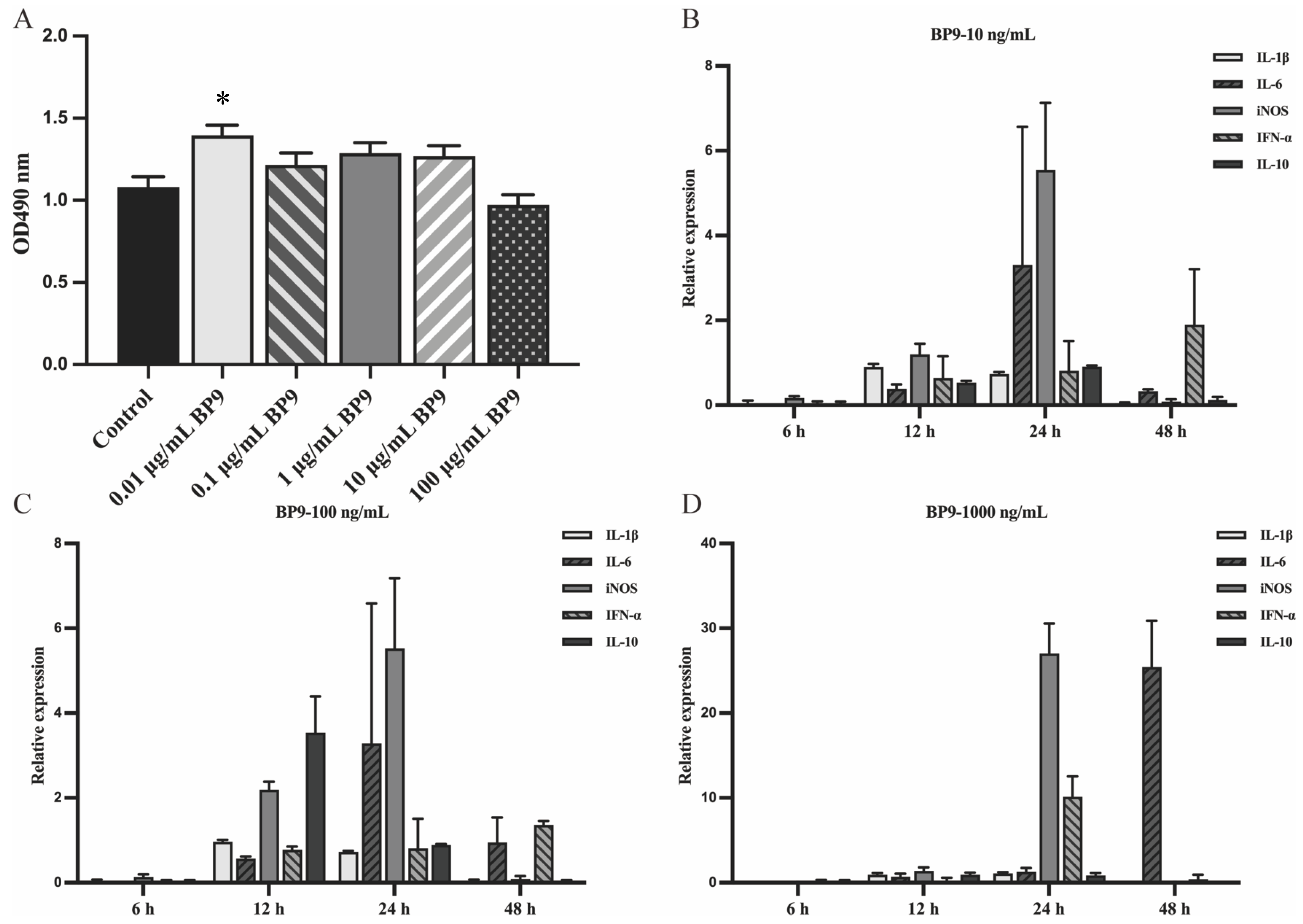
3.1. BP9 Enhanced the Antibody Production and the Proliferation of Splenic Lymphocytes In Vivo
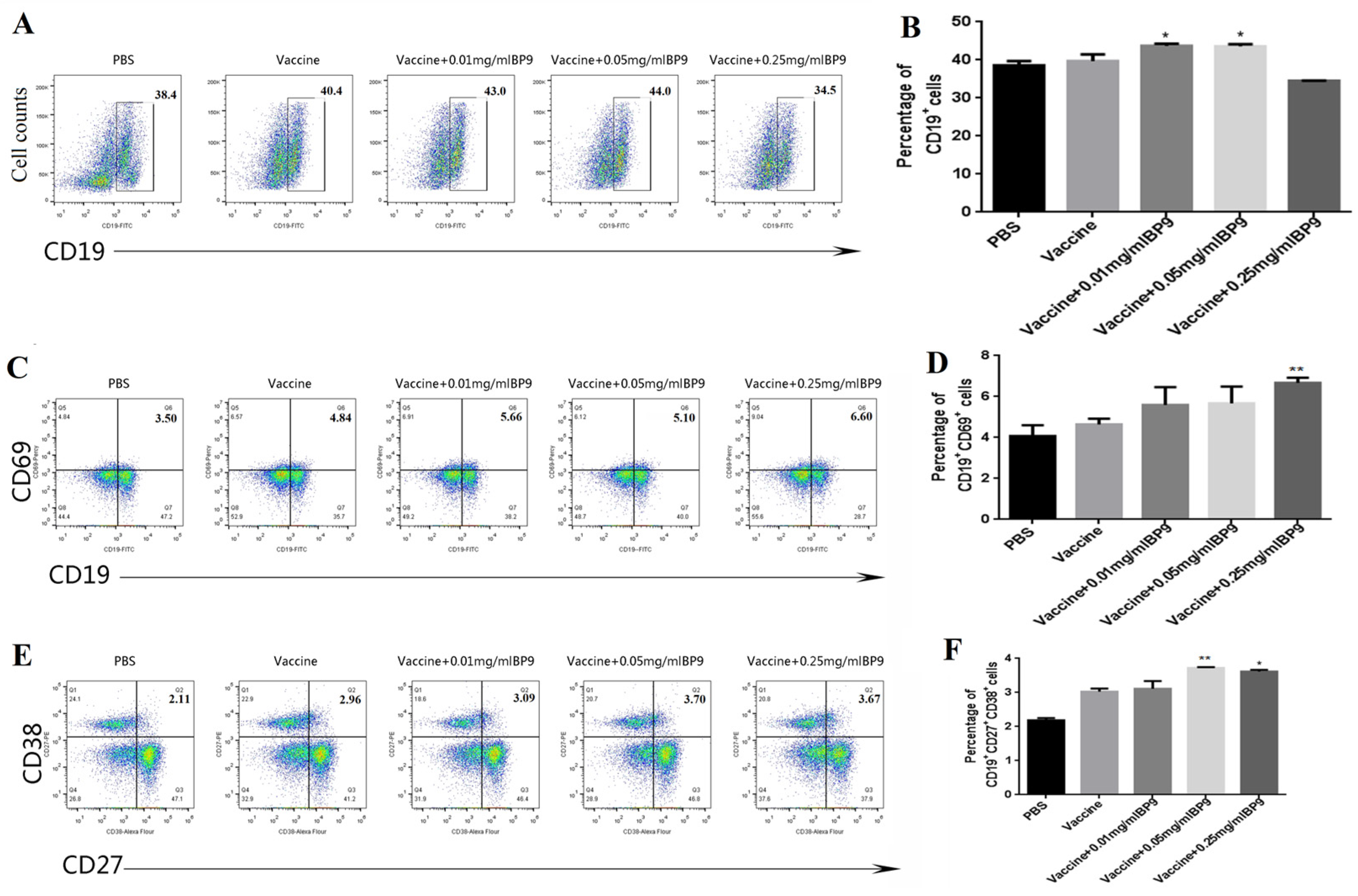
3.2. BP9 Stimulated the Differentiation of B Cells In Vivo
3.3. BP9 Induced Differential Expressions of Multiple Genes and Transcription Factors
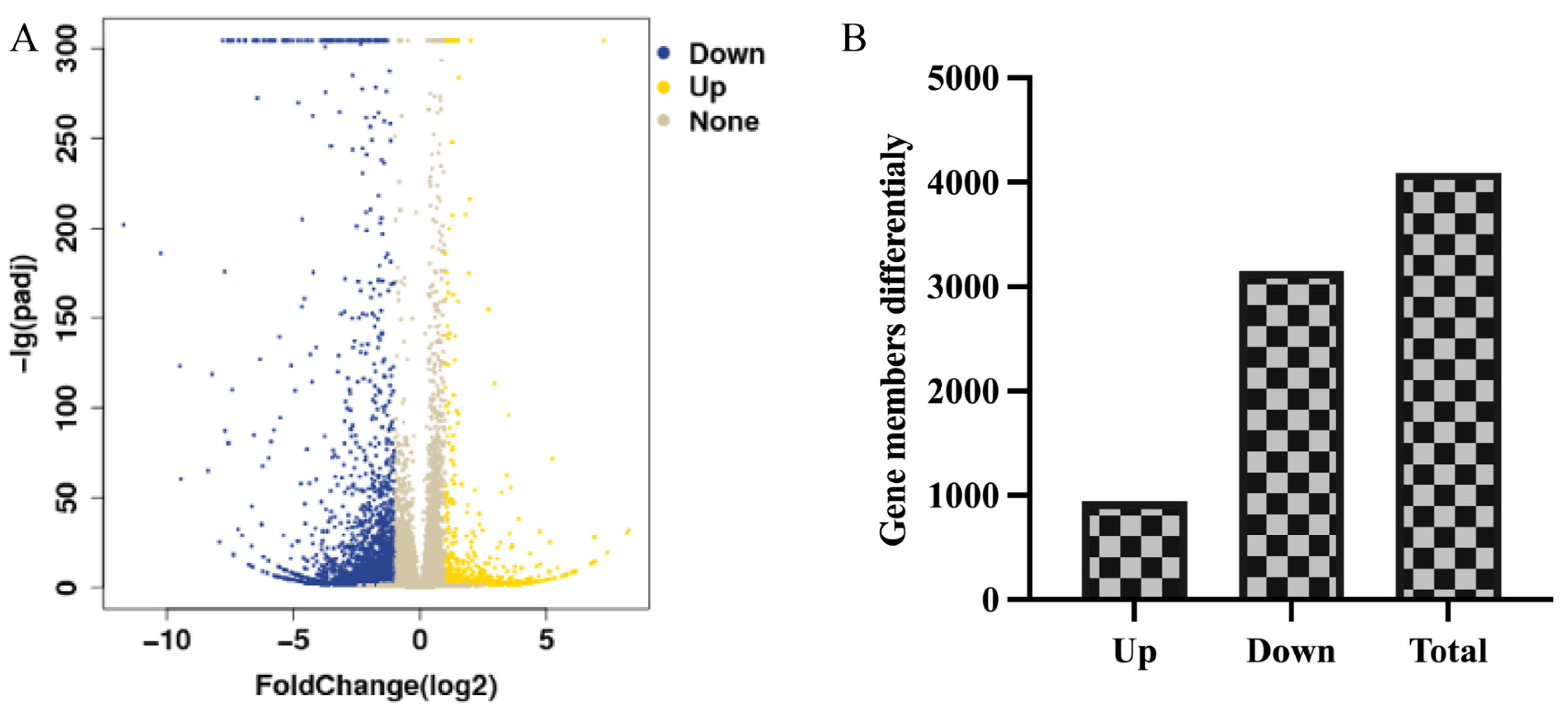
3.3.1. Statistical Assessment of Differentially Expressed Genes
3.3.2. BP9 Enhanced Various Antigen Presentation Pathways
3.3.3. BP9 Activated the Multiple Biological Processes in B Cells
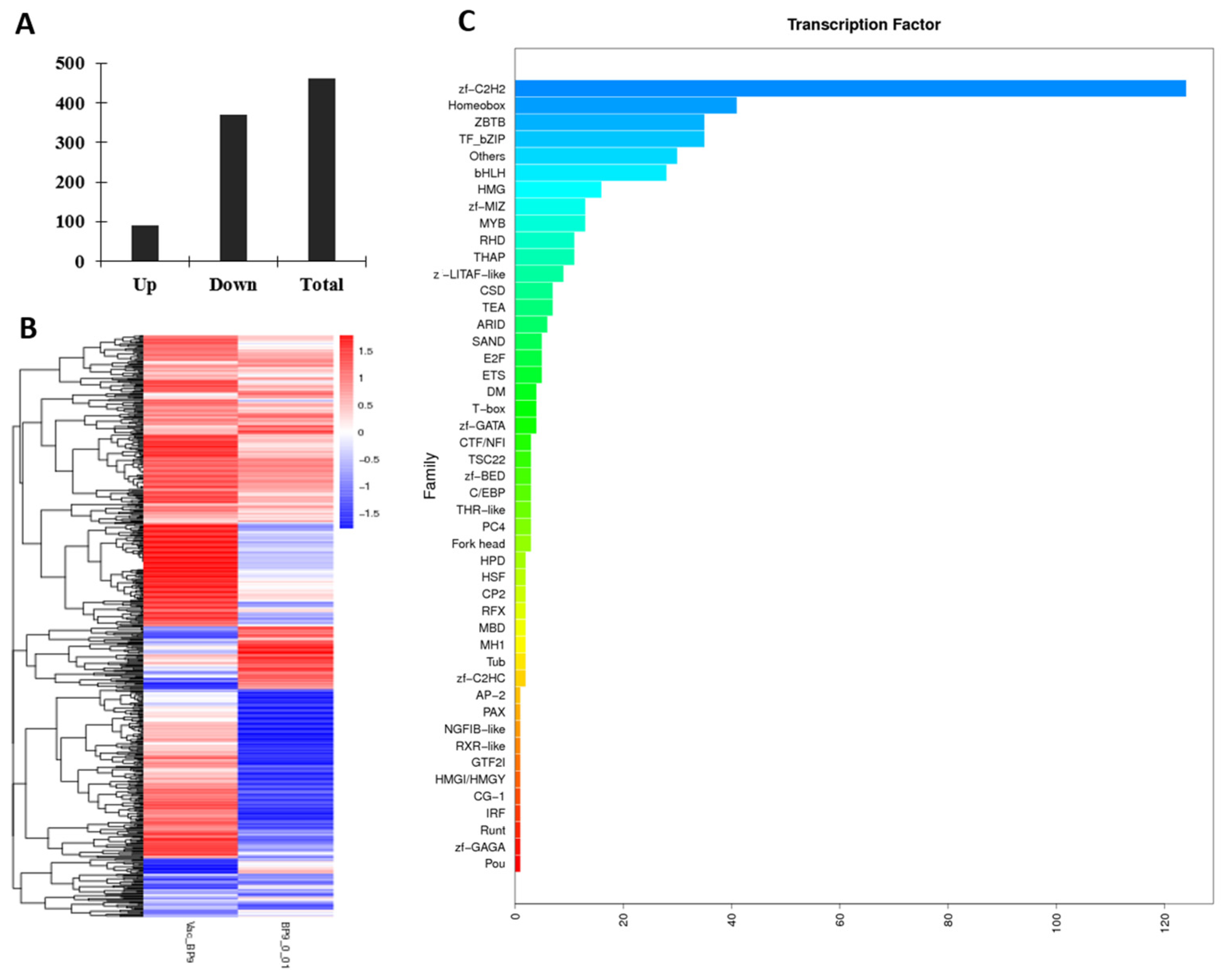
3.3.4. BP9 Activated the Multiple Transcript Factors in B Cells
3.4. BP9 Stimulated the Cytokine Expressions in Chicken Macrophages
3.5. BP9 Promoted Various Signal and Biological Processes in Chicken Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Wang, W.; Qin, Y.; Zou, J.; Yu, H. Global Epidemiology of Human Infections with Variant Influenza Viruses, 1959–2021: A Descriptive Study. Clin. Infect. Dis. 2022, 75, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, M.J.; Lassila, O.; Pink, J.R.L.; Vainio, O. Avian B cell precursors: Surface immunoglobulin expression is an early, possibly bursa-independent event. Eur. J. Immunol. 1986, 16, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Bäck, O.; Bäck, R.; Hemmingsson, E.J.; Liden, S.; Linna, T.J. Migration of bone marrow cells to the bursa of Fabricius and the spleen in the chicken. Scand. J. Immunol. 1973, 2, 357–366. [Google Scholar] [CrossRef]
- Nagy, N.; Busalt, F.; Halasy, V.; Kohn, M.; Schmieder, S.; Fejszak, N.; Härtle, S. In and out of the bursa—The role of CXCR4 in chicken B cell development. Front. Immunol. 2020, 11, 1468. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.; Gilmour, D.G.; Goldstein, G. Lymphocyte-differentiating hormone of bursa of Fabricius. Science 1976, 193, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, X.; Wu, T.; Li, D.; Niu, M.; Wang, Y.; Chen, P. Bursin-like peptide (BLP) enhances H9N2 influenza vaccine induced humoral and cell mediated immune responses. Cell. Immunol. 2014, 292, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cai, J.X.; Hao, S.S.; Li, C.F.; Chen, J.J.; Li, T.T.; Feng, X.L. Transcriptomic analysis of spleen B cell revealed the molecular basis of bursopentin on B cell differentiation. Vet. Res. 2022, 53, 109. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qin, T.; Yu, Q.; Yang, Q. Bursopentin (BP5) from Chicken Bursa of Fabricius Attenuates the Immune Function of Dendritic Cells. Amino Acids 2014, 46, 1763–1774. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, Z.; Li, C.F.; Hao, S.S.; Lu, A.R.; Huang, X.Y.; Feng, X.L. Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells. Vaccines 2022, 10, 1960. [Google Scholar] [CrossRef]
- Caldwell, D.J.; Dean, C.E.; McElroy, A.P.; Caldwell, D.Y.; Hargis, B.M. Bursal anti-steroidogenic peptide (BASP): Modulation of mitogen-stimulated bursal-lymphocyte DNA synthesis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1999, 123, 385–391. [Google Scholar] [CrossRef]
- García-Espinosa, G.; Moore, R.W.; Berghman, L.R.; Hargis, B.M. Relationship of Bursal Anti-Steroidogenic Peptide (BASP) and Histone H1. Life Sci. 2002, 71, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, J.; Li, S.; Cai, K.; Guo, X.; Liao, C.; Wang, C. Bursal Hexapeptide, a Potential Immunomodulator, Inhibits Tumor Cells Proliferation via p53 Signaling Pathway. Anti-Cancer Agents Med. Chem. 2018, 18, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Bie, J.; Liu, X. Research Note: Isolation and Immunomodulatory Activity of Bursal Peptide, a Novel Peptide from Avian Immune System Developments. Poult. Sci. 2024, 103, 103294. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.L.; Zong, M.M.; Zhou, G.F.; Zheng, Y.; Yu, Y.N.; Cao, R.B.; Chen, P.Y.; Yang, M. The functions and mechanism of a new oligopeptide BP9 from avian bursa on antibody responses, immature B cell, and autophagy. J. Immunol. Res. 2019, 2019, 1574383. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zong, M.M.; Chen, B.Y.; Zhou, X.H.; Liu, Z.N.; Zhou, G.F.; Chen, P.Y.; Feng, X.L. The roles of bursal nonapeptide (BP9) on AIV vaccine immune response in chick immunization and on avian immature B cell. Protein Pept. Lett. 2019, 26, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.N.; Zheng, Y.; Hao, S.S.; Zhang, Z.; Cai, J.X.; Zong, M.M.; Feng, X.L.; Liu, Q.T. The molecular evolutionary characteristics of new isolated H9N2 AIV from East China and the function of vimentin on virus replication in MDCK cells. Virol. J. 2020, 17, 78. [Google Scholar] [CrossRef] [PubMed]
- Twentyman, P.R.; Luscombe, M. A Study of Some Variables in a Tetrazolium Dye (MTT) Based Assay for Cell Growth and Chemosensitivity. Br. J. Cancer 1987, 56, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA Transcriptome of Breast Muscle from the Embryonic to Post-Hatching Periods in Chickens. BMC Genom. 2021, 22, 64. [Google Scholar] [CrossRef]
- Wang, X.; Yan, P.Q.; Liu, L.; Luo, Y.; Zhao, L.; Liu, H.F.; Tang, Q.Z.; Long, K.R.; Jin, L.; Ma, J.D.; et al. MicroRNA Expression Profiling Reveals Potential Roles for MicroRNA in the Liver during Pigeon (Columba livia) Development. Poult. Sci. 2020, 99, 6378–6389. [Google Scholar] [CrossRef]
- Karen, S.; Young, J.R.; Colin, B.; Charles, M.R. Expression of chicken dec205 reflects the unique structure and function of the avian immune system. PLoS ONE 2013, 8, e51799. [Google Scholar]
- Davison, F. The Importance of the Avian Immune System and Its Unique Features. In Avian Immunology; Academic Press: Cambridge, MA, USA, 2022; pp. 1–9. [Google Scholar]
- Garcia, P.; Wang, Y.; Viallet, J.; Macek Jilkova, Z. The Chicken Embryo Model: A Novel and Relevant Model for Immune-Based Studies. Front. Immunol. 2021, 12, 791081. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Burrows, P.D.; Wang, J.Y. B Cell Development and Maturation. In B Cells in Immunity and Tolerance. Advances in Experimental Medicine and Biology; Springer: Singapore, 2020; Volume 1254, pp. 1–22. [Google Scholar]
- Pape, K.A.; Taylor, J.J.; Maul, R.W.; Gearhart, P.J.; Jenkins, M.K. Different B cell populations mediate early and late memory during an endogenous immune response. Science 2011, 331, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, M.J.; Härtle, S. B Cells, the Bursa of Fabricius, and the Generation of Antibody Repertoires. In Avian Immunology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 71–99. [Google Scholar]
- Laidlaw, B.J.; Cyster, J.G. Transcriptional regulation of memory B cell differentiation. Nat. Rev. Immunol. 2021, 21, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Vera, M.; Gandin, V.; Singer, R.H.; Tutucci, E. Intracellular mRNA Transport and Localized Translation. Nat. Rev. Mol. Cell Biol. 2021, 22, 483–504. [Google Scholar] [CrossRef] [PubMed]
- Slobodin, B.; Dikstein, R. So Close, No Matter How Far: Multiple Paths Connecting Transcription to mRNA Translation in Eukaryotes. EMBO Rep. 2020, 21, e50799. [Google Scholar] [CrossRef] [PubMed]
- Roberto, M.P.; Varano, G.; Vinas-Castells, R.; Holmes, A.B.; Kumar, R.; Pasqualucci, L.; Farinha, P.; Scott, D.W.; Dominguez-Sola, D. Mutations in the transcription factor FOXO1 mimic positive selection signals to promote germinal center B cell expansion and lymphomagenesis. Immunity 2021, 10, 1807–1824. [Google Scholar] [CrossRef] [PubMed]
- Passmore, L.A.; Coller, J. Roles of mRNA Poly (A) Tails in Regulation of Eukaryotic Gene Expression. Nat. Rev. Mol. Cell Biol. 2022, 23, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Craft, J. T Follicular Helper Cell Heterogeneity. Annu. Rev. Immunol. 2023, 42, 1. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.S.; Sun, X.H.; Zhang, X.; Zhou, Z.H.; Yu, Z.; Zhu, X.J.; Huang, L.Y.; Fang, M.; Gao, Y.T.; Li, M.; et al. Mechanism of Follicular Helper T Cell Differentiation Regulated by Transcription Factors. J. Immunol. Res. 2020, 2020, 1826587. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, X.H.; Liu, X.D.; Zhong, B.; Tang, H.; Jin, W.; Clevers, H.; Wang, H.; Wang, X.H.; Dong, C. Transcription factor Ascl2 promotes germinal center B cell responses by directly regulating AID transcription. Cell Rep. 2021, 35, 109188. [Google Scholar] [CrossRef]
- Muntjewerff, E.M.; Meesters, L.D.; Van den Bogaart, G. Antigen Cross-Presentation by Macrophages. Front. Immunol. 2020, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- Borbet, T.C.; Hines, M.J.; Koralov, S.B. MicroRNA regulation of B cell receptor signaling. Immunol. Rev. 2021, 304, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Cheng, A.H.; Wang, M.S.; Jia, R. Y The Influence of Host miRNA Binding to RNA within RNA Viruses on Virus Multiplication. Front. Cell. Infect. Microbiol. 2022, 12, 802149. [Google Scholar] [CrossRef] [PubMed]
- Schell, S.L.; Rahman, Z.S.M. miRNA-Mediated Control of B Cell Responses in Immunity and SLE. Front. Immunol. 2021, 12, 683710. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, I.; Jeon, D.; Moseman, J.E.; Muralidhar, A.; Potluri, H.K.; McNeel, D.G. Role of B Cells as Antigen Presenting Cells. Front. Immunol. 2022, 13, 954936. [Google Scholar] [CrossRef]
- Wu, Z.; Harne, R.; Chintoan-Uta, C.; Hu, T.J.; Wallace, R.; MacCallum, A.; Stevens, M.P.; Kaiser, P.; Balic, A.; Hume, D.A. Regulation and Function of Macrophage Colony-Stimulating Factor (CSF1) in the Chicken Immune System. Dev. Comp. Immunol. 2020, 105, 103586. [Google Scholar] [CrossRef]
- Kaspers, B.; Schat, K.A.; Göbel, T.; Vervelde, L. Avian Immunology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]

| Primer Name | NCBI Reference | Sequence (5′-3′) |
|---|---|---|
| IL-1β-F | NM_204524.2 | ACCCGCTTCATCTTCTACCG |
| IL-1β-R | TCAGCGCCCACTTAGCTTG | |
| IL-6-F | NM_204628.2 | AGGACGAGATGTGCAAGAAGTTC |
| IL-6-R | TTGGGCAGGTTGAGGTTGTT | |
| IL-10-F | NM_001004414.4 | CGCTGTCACCGCTTCTTCA |
| IL-10-R | CGTCTCCTTGATCTGCTTGATG | |
| iNOS-F | U46504.1 | AGGCCAAACATCCTGGAGGTC |
| iNOS-R | TCATAGAGACGCTGCTGCCAG | |
| IFN-α-F | GU119896.1 | GGACATGGCTCCCACACTAC |
| IFN-α-R | GGCTGCTGAGGATTTTGAAGA | |
| β-actin-F | NM_205518.2 | AGACATCAGGGTGTGATGGTTGGT |
| β-actin-R | TGGTGACAATACCGTGTTCAATGG |
| Primer Name | Sequence (5′-3′) |
|---|---|
| gga-miR-26a-5p | TCCAGCTGGGTTCAAGTAATCCAGG |
| gga-miR-210a-3p | CTGTGCGTGTGACAGCGGCTAA |
| gga-miR-218-5p | TTGTGCTTGATCTAACCATGT |
| 5sR-F | GTCTACGGCCATACCACCCTGAAC |
| Name | Q-Value | Up Count | Down Count |
|---|---|---|---|
| Hematopoietic cell lineage | 5.0588 × 10−9 | 3 | 30 |
| Cytokine–cytokine receptor interaction | 1.4634 × 10−8 | 13 | 53 |
| ECM–receptor interaction | 6.8286 × 10−6 | 6 | 20 |
| Cell adhesion molecules (CAMs) | 3.2955 × 10−5 | 5 | 32 |
| Protein digestion and absorption | 0.00014185 | 3 | 23 |
| Tryptophan metabolism | 0.00014185 | 3 | 14 |
| ABC transporters | 0.00020629 | 1 | 17 |
| Malaria | 0.00027825 | 4 | 13 |
| Steroid biosynthesis | 0.00059044 | 0 | 10 |
| Glycine, serine, and threonine metabolism | 0.00158069 | 3 | 17 |
| African trypanosomiasis | 0.00225022 | 3 | 9 |
| Nitrogen metabolism | 0.00231943 | 2 | 8 |
| Glycerolipid metabolism | 0.0034593 | 2 | 18 |
| Osteoclast differentiation | 0.00443348 | 8 | 21 |
| Axon guidance | 0.0054491 | 8 | 31 |
| Pathways in cancer | 0.005574 | 14 | 92 |
| Porphyrin and chlorophyll metabolism | 0.00712266 | 0 | 13 |
| Glutathione metabolism | 0.00831065 | 2 | 22 |
| Complement and coagulation cascades | 0.00896268 | 7 | 14 |
| Ether lipid metabolism | 0.00942156 | 3 | 12 |
| Valine, leucine, and isoleucine degradation | 0.01069113 | 3 | 12 |
| Alanine, aspartate, and glutamate metabolism | 0.01326763 | 1 | 12 |
| Glycerophospholipid metabolism | 0.01326763 | 4 | 19 |
| Metabolism of xenobiotics by cytochrome P450 | 0.01326763 | 2 | 19 |
| Pancreatic secretion | 0.01326763 | 1 | 24 |
| cAMP signaling pathway | 0.02294556 | 6 | 33 |
| Fat digestion and absorption | 0.02354943 | 0 | 12 |
| MAPK signaling pathway | 0.02362328 | 9 | 44 |
| microRNAs in cancer | 0.025189 | 6 | 27 |
| Neuroactive ligand–receptor interaction | 0.04037019 | 12 | 39 |
| Fluid shear stress and atherosclerosis | 0.04050935 | 7 | 33 |
| Relaxin signaling pathway | 0.04050935 | 4 | 22 |
| Mineral absorption | 0.04379525 | 0 | 13 |
| Bile secretion | 0.04465393 | 0 | 17 |
| Vitamin digestion and absorption | 0.04546349 | 2 | 6 |
| Glyoxylate and dicarboxylate metabolism | 0.04715224 | 1 | 11 |
| Adrenergic signaling in cardiomyocytes | 0.04788709 | 8 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Zhang, Z.; Cai, J.; Hao, S.; Li, C.; Feng, X. The Functional Mechanism of BP9 in Promoting B Cell Differentiation and Inducing Antigen Presentation. Vaccines 2024, 12, 607. https://doi.org/10.3390/vaccines12060607
Hu J, Zhang Z, Cai J, Hao S, Li C, Feng X. The Functional Mechanism of BP9 in Promoting B Cell Differentiation and Inducing Antigen Presentation. Vaccines. 2024; 12(6):607. https://doi.org/10.3390/vaccines12060607
Chicago/Turabian StyleHu, Jianing, Ze Zhang, Jiaxi Cai, Shanshan Hao, Chenfei Li, and Xiuli Feng. 2024. "The Functional Mechanism of BP9 in Promoting B Cell Differentiation and Inducing Antigen Presentation" Vaccines 12, no. 6: 607. https://doi.org/10.3390/vaccines12060607






