Identifying Key Drivers of Efficient B Cell Responses: On the Role of T Help, Antigen-Organization, and Toll-like Receptor Stimulation for Generating a Neutralizing Anti-Dengue Virus Response
Abstract
1. Introduction
2. Materials and Methods
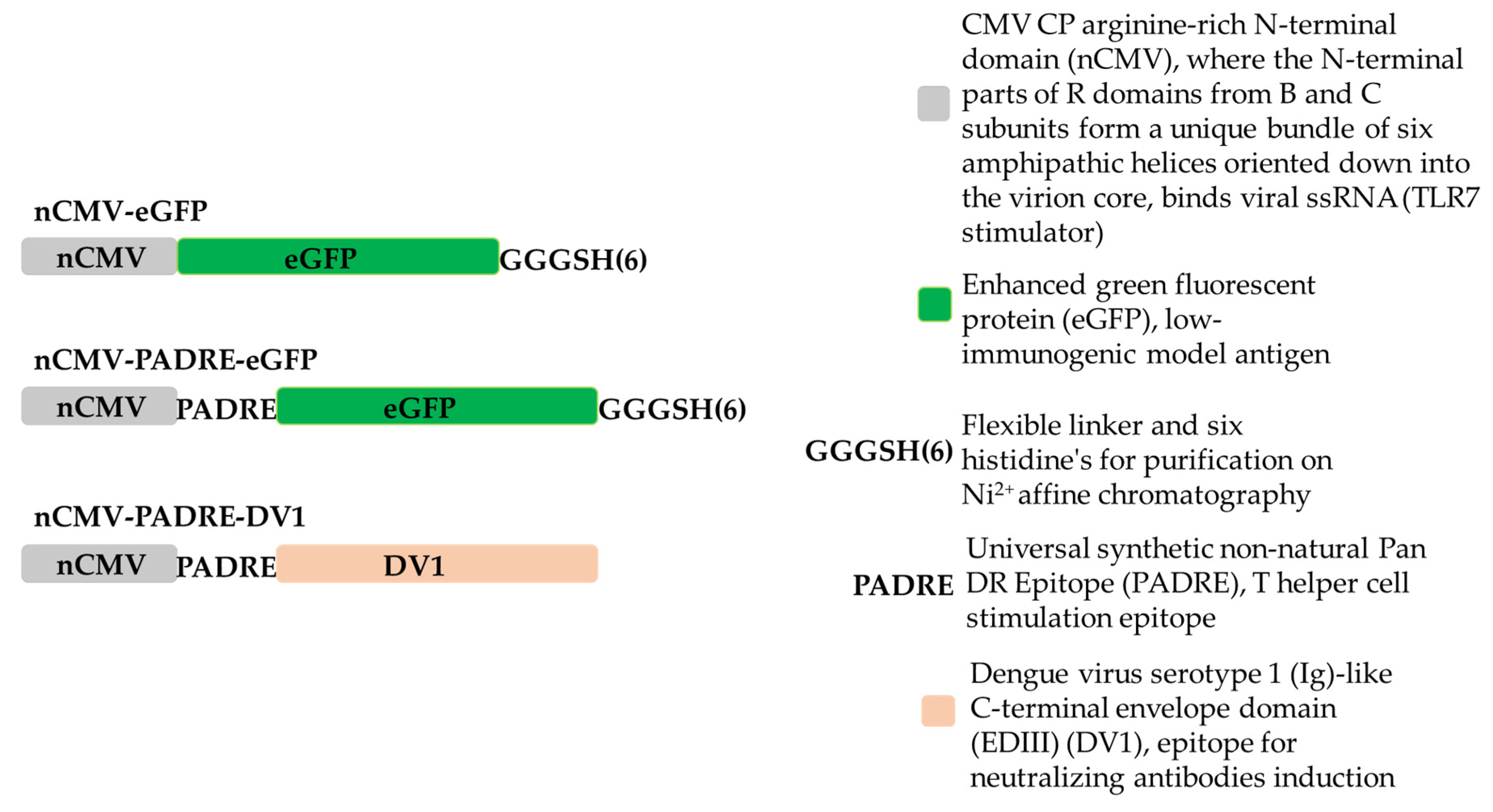
2.1. Cloning of “Immune-Tag” nCMV-eGFP, nCMV-PADRE-eGFP, nCMV-PADRE-DV1 Variants
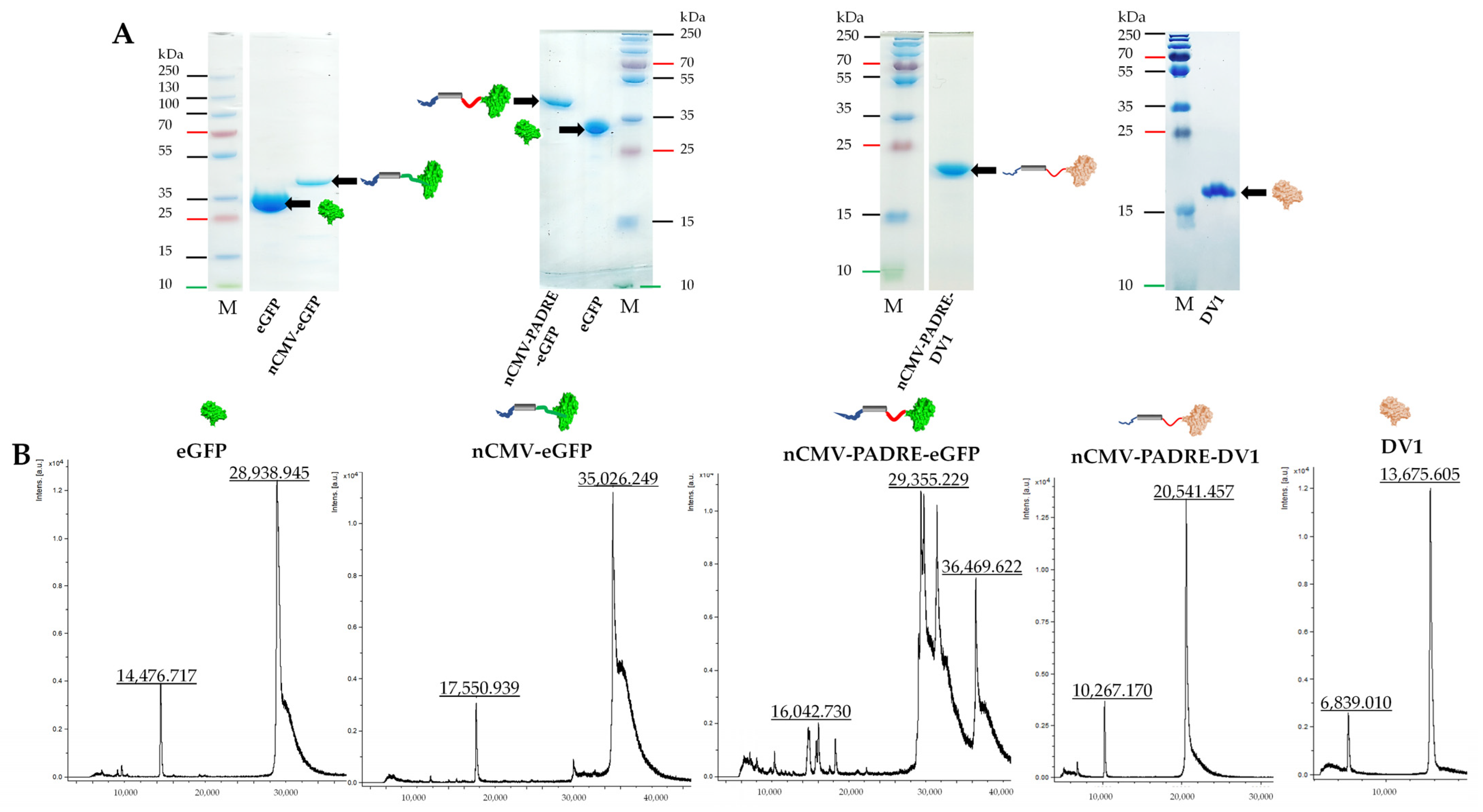
2.2. Protein Production, Purification, and Analysis
2.3. Expression and Purification of CuMV and CuMVTT VLPs
2.4. Isolation and Quantification of RNA from CuMV Samples
2.5. CMV CP mRNA Transcription
2.6. nCMV Binding to Nucleic Acid
2.7. Electrophoretic Nucleic Acid Mobility Shift Assay (Gel Shift)
2.8. Development of CuMV-eGFP and CuMVTT-DV1 Vaccines
2.9. Dynamic Light Scattering Measurement (DLS)
2.10. Sample Analysis by Mass Spectrometry (MS)
2.11. Transmission Electron Microscopy (TEM)
2.12. Mice
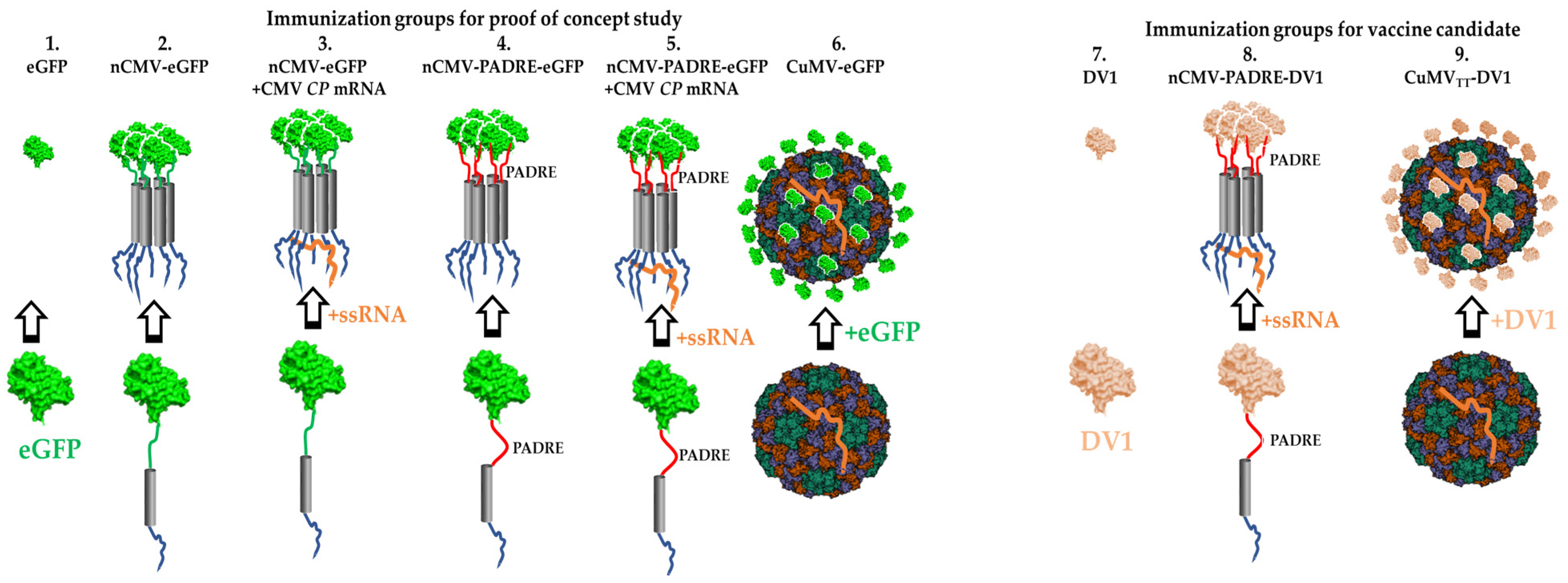
2.13. Immunization Regimen
2.14. The Enzyme-Linked Immunosorbent Assay (ELISA)
2.15. DENV-1 Focus Reduction Virus Neutralization Test (FRNT)
2.16. Data Analysis
3. Results
3.1. Development of “IMMUNE-TAG” as a Vaccine Platform
3.2. Chemically Modified VLP-Based Platform
3.3. Characterization of the Immunogenic Potential of nCMV–eGFP Using “Immune-Tag” Technology
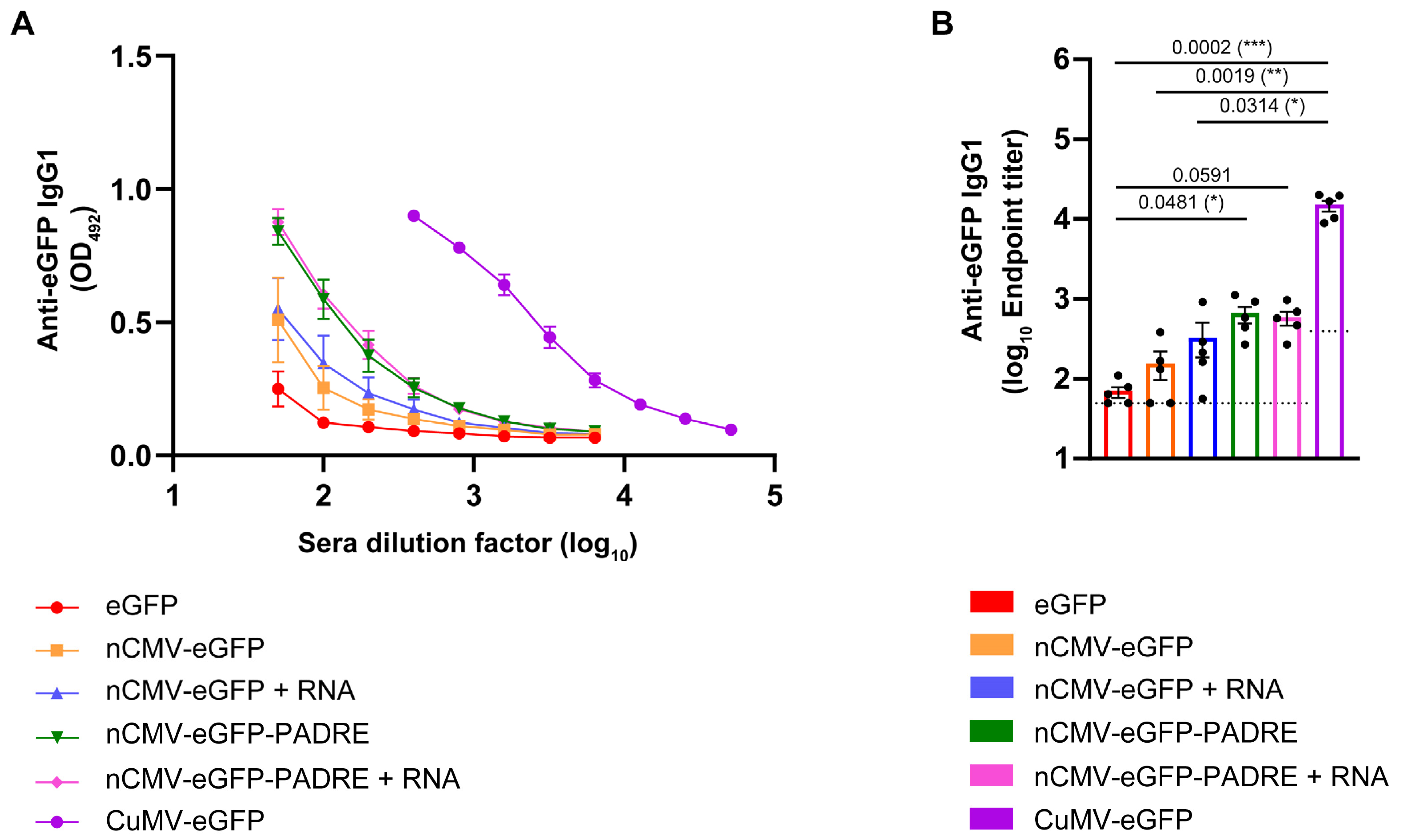
3.4. Total Levels of Anti-eGFP IgG
3.5. Subclass-Specific Anti-eGFP IgG Antibodies
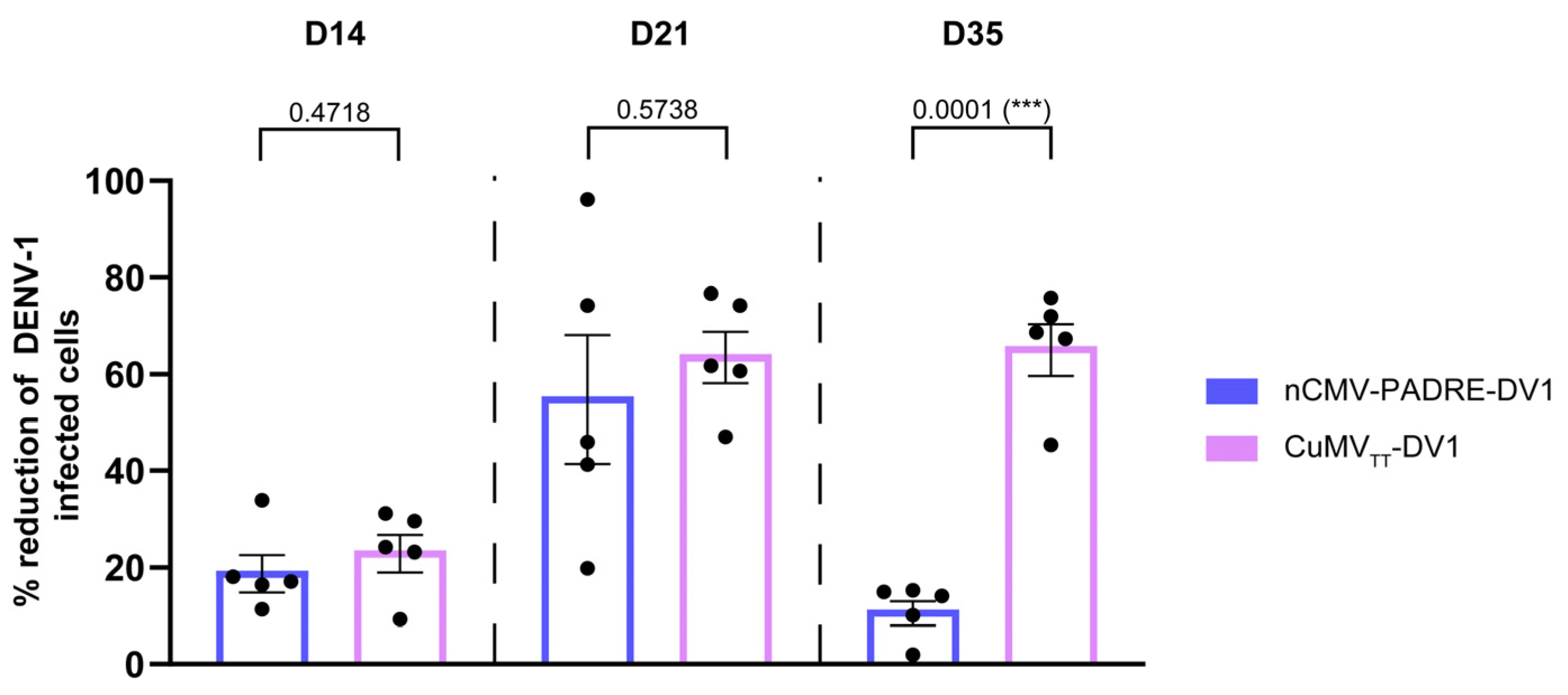
3.6. nCMV-PADRE “Immune-Tag” as a Dengue Vaccine Candidate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2,5-DHAP | 2,5-dihydroxyacetophenone |
| Ab/Abs | antibody/antibodies |
| AEC | addition of 3-amino-9-ethylcarbazole |
| APCs | antigen-presenting cells |
| AI | avidity index |
| CMV | cucumber mosaic virus |
| CP | capsid or coat protein |
| CuMV | cucumber mosaic virus-derived virus-like particles |
| CuMVTT | immunologically optimized cucumber mosaic virus-derived VLPs with incorporated universal Th-cell epitope from tetanus toxin |
| DENV | dengue virus |
| DLS | dynamic light scattering |
| DV1 EDIII | third domain of dengue virus 1 envelope protein |
| eGFP | enhanced green fluorescent protein |
| ELISA | enzyme-linked immunosorbent assay |
| FRNT | focus reduction virus neutralization test |
| IB | inclusion bodies |
| MS | mass spectrometry |
| NAGE | native agarose gel electrophoresis |
| nCMV | N-terminal fragment of the cucumber mosaic virus capsid protein containing the functional R domain |
| ON | overnight |
| PADRE | universal synthetic non-natural Pan DR Epitope |
| PAMPs | pathogen-associated molecular patterns |
| PASPs | Pathogen-associated structural patterns |
| PMSF | phenylmethylsulfonyl fluoride |
| RT | room temperature |
| SATA | N-succinimidyl S-acetylthioacetate |
| s.c. | subcutaneous |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SMPH | succinimidyl 6-((β-maleimidopropionamido) hexanoate |
| TEM | transmission electron microscopy |
| Th | T-cell help |
| Th cell | T-helper cell |
| TLR | toll-like receptor |
| TMB | tetramethylbenzidine |
| TT | tetanus toxin |
| VLPs | virus-like particles |
| WT | wild type |
References
- Feldmann, M.; Easten, A. The relationship between antigenic structure and the requirement for thymus-derived cells in the immune response. J. Exp. Med. 1971, 134, 103–119. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics, and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Vogel, M.; Bachmann, M.F. Immunogenicity and immunodominance in antibody responses. Curr. Top. Microbiol. Immunol. 2020, 428, 89–102. [Google Scholar]
- Pushko, P.; Pumpens, P.; Grens, E. Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures. Intervirology 2013, 56, 141–165. [Google Scholar] [CrossRef]
- Zinkhan, S.; Ogrina, A.; Balke, I.; Reseviča, G.; Zeltins, A.; de Brot, S.; Lipp, C.; Chang, X.; Zha, L.; Vogel, M.; et al. The impact of size on particle drainage dynamics and antibody response. J. Control. Release 2021, 331, 296–308. [Google Scholar] [CrossRef]
- Evtushenko, E.A.; Ryabchevskaya, E.M.; Nikitin, N.A.; Atabekov, J.G.; Karpova, O.V. Plant virus particles with various shapes as potential adjuvants. Sci. Rep. 2020, 10, 10365. [Google Scholar] [CrossRef]
- Hilleman, M.R.; Bertland, A.U.; Buynak, E.B.; Lampson, G.P.; McAleer, W.J.; McLean, A.A.; Roehm, R.R.; Tytell, A.A. Clinical and laboratory studies of HBsAg vaccine; Vyas, G.N., Cohen, S.N., Schmid, R., Eds.; Franklin Institute Press: Philadelphia, PA, USA, 1978; pp. 525–537. [Google Scholar]
- Hilleman, M.R. Yeast recombinant hepatitis B vaccine. Infection 1987, 15, 3–7. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Bachmann, M.F. Virus-like particle vaccinology, from bench to bedside. Cell. Mol. Immunol. 2022, 19, 993–1011. [Google Scholar] [CrossRef]
- Kheirvari, M.; Liu, H.; Tumban, E. Virus-like Particle Vaccines and Platforms for Vaccine Development. Viruses 2023, 15, 1109. [Google Scholar] [CrossRef]
- Schiller, J.T.; Lowy, D.R. Papillomavirus-like particles and HPV vaccine development. Semin. Cancer Biol. 1996, 7, 373–382. [Google Scholar] [CrossRef]
- Worm, H.C.; Wirnsberger, G. Hepatitis E vaccines: Progress and prospects. Drugs 2004, 64, 1517–1531. [Google Scholar] [CrossRef]
- Xia, M.; Farkas, T.; Jiang, X. Norovirus capsid protein expressed in yeast forms virus-like particles and stimulates systemic and mucosal immunity in mice following an oral administration of raw yeast extracts. J. Med. Virol. 2007, 79, 74–83. [Google Scholar] [CrossRef]
- Ross, T.M.; Mahmood, K.; Crevar, C.J.; Schneider-Ohrum, K.; Heaton, P.M.; Bright, R.A. A trivalent virus-like particle vaccine elicits protective immune responses against seasonal influenza strains in mice and ferrets. PLoS ONE 2009, 4, e6032. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Yang, H.; Qian, Y.; Zhang, Y.; Fang, R.; Chen, X. Immunogenicity and virus-like particle formation of rotavirus capsid proteins produced in transgenic plants. Sci. China Life Sci. 2011, 54, 82–89. [Google Scholar] [CrossRef]
- Akahata, W.; Yang, Z.-Y.; Andersen, H.; Sun, S.; Holdaway, H.A.; Kong, W.-P.; Lewis, M.G.; Higgs, S.; Rossmann, M.G.; Rao, S. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010, 16, 334–338. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Sahly, H.M.E.; Keitel, W.A.; Wolff, M.; Simone, G.; Segawa, C.; Wong, S.; Shelly, D.; Young, N.S.; Dempsey, W. Safety and immunogenicity of a candidate parvovirus B19 vaccine. Vaccine 2011, 29, 7357–7363. [Google Scholar] [CrossRef]
- Li, H.Y.; Han, J.F.; Qin, C.F.; Chen, R. Virus-like particles for enterovirus 71 produced from Saccharomyces cerevisiae potently elicits protective immune responses in mice. Vaccine 2013, 31, 3281–3287. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.Y.; Han, J.F.; Deng, Y.Q.; Li, Y.X.; Zhu, S.Y.; He, Y.L.; Qin, E.D.; Chen, R.; Qin, C.F. Virus-like particles produced in Saccharomyces cerevisiae elicit protective immunity against Coxsackievirus A16 in mice. Appl. Microbiol. Biotechnol. 2013, 97, 10445–10452. [Google Scholar] [CrossRef]
- Chang, L.J.; Dowd, K.A.; Mendoza, F.H.; Saunders, J.G.; Sitar, S.; Plummer, S.H.; Yamshchikov, G.; Sarwar, U.N.; Hu, Z.; Enama, M.E.; et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet 2014, 384, 2046–2052. [Google Scholar] [CrossRef]
- Walpita, P.; Cong, Y.; Jahrling, P.B.; Rojas, O.; Postnikova, E.; Yu, S.; Johns, L.; Holbrook, M. A VLP-based vaccine provides complete protection against Nipah virus challenge following multiple-dose or single-dose vaccination schedules in a hamster model. NPJ Vaccines 2017, 2, 21. [Google Scholar] [CrossRef]
- Sunay, M.M.E.; Martins, K.A.O.; Steffens, J.T.; Gregory, M.; Vantongeren, S.A.; Van Hoeven, N.; Garnes, P.G.; Bavari, S. Glucopyranosyl lipid adjuvant enhances immune response to Ebola virus-like particle vaccine in mice. Vaccine 2019, 37, 3902–3910. [Google Scholar] [CrossRef]
- Chang, Y.H.; Chiao, D.J.; Hsu, Y.L.; Lin, C.C.; Wu, H.L.; Shu, P.Y.; Chang, S.F.; Chang, J.H.; Kuo, S.C. Mosquito cell-derived Japanese encephalitis virus-like particles induce specific humoral and cellular immune responses in mice. Viruses 2020, 12, 336. [Google Scholar] [CrossRef]
- Vang, L.; Morello, C.S.; Mendy, J.; Thompson, D.; Manayani, D.; Guenther, B.; Julander, J.; Sanford, D.; Jain, A.; Patel, A.; et al. Zika virus-like particle vaccine protects AG129 mice and rhesus macaques against Zika virus. PLoS Negl. Trop. Dis. 2021, 15, e0009195. [Google Scholar] [CrossRef]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.Y.; Couture, M.; D’Aoust, M.A.; Dhaliwall, J.; Finkle, C. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Brown, C.S.; Welling-Wester, S.; Feijlbrief, M.; Van Lent, J.W.; Spaan, W.J. Chimeric parvovirus B19 capsids for the presentation of foreign epitopes. Virology 1994, 198, 477–488. [Google Scholar] [CrossRef]
- Tan, M.; Xia, M.; Huang, P.; Wang, L.; Zhong, W.; McNeal, M.; Wei, C.; Jiang, X. Norovirus P particle as a platform for antigen presentation. Procedia Vaccinol. 2011, 4, 19–26. [Google Scholar] [CrossRef]
- Jiang, L.; Fan, R.; Sun, S.; Fan, P.; Su, W.; Zhou, Y.; Gao, F.; Xu, F.; Kong, W.; Jiang, C. A new EV71 VP3 epitope in norovirus P particle vector displays neutralizing activity and protection in vivo in mice. Vaccine 2015, 33, 6596–6603. [Google Scholar] [CrossRef]
- Pascual, E.; Mata, C.P.; Gómez-Blanco, J.; Moreno, N.; Bárcena, J.; Blanco, E.; Rodríguez-Frandsen, A.; Nieto, A.; Carrascosa, J.L.; Castón, J.R. Structural basis for the development of avian virus capsids that display influenza virus proteins and induce protective immunity. J. Virol. 2015, 89, 2563–2574. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, X.; Gai, W.; Wong, G.; Wang, H.; Jin, H.; Feng, N.; Zhao, Y.; Zhang, W.; Li, N. Novel chimeric virus-like particles vaccine displaying MERS-CoV receptor-binding domain induce specific humoral and cellular immune response in mice. Antivir. Res. 2017, 140, 55–61. [Google Scholar] [CrossRef]
- Czarnota, A.; Offersgaard, A.; Pihl, A.F.; Prentoe, J.; Bukh, J.; Gottwein, J.M.; Bieńkowska-Szewczyk, K.; Grzyb, K. Specific Antibodies Induced by Immunization with Hepatitis B Virus-Like Particles Carrying Hepatitis C Virus Envelope Glycoprotein 2 Epitopes Show Differential Neutralization Efficiency. Vaccines 2020, 8, 294. [Google Scholar] [CrossRef]
- Kotiw, M.; Johnson, M.; Pandey, M.; Fry, S.; Hazell, S.L.; Netter, H.J.; Good, M.F.; Olive, C. Immunological response to parenteral vaccination with recombinant hepatitis B virus surface antigen virus-like particles expressing Helicobacter pylori KatA epitopes in a murine H. pylori challenge model. Clin. Vaccine Immunol. 2012, 19, 268–276. [Google Scholar] [CrossRef]
- Rashidijahanabad, Z.; Kelly, M.; Kamruzzaman, M.; Qadri, F.; Bhuiyan, T.R.; McFall-Boegeman, H.; Wu, D.; Piszczek, G.; Xu, P.; Ryan, E.T.; et al. Virus-like Particle Display of Vibrio cholerae O-Specific Polysaccharide as a Potential Vaccine against Cholera. ACS Infect. Dis. 2022, 8, 574–583. [Google Scholar] [CrossRef]
- Rutgers, T.; Gordon, O.M.; Gathoye, A.M.; Hockmeyer, W.T.; De Wilde, M.; Rosenberg, M. Hepatitis B surface antigen as a carrier matrix for the repetitive epitope of the circumsporozoite protein of Plasmodium falciparum. Biotechnology 1988, 6, 1065–1070. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, S.H.; Kim, A.R.; Quan, F.S. Virus-Like Nanoparticle Vaccine Confers Protection against Toxoplasma gondii. PLoS ONE 2016, 11, e0161231. [Google Scholar] [CrossRef]
- Collins, K.A.; Snaith, R.; Cottingham, M.G.; Gilbert, S.C.; Hill, A.V.S. Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci. Rep. 2017, 7, 46621. [Google Scholar] [CrossRef]
- Cecílio, P.; Pérez-Cabezas, B.; Fernández, L.; Moreno, J.; Carrillo, E.; Requena, J.M.; Fichera, E.; Reed, S.G.; Coler, R.N.; Kamhawi, S.; et al. Pre-clinical antigenicity studies of an innovative multivalent vaccine for human visceral leishmaniasis. PLoS Negl. Trop. Dis. 2017, 11, e0005951. [Google Scholar] [CrossRef]
- Moura, A.P.V.; Santos, L.C.B.; Brito, C.R.N.; Valencia, E.; Junqueira, C.; Filho, A.A.P.; Sant’Anna, M.R.V.; Gontijo, N.F.; Bartholomeu, D.C.; Fujiwara, R.T.; et al. Virus-like Particle Display of the α-Gal Carbohydrate for Vaccination against Leishmania Infection. ACS Cent. Sci. 2017, 3, 1026–1031. [Google Scholar] [CrossRef]
- Chackerian, B.; Lowy, D.R.; Schiller, J.T. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Investig. 2001, 108, 415–423. [Google Scholar] [CrossRef]
- Di Bonito, P.; Grasso, F.; Mochi, S.; Petrone, L.; Fanales-Belasio, E.; Mei, A.; Cesolini, A.; Laconi, G.; Conrad, H.; Bernhard, H.; et al. Anti-tumor CD8+ T cell immunity elicited by HIV-1-based virus-like particles incorporating HPV-16 E7 protein. Virology 2009, 395, 45–55. [Google Scholar] [CrossRef]
- Cubas, R.; Zhang, S.; Li, M.; Chen, C.; Yao, Q. Chimeric Trop2 virus-like particles: A potential immunotherapeutic approach against pancreatic cancer. J. Immunother. 2011, 34, 251–263. [Google Scholar] [CrossRef]
- Spohn, G.; Arenas-Ramirez, N.; Bouchaud, G.; Boyman, O. Endogenous polyclonal anti-IL-1 antibody responses potentiate IL-1 activity during pathogenic inflammation. J. Allergy Clin. Immunol. 2017, 139, 1957–1965.e3. [Google Scholar] [CrossRef]
- Zeltins, A.; West, J.; Zabel, F.; El Turabi, A.; Balke, I.; Haas, S.; Maudrich, M.; Storni, F.; Engeroff, P.; Jennings, G.T.; et al. Incorporation of tetanus-epitope into virus-like particles achieves vaccine responses even in older recipients in models of psoriasis, alzheimer’s and cat allergy. NPJ Vaccines 2017, 2, 30. [Google Scholar] [CrossRef]
- Palladini, A.; Thrane, S.; Janitzek, C.M.; Pihl, J.; Clemmensen, S.B.; de Jongh, W.A.; Clausen, T.M.; Nicoletti, G.; Landuzzi, L.; Penichet, M.L.; et al. Virus-like particle display of HER2 induces potent anti-cancer responses. Oncoimmunology 2018, 7, e1408749. [Google Scholar] [CrossRef]
- Schumacher, J.; Bacic, T.; Staritzbichler, R.; Daneschdar, M.; Klamp, T.; Arnold, P.; Jägle, S.; Türeci, Ö.; Markl, J.; Sahin, U. Enhanced stability of a chimeric hepatitis B core antigen virus-like-particle (HBcAg-VLP) by a C-terminal linker-hexahistidine-peptide. J. Nanobiotechnol. 2018, 16, 39. [Google Scholar] [CrossRef]
- Storni, F.; Zeltins, A.; Balke, I.; Heath, M.D.; Kramer, M.F.; Skinner, M.A.; Zha, L.; Roesti, E.; Engeroff, P.; Muri, L.; et al. Vaccine against peanut allergy based on engineered virus-like particles displaying single major peanut allergens. J. Allergy Clin. Immunol. 2020, 145, 1240–1253.e3. [Google Scholar] [CrossRef]
- Cheng, K.; Du, T.; Li, Y.; Qi, Y.; Min, H.; Wang, Y.; Zhang, Q.; Wang, C.; Zhou, Y.; Li, L.; et al. Dual-Antigen-Loaded Hepatitis B Virus Core Antigen Virus-like Particles Stimulate Efficient Immunotherapy against Melanoma. ACS Appl. Mater. Interfaces 2020, 12, 53682–53690. [Google Scholar] [CrossRef]
- Rolih, V.; Caldeira, J.; Bolli, E.; Salameh, A.; Conti, L.; Barutello, G.; Riccardo, F.; Magri, J.; Lamolinara, A.; Parra, K.; et al. Development of a VLP-Based Vaccine Displaying an xCT Extracellular Domain for the Treatment of Metastatic Breast Cancer. Cancers 2020, 12, 1492. [Google Scholar] [CrossRef]
- Klimek, L.; Kundig, T.; Kramer, M.F.; Guethoff, S.; Jensen-Jarolim, E.; Schmidt-Weber, C.B.; Palomares, O.; Mohsen, M.O.; Jakob, T.; Bachmann, M. Virus-like particles (VLP) in prophylaxis and immunotherapy of allergic diseases. Allergo J. Int. 2018, 27, 245–255. [Google Scholar] [CrossRef]
- Balke, I.; Zeltins, A. Use of plant viruses and virus-like particles for the creation of novel vaccines. Adv. Drug Deliv. Rev. 2019, 145, 119–129. [Google Scholar] [CrossRef]
- Balke, I.; Zeltins, A. Recent advances in the use of plant virus-like particles as vaccines. Viruses 2020, 12, 270. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Speiser, D.E.; Knuth, A.; Bachmann, M.F. Virus-like particles for vaccination against cancer. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1579. [Google Scholar] [CrossRef]
- Liu, X.; Chang, X.; Rothen, D.; Derveni, M.; Krenger, P.; Roongta, S.; Wright, E.; Vogel, M.; Tars, K.; Mohsen, M.O.; et al. AP205 VLPs Based on Dimerized Capsid Proteins Accommodate RBM Domain of SARS-CoV-2 and Serve as an Attractive Vaccine Candidate. Vaccines 2021, 9, 403. [Google Scholar] [CrossRef]
- Chang, X.; Zeltins, A.; Mohsen, M.O.; Gharailoo, Z.; Zha, L.; Liu, X.; Walton, S.; Vogel, M.; Bachmann, M.F. A novel double mosaic virus-like particle-based vaccine against SARS-CoV-2 incorporates both receptor binding motif (RBM) and fusion domain. Vaccines 2021, 9, 1287. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Rothen, D.; Balke, I.; Martina, B.; Zeltina, V.; Inchakalody, V.; Gharailoo, Z.; Nasrallah, G.; Dermime, S.; Tars, K. Neutralization of MERS coronavirus through a scalable nanoparticle vaccine. NPJ Vaccines 2021, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Balke, I.; Zinkhan, S.; Zeltina, V.; Liu, X.; Chang, X.; Krenger, P.S.; Plattner, K.; Gharailoo, Z.; Vogt, A.C.S.; et al. A scalable and highly immunogenic virus-like particle-based vaccine against SARS-CoV-2. Allergy 2022, 77, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Ogrina, A.; Skrastina, D.; Balke, I.; Kalnciema, I.; Jansons, J.; Bachmann, M.F.; Zeltins, A. Comparison of bacterial expression systems based on potato virus Y-like particles for vaccine generation. Vaccines 2022, 10, 485. [Google Scholar] [CrossRef]
- Ogrina, A.; Balke, I.; Kalnciema, I.; Skrastina, D.; Jansons, J.; Bachmann, M.F.; Zeltins, A. Bacterial expression systems based on Tymovirus-like particles for the presentation of vaccine antigens. Front. Microbiol. 2023, 14, 1154990. [Google Scholar] [CrossRef] [PubMed]
- Sobczak, J.M.; Krenger, P.S.; Storni, F.; Mohsen, M.O.; Balke, I.; Reseviča, G.; Heath, M.D.; Carreno Velazquez, T.L.; Kramer, M.F.; Scott, C.J.W.; et al. The next generation virus-like particle platform for the treatment of peanut allergy. Allergy 2023, 78, 1980–1996. [Google Scholar] [CrossRef]
- Cabral-Miranda, G.; Lim, S.M.; Mohsen, M.O.; Pobelov, I.V.; Roesti, E.S.; Heath, M.D.; Skinner, M.A.; Kramer, M.F.; Martina, B.E.E.; Bachmann, M.F. Zika virus-derived E-DIII protein displayed on immunologically optimized VLPs induces neutralizing antibodies without causing enhancement of dengue virus infection. Vaccines 2019, 7, 72. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Heath, M.D.; Cabral-Miranda, G.; Lipp, C.; Zeltins, A.; Sande, M.; Stein, J.V.; Riether, C.; Roesti, E.; Zha, L.; et al. Vaccination with nanoparticles combined with micro-adjuvants protects against cancer. J. Immunother. Cancer 2019, 7, 114. [Google Scholar] [CrossRef]
- Thoms, F.; Jennings, G.T.; Maudrich, M.; Vogel, M.; Haas, S.; Zeltins, A.; Hofmann-Lehmann, R.; Riond, B.; Grossmann, J.; Hunziker, P. Immunization of cats to induce neutralizing antibodies against Fel d 1, the major feline allergen in human subjects. J. Allergy Clin. Immunol. 2019, 144, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Olomski, F.; Fettelschoss, V.; Jonsdottir, S.; Birkmann, K.; Thoms, F.; Marti, E.; Bachmann, M.F.; Kundig, T.M.; Fettelschoss-Gabriel, A. Interleukin 31 in insect bite hypersensitivity—Alleviating clinical symptoms by active vaccination against itch. Allergy 2020, 75, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Liekniņa, I.; Sutov, G.; Morris, A.R.; Jovicevic, D.; Kalniņš, G.; Kazāks, A.; Kluga, R.; Kastaljana, S.; Zajakina, A.; et al. N-Terminal Modification of Gly-His-Tagged Proteins with Azidogluconolactone. Chembiochem 2021, 22, 3199–3207. [Google Scholar] [CrossRef] [PubMed]
- Zha, L.; Chang, X.; Zhao, H.; Mohsen, M.O.; Hong, L.; Zhou, Y.; Chen, H.; Liu, X.; Zhang, J.; Li, D.; et al. Development of a Vaccine against SARS-CoV-2 Based on the Receptor-Binding Domain Displayed on Virus-Like Particles. Vaccines 2021, 9, 395. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-display: Decoration of virus-like particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef] [PubMed]
- Thérien, A.; Bédard, M.; Carignan, D.; Rioux, G.; Gauthier-Landry, L.; Laliberté-Gagné, M.; Bolduc, M.; Savard, P.; Leclerc, D. A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. J. Nanobiotechnol. 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.M.C.; Buldun, C.M.; Pattinson, D.J.; Draper, S.J.; Howarth, M. Snoopligase peptide-peptide conjugation enables modular vaccine assembly. Sci. Rep. 2019, 9, 4625. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Yang, Z.; Gnanapragasam, P.N.P.; Ou, S.; Dam, K.A.; Wang, H.; Bjorkman, P.J. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS ONE 2021, 16, e0247963. [Google Scholar] [CrossRef]
- Pejawar-Gaddy, S.; Rajawat, Y.; Hilioti, Z.; Xue, J.; Gaddy, D.F.; Finn, O.J.; Viscidi, R.P.; Bossis, I. Generation of a tumor vaccine candidate based on conjugation of a MUC1 peptide to polyionic papillomavirus virus-like particles. Cancer Immunol. Immunother. 2010, 59, 1685–1696. [Google Scholar] [CrossRef]
- van Eldijk, M.B.; Wang, J.C.; Minten, I.J.; Li, C.; Zlotnick, A.; Nolte, R.J.; Cornelissen, J.J.; van Hest, J.C. Designing two self-assembly mechanisms into one viral capsid protein. J. Am. Chem. Soc. 2012, 134, 18506–18509. [Google Scholar] [CrossRef]
- Trifonova, E.A.; Zenin, V.A.; Nikitin, N.A.; Yurkova, M.S.; Ryabchevskaya, E.M.; Putlyaev, E.V.; Donchenko, E.K.; Kondakova, O.A.; Fedorov, A.N.; Atabekov, J.G. Study of rubella candidate vaccine based on a structurally modified plant virus. Antivir. Res. 2017, 144, 27–33. [Google Scholar] [CrossRef]
- Zapata-Cuellar, L.; Gaona-Bernal, J.; Manuel-Cabrera, C.A.; Martínez-Velázquez, M.; Sánchez-Hernández, C.; Elizondo-Quiroga, D.; Camacho-Villegas, T.A.; Gutiérrez-Ortega, A. Development of a platform for noncovalent coupling of full antigens to tobacco etch virus-like particles by means of coiled-coil oligomerization motifs. Molecules 2021, 26, 4436. [Google Scholar] [CrossRef]
- Choi, B.; Kim, H.; Choi, H.; Kang, S. Protein cage nanoparticles as delivery nanoplatforms. Adv. Exp. Med. Biol. 2018, 1064, 27–43. [Google Scholar]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Hadj Hassine, I.; Ben M’hadheb, M.; Almalki, M.A.; Gharbi, J. Virus-like particles as powerful vaccination strategy against human viruses. Rev. Med. Virol. 2024, 34, e2498. [Google Scholar] [CrossRef] [PubMed]
- Tumban, E. Bacteriophage virus-like particles: Platforms for vaccine design. Methods Mol. Biol. 2024, 2738, 411–423. [Google Scholar] [PubMed]
- Matsuura, K.; Watanabe, K.; Matsuzaki, T.; Sakurai, K.; Kimizuka, N. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Mizuguchi, Y.; Kimizuka, N. Peptide nanospheres self-assembled from a modified β-annulus peptide of sesbania mosaic virus. Biopolymers 2016, 106, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Lainšček, D.; Fink, T.; Forstnerič, V.; Hafner-Bratkovič, I.; Orehek, S.; Strmšek, Ž.; Manček-Keber, M.; Pečan, P.; Esih, H.; Malenšek, Š.; et al. A nanoscaffolded spike-rbd vaccine provides protection against sars-cov-2 with minimal anti-scaffold response. Vaccines 2021, 9, 431. [Google Scholar] [CrossRef]
- Han, J.A.; Kang, Y.J.; Shin, C.; Ra, J.S.; Shin, H.H.; Hong, S.Y.; Do, Y.; Kang, S. Ferritin protein cage nanoparticles as versatile antigen delivery nanoplatforms for dendritic cell (DC)-based vaccine development. Nanomedicine 2014, 10, 561–569. [Google Scholar] [CrossRef]
- Khoshnejad, M.; Parhiz, H.; Shuvaev, V.V.; Dmochowski, I.J.; Muzykantov, V.R. Ferritin-based drug delivery systems: Hybrid nanocarriers for vascular immunotargeting. J. Control. Release 2018, 282, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Cho, S.; Kim, I.S. Ferritin—A multifaceted protein scaffold for biotherapeutics. Exp. Mol. Med. 2022, 54, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Obozina, A.S.; Komedchikova, E.N.; Kolesnikova, O.A.; Iureva, A.M.; Kovalenko, V.L.; Zavalko, F.A.; Rozhnikova, T.V.; Tereshina, E.D.; Mochalova, E.N.; Shipunova, V.O. Genetically encoded self-assembling protein nanoparticles for the targeted delivery in vitro and in vivo. Pharmaceutics 2023, 15, 231. [Google Scholar] [CrossRef] [PubMed]
- Reutovich, A.A.; Srivastava, A.K.; Arosio, P.; Bou-Abdallah, F. Ferritin nanocages as efficient nanocarriers and promising platforms for COVID-19 and other vaccines development. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130288. [Google Scholar] [CrossRef]
- Moon, H.; Lee, J.; Min, J.; Kang, S. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules 2014, 15, 3794–3801. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Lee, J.; Kim, H.; Heo, S.; Min, J.; Kang, S. Genetically engineering encapsulin protein cage nanoparticle as a SCC-7 cell targeting optical nanoprobe. Biomater. Res. 2014, 18, 21. [Google Scholar] [CrossRef]
- Choi, B.; Moon, H.; Hong, S.J.; Shin, C.; Do, Y.; Ryu, S.; Kang, S. Effective delivery of antigen-encapsulin nanoparticle fusions to dendritic cells leads to antigen-specific cytotoxic T cell activation and tumor rejection. ACS Nano 2016, 10, 7339–7350. [Google Scholar] [CrossRef]
- Van de Steen, A.; Khalife, R.; Colant, N.; Khan, H.M.; Deveikis, M.; Charalambous, S.; Robinson, C.M.; Dabas, R.; Esteban Serna, S.; Catana, D.A.; et al. Bioengineering bacterial encapsulin nanocompartments as targeted drug delivery system. Synth. Syst. Biotechnol. 2021, 6, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y.; Edwardson, T.G.W.; Hilvert, D. Tailoring lumazine synthase assemblies for bionanotechnology. Chem. Soc. Rev. 2018, 47, 3543–3557. [Google Scholar] [CrossRef]
- Kang, Y.-F.; Zhang, X.; Yu, X.-H.; Zheng, Q.; Liu, Z.; Li, J.-P.; Sun, C.; Kong, X.-W.; Zhu, Q.-Y.; Chen, H.-W.; et al. Immunization with a self-assembled nanoparticle vaccine elicits potent neutralizing antibody responses against EBV infection. Nano Lett. 2021, 21, 2476–2486. [Google Scholar] [CrossRef]
- Malonis, R.J.; Georgiev, G.I.; Haslwanter, D.; VanBlargan, L.A.; Fallon, G.; Vergnolle, O.; Cahill, S.M.; Harris, R.; Cowburn, D.; Chandran, K.; et al. A Powassan virus domain III nanoparticle immunogen elicits neutralizing and protective antibodies in mice. PLoS Pathog. 2022, 18, e1010573. [Google Scholar] [CrossRef]
- Ulbrich, K.; Hekmatara, T.; Herbert, E.; Kreuter, J. Transferrin- and transferrin-receptor-antibody-modified nanoparticles enable drug delivery across the blood-brain barrier (BBB). Eur. J. Pharm. Biopharm. 2009, 71, 251–256. [Google Scholar] [CrossRef]
- Ferris, D.P.; Lu, J.; Gothard, C.; Yanes, R.; Thomas, C.R.; Olsen, J.-C.; Stoddart, J.F.; Tamanoi, F.; Zink, J.I. Synthesis of biomolecule-modified mesoporous silica nanoparticles for targeted hydrophobic drug delivery to cancer cells. Small 2011, 7, 1816–1826. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, M.J.; Bravo, M.; Loureiro, J.A.; Lima, J.; Pereira, M.C. Transferrin-modified nanoparticles for targeted delivery of asiatic acid to glioblastoma cells. Life Sci. 2022, 296, 120435. [Google Scholar] [CrossRef] [PubMed]
- Golla, K.; Cherukuvada, B.; Ahmed, F.; Kondapi, A.K. Efficacy, safety and anticancer activity of protein nanoparticle-based delivery of doxorubicin through intravenous administration in rats. PLoS ONE 2012, 7, e51960. [Google Scholar] [CrossRef]
- Desoize, B.; Jardillier, J.C.; Kanoun, K.; Guerin, D.; Levy, M.C. In-vitro cytotoxic activity of cross-linked protein microcapsules. J. Pharm. Pharmacol. 1986, 38, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Willmott, N.; Anderson, J.; Florence, A.T. Comparison of albumin and casein microspheres as a carrier for doxorubicin. J. Pharm. Pharmacol. 1987, 39, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wei, J.; An, M.; Zhang, L.; Lin, S.; Shu, G.; Yuan, Z.; Lin, J.; Peng, G.; Liang, X.; et al. Casein nanoparticles as oral delivery carriers of mequindox for the improved bioavailability. Colloids Surf. B Biointerfaces 2020, 195, 111221. [Google Scholar] [CrossRef]
- Caivano, A.; Doria-Rose, N.A.; Buelow, B.; Sartorius, R.; Trovato, M.; D’Apice, L.; Domingo, G.J.; Sutton, W.F.; Haigwood, N.L.; De Berardinis, P. HIV-1 gag p17 presented as virus-like particles on the E2 scaffold from Geobacillus stearothermophilus induces sustained humoral and cellular immune responses in the absence of IFNγ production by CD4+ T cells. Virology 2010, 407, 296–305. [Google Scholar] [CrossRef]
- Jaworski, J.P.; Krebs, S.J.; Trovato, M.; Kovarik, D.N.; Brower, Z.; Sutton, W.F.; Waagmeester, G.; Sartorius, R.; D’Apice, L.; Caivano, A.; et al. Co-immunization with multimeric scaffolds and DNA rapidly induces potent autologous HIV-1 neutralizing antibodies and CD8+ T cells. PLoS ONE 2012, 7, e31464. [Google Scholar] [CrossRef]
- Molino, N.M.; Neek, M.; Tucker, J.A.; Nelson, E.L.; Wang, S.W. Viral-mimicking protein nanoparticle vaccine for eliciting anti-tumor responses. Biomaterials 2016, 86, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Melchers, M.; Matthews, K.; de Vries, R.P.; Eggink, D.; van Montfort, T.; Bontjer, I.; van de Sandt, C.; David, K.; Berkhout, B.; Moore, J.P.; et al. A stabilized HIV-1 envelope glycoprotein trimer fused to CD40 ligand targets and activates dendritic cells. Retrovirology 2011, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; van Montfort, T.; Melchers, M.; Isik, G.; Sanders, R.W. Immunosilencing a highly immunogenic protein trimerization domain. J. Biol. Chem. 2015, 290, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Hu, K.; Deng, X.; Guan, X.; Luo, S.; Tong, L.; Du, T.; Fu, M.; Zhang, M.; Liu, Y.; et al. Immunization with HSV-2 gB-CCL19 fusion constructs protects mice against lethal vaginal challenge. J. Immunol. 2015, 195, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Jones, G.B.E.; Thomas, P.V.; Chen, M.; Druz, A.; Joyce, M.G.; Kong, W.-P.; Sastry, M.; Soto, C.; Yang, Y.; Zhang, B.; et al. A cysteine zipper stabilizes a pre-fusion F glycoprotein vaccine for respiratory syncytial virus. PLoS ONE 2015, 10, e0128779. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, J.; Peña, D.G.; Barka, G.; Sciortino, G.; Maréchal, J.-D.; Vázquez López, M.; Vázquez, M.E. Directed self-assembly of trimeric DNA-binding chiral miniprotein helicates. Front. Chem. 2018, 6, 520. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Yanase, K.; Sato, T.; Unzai, S.; Hecht, M.H.; Arai, R. Self-assembling nano-architectures created from a protein nano-building block using an intermolecularly folded dimeric de novo protein. J. Am. Chem. Soc. 2015, 137, 11285–11293. [Google Scholar] [CrossRef]
- Kobayashi, N.; Arai, R. Protein cages and nanostructures constructed from protein nanobuilding blocks. Methods Mol. Biol. 2023, 2671, 79–94. [Google Scholar] [PubMed]
- Wang, L.; Prozorov, T.; Palo, P.E.; Liu, X.; Vaknin, D.; Prozorov, R.; Mallapragada, S.; Nilsen-Hamilton, M. Self-assembly and biphasic iron-binding characteristics of mms6, a bacterial protein that promotes the formation of superparamagnetic magnetite nanoparticles of uniform size and shape. Biomacromolecules 2012, 13, 98–105. [Google Scholar] [CrossRef]
- Kotelnikova, P.A.; Shipunova, V.O.; Aghayeva, U.F.; Stremovskiy, O.A.; Nikitin, M.P.; Novikov, I.A.; Schulga, A.A.; Deyev, S.M.; Petrov, R.V. Synthesis of magnetic nanoparticles stabilized by magnetite-binding protein for targeted delivery to cancer cells. Dokl. Biochem. Biophys. 2018, 481, 198–200. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Kotelnikova, P.A.; Aghayeva, U.F.; Stremovskiy, O.A.; Novikov, I.A.; Schulga, A.A.; Nikitin, M.P.; Deyev, S.M. Self-assembling nanoparticles biofunctionalized with magnetite-binding protein for the targeted delivery to HER2/neu overexpressing cancer cells. J. Magn. Magn. Mater. 2019, 469, 450–455. [Google Scholar] [CrossRef]
- Clem, A.S. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011, 3, 73–78. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Augusto, G.; Bachmann, M.F. The 3Ds in virus-like particle based-vaccines: “Design, Delivery and Dynamics”. Immunol. Rev. 2020, 296, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Ochsenbein, A.F.; Fehr, T.; Lutz, C.; Suter, M.; Brombacher, F.; Hengartner, H.; Zinkernagel, R.M. Control of early viral and bacterial distribution and disease by natural antibodies. Science 1999, 286, 2156–2159. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Dintzis, R.Z.; Dintzis, H.M. Specific cellular stimulation in the primary immune response: A quantized model. Proc. Natl. Acad. Sci. USA 1982, 79, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Jobsri, J.; Allen, A.; Rajagopal, D.; Shipton, M.; Kanyuka, K.; Lomonossoff, G.P.; Ottensmeier, C.; Diebold, S.S.; Stevenson, F.K.; Savelyeva, N. Plant virus particles carrying tumour antigen activate TLR7 and induce high levels of protective antibody. PLoS ONE 2015, 10, e0118096. [Google Scholar] [CrossRef] [PubMed]
- Terhuja, M.; Saravanan, P.; Tamilselvan, R.P. Comparative efficacy of virus-like particle (VLP) vaccine of foot-and-mouth disease virus (FMDV) type O adjuvanted with poly I:C or CpG in guinea pigs. Biologicals 2015, 43, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Gomes, A.C.; Vogel, M.; Bachmann, M.F. Interaction of viral capsid-derived virus-like particles (VLPs) with the innate immune system. Vaccines 2018, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chi, Y.; Bao, J.; Zhao, X.; Zhang, J.; Wang, L. Virus-like Particles for TEM Regulation and Antitumor Therapy. J. Funct. Biomater. 2022, 13, 304. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Roesti, E.S.; El-Turabi, A.; Bachmann, M.F. Type of RNA packed in VLPs impacts IgG class switching—Implications for an influenza vaccine design. Vaccines 2019, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Krueger, C.C.; Thoms, F.; Keller, E.; Leoratti, F.M.S.; Vogel, M.; Bachmann, M.F. RNA and toll-like receptor 7 license the generation of superior secondary plasma cells at multiple levels in a B cell intrinsic fashion. Front. Immunol. 2019, 10, 736. [Google Scholar] [CrossRef] [PubMed]
- Markine-Goriaynoff, D.; Coutelier, J.P. Increased efficacy of the immunoglobulin G2a subclass in antibody-mediated protection against lactate dehydrogenase-elevating virus-induced polioencephalomyelitis revealed with switch mutants. J. Virol. 2002, 76, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.S.; Ducry, J.; Oxenius, A. Dissecting the contribution of IgG subclasses in restricting airway infection with Legionella pneumophila. J. Immunol. 2014, 193, 4053–4059. [Google Scholar] [CrossRef]
- Novák, J.; Panská, L.; Macháček, T.; Kolářová, L.; Horák, P. Humoral response of mice infected with Toxocara canis following different infection schemes. Acta Parasitol. 2017, 62, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.R.; Crill, W.D.; Davis, B.S.; Chang, G.-J.J. A West Nile virus CD4 T cell epitope improves the immunogenicity of Dengue virus serotype 2 vaccines. Virology 2012, 424, 129–137. [Google Scholar] [CrossRef]
- Del Guercio, M.F.; Alexander, J.; Kubo, R.T.; Arrhenius, T.; Maewal, A.; Appella, E.; Hoffman, S.L.; Jones, T.; Valmori, D.; Sakaguchi, K.; et al. Potent immunogenic short linear peptide constructs composed of B cell epitopes and pan DR T helper epitopes (PADRE) for antibody responses in vivo. Vaccine 1997, 15, 441–448. [Google Scholar] [CrossRef]
- Ressing, M.E.; van Driel, W.J.; Brandt, R.M.; Kenter, G.G.; de Jong, J.H.; Bauknecht, T.; Fleuren, G.J.; Hoogerhout, P.; Offringa, R.; Sette, A.; et al. of T helper responses, but not of human papillomavirus-specific cytotoxic T lymphocyte responses, after peptide vaccination of patients with cervical carcinoma. J. Immunother. 2000, 23, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Zhang, W.; Li, J.; Wan, Y.; Wang, Z.; Duan, R.; Yu, P.; Zhao, N.; Zhang, K.; Wang, S.; et al. A single codon optimization enhances recombinant human TNF-α vaccine expression in Escherichia coli. BioMed. Res. Int. 2018, 2018, 3025169. [Google Scholar] [CrossRef] [PubMed]
- Micoli, F.; Adamo, R.; Costantino, P. Protein carriers for glycoconjugate vaccines: History, selection criteria, characterization and new trends. Molecules 2018, 23, 1451. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kelly, S.H.; Sanchez-Perez, L.; Sampson, J.H.; Collier, J.H. Comparative study of α-helical and β-sheet self-assembled peptide nanofiber vaccine platforms: Influence of integrated T-cell epitopes. Biomater. Sci. 2020, 8, 3522–3535. [Google Scholar] [CrossRef] [PubMed]
- Panina-Bordignon, P.; Tan, A.; Termijtelen, A.; Demotz, S.; Corradin, G.; Lanzavecchia, A. Universally immunogenic T cell epitopes: Promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 1989, 19, 2237–2242. [Google Scholar] [CrossRef] [PubMed]
- Demotz, S.; Lanzavecchia, A.; Eisel, U.; Niemann, H.; Widmann, C.; Corradin, G. Delineation of several DR-restricted tetanus toxin T cell epitopes. J. Immunol. 1989, 142, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Chianese-Bullock, K.A.; Irvin, W.P., Jr.; Petroni, G.R.; Murphy, C.; Smolkin, M.; Olson, W.C.; Coleman, E.; Boerner, S.A.; Nail, C.J.; Neese, P.Y.; et al. A multipeptide vaccine is safe and elicits T-cell responses in participants with advanced stage ovarian cancer. J. Immunother. 2008, 31, 420–430. [Google Scholar] [CrossRef]
- Rechtsteiner, G.; Warger, T.; Osterloh, P.; Schild, H.; Radsak, M.P. Cutting edge: Priming of CTL by transcutaneous peptide immunization with imiquimod. J. Immunol. 2005, 174, 2476–2480. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Sidney, J.; Southwood, S.; Ruppert, J.; Oseroff, C.; Maewal, A.; Snoke, K.; Serra, H.M.; Kubo, R.T.; Sette, A.; et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity 1994, 1, 751–761. [Google Scholar] [CrossRef]
- Alexander, J.; Fikes, J.; Hoffman, S.; Franke, E.; Sacci, J.; Appella, E.; Chisari, F.V.; Guidotti, L.G.; Chesnut, R.W.; Livingston, B.; et al. The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunol. Res. 1998, 18, 79–92. [Google Scholar] [CrossRef]
- Ochoa, R.; Lunardelli, V.A.S.; Rosa, D.S.; Laio, A.; Cossio, P. Multiple-Allele MHC Class II Epitope Engineering by a Molecular Dynamics-Based Evolution Protocol. Front. Immunol. 2022, 13, 862851. [Google Scholar] [CrossRef]
- Wu, C.Y.; Monie, A.; Pang, X.; Hung, C.F.; Wu, T.C. Improving therapeutic HPV peptide-based vaccine potency by enhancing CD4+ T help and dendritic cell activation. J. Biomed. Sci. 2010, 17, 88. [Google Scholar] [CrossRef]
- Li, S.; Peng, L.; Zhao, W.; Zhong, H.; Zhang, F.; Yan, Z.; Cao, H. Synthetic peptides containing B- and T-cell epitope of dengue virus-2 E domain III provoked B- and T-cell responses. Vaccine 2011, 29, 3695–3702. [Google Scholar] [CrossRef]
- Wang, H.; Su, X.; Zhang, P.; Liang, J.; Wei, H.; Wan, M.; Wu, X.; Yu, Y.; Wang, L. Recombinant heat shock protein 65 carrying PADRE and HBV epitopes activates dendritic cells and elicits HBV-specific CTL responses. Vaccine 2011, 29, 2328–2335. [Google Scholar] [CrossRef]
- La Rosa, C.; Longmate, J.; Lacey, S.F.; Kaltcheva, T.; Sharan, R.; Marsano, D.; Kwon, P.; Drake, J.; Williams, B.; Denison, S.; et al. Clinical evaluation of safety and immunogenicity of PADRE-cytomegalovirus (CMV) and tetanus-CMV fusion peptide vaccines with or without PF03512676 adjuvant. J. Infect. Dis. 2012, 205, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.-d.; Xue, X.-c.; Gao, M.-l.; Wang, X.-f.; Shu, Z.; Mu, N.; Gao, Y.; Wang, Z.-l.; Hao, Q.; Li, W.-n.; et al. Therapeutic effects of PADRE-BAFF autovaccine on rat adjuvant arthritis. BioMed. Res. Int. 2014, 2014, 854954. [Google Scholar] [CrossRef] [PubMed]
- El Bissati, K.; Chentoufi, A.A.; Krishack, P.A.; Zhou, Y.; Woods, S.; Dubey, J.P.; Vang, L.; Lykins, J.; Broderick, K.E.; Mui, E.; et al. Adjuvanted multi-epitope vaccines protect HLA-A*11, 01 transgenic mice against Toxoplasma gondii. JCI Insight 2016, 1, e85955. [Google Scholar] [CrossRef] [PubMed]
- Snook, A.E.; Baybutt, T.R.; Hyslop, T.; Waldman, S.A. Preclinical evaluation of a replication-deficient recombinant adenovirus serotype 5 vaccine expressing guanylate cyclase C and the PADRE T-helper epitope. Hum. Gene Ther. Methods 2016, 27, 238–250. [Google Scholar] [CrossRef]
- El Bissati, K.; Zhou, Y.; Paulillo, S.M.; Raman, S.K.; Karch, C.P.; Roberts, C.W.; Lanar, D.E.; Reed, S.; Fox, C.; Carter, D.; et al. Protein nanovaccine confers robust immunity against toxoplasma. NPJ Vaccines 2017, 2, 24. [Google Scholar] [CrossRef]
- Safavi, A.; Kefayat, A.; Mahdevar, E.; Abiri, A.; Ghahremani, F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine 2020, 38, 7612–7628. [Google Scholar] [CrossRef]
- Alexander, J.; del Guercio, M.F.; Maewal, A.; Qiao, L.; Fikes, J.; Chesnut, R.W.; Paulson, J.; Bundle, D.R.; DeFrees, S.; Sette, A. Linear PADRE T helper epitope and carbohydrate B cell epitope conjugates induce specific high titer IgG antibody responses. J. Immunol. 2000, 164, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; del Guercio, M.F.; Frame, B.; Maewal, A.; Sette, A.; Nahm, M.H.; Newman, M.J. Development of experimental carbohydrate-conjugate vaccines composed of Streptococcus pneumoniae capsular polysaccharides and the universal helper T-lymphocyte epitope (PADRE). Vaccine 2004, 22, 2362–2367. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.F.; Tsai, Y.C.; He, L.; Wu, T.C. DNA vaccines encoding Ii-PADRE generates potent PADRE-specific CD4+ T-cell immune responses and enhances vaccine potency. Mol. Ther. 2007, 15, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari-Nazari, H.; Tavakkol-Afshari, J.; Jaafari, M.R.; Tahaghoghi-Hajghorbani, S.; Masoumi, E.; Jalali, S.A. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PLoS ONE 2015, 10, e0142563. [Google Scholar] [CrossRef] [PubMed]
- Zamani, P.; Teymouri, M.; Nikpoor, A.R.; Gholizadeh Navashenaq, J.; Gholizadeh, Z.; Amiri Darban, S.; Jaafari, M.R. Nanoliposomal vaccine containing long multi-epitope peptide E75-AE36 pulsed PADRE-induced effective immune response in mice TuBo model of breast cancer. Eur. J. Cancer 2020, 129, 80–96. [Google Scholar] [CrossRef]
- Pompano, R.R.; Chen, J.; Verbus, E.A.; Han, H.; Fridman, A.; McNeely, T.; Collier, J.H.; Chong, A.S. Titrating T-cell epitopes within self-assembled vaccines optimizes CD4+ helper T cell and antibody outputs. Adv. Health Mater. 2014, 3, 1898–1908. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Norberg, P.K.; Reap, E.A.; Congdon, K.L.; Fries, C.N.; Kelly, S.H.; Sampson, J.H.; Conticello, V.P.; Collier, J.H. A Supramolecular Vaccine Platform Based on α-Helical Peptide Nanofibers. ACS Biomater. Sci. Eng. 2017, 3, 3128–3132. [Google Scholar] [CrossRef] [PubMed]
- Shores, L.S.; Kelly, S.H.; Hainline, K.M.; Suwanpradid, J.; MacLeod, A.S.; Collier, J.H. Multifactorial design of a supramolecular peptide anti-IL-17 vaccine toward the treatment of psoriasis. Front. Immunol. 2020, 11, 1855. [Google Scholar] [CrossRef] [PubMed]
- Krenger, P.S.; Josi, R.; Sobczak, J.; Velazquez, T.L.C.; Balke, I.; Skinner, M.A.; Kramer, M.F.; Scott, C.J.W.; Hewings, S.; Heath, M.D. Influence of antigen density and TLR ligands on preclinical efficacy of a VLP-based vaccine against peanut allergy. Allergy 2024, 79, 184–199. [Google Scholar] [CrossRef]
- Kozlovska, T.M.; Cielens, I.; Dreilina, D.; Dislers, A.; Baumanis, V.; Ose, V.; Pumpens, P. Recombinant RNA phage Q beta capsid particles synthesized and self-assembled in Escherichia coli. Gene 1993, 137, 133–137. [Google Scholar] [CrossRef]
- Fettelschoss-Gabriel, A.; Fettelschoss, V.; Olomski, F.; Birkmann, K.; Thoms, F.; Buhler, M.; Kummer, M.; Zeltins, A.; Kundig, T.M.; Bachmann, M.F. Active vaccination against interleukin-5 as long-term treatment for insect-bite hypersensitivity in horses. Allergy 2019, 74, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Liu, X.; Mohsen, M.O.; Zeltins, A.; Martina, B.; Vogel, M.; Bachmann, M.F. Induction of Broadly Cross-Reactive Antibodies by Displaying Receptor Binding Domains of SARS-CoV-2 on Virus-like Particles. Vaccines 2022, 10, 307. [Google Scholar] [CrossRef]
- Rothen, D.A.; Krenger, P.S.; Nonic, A.; Balke, I.; Vogt, A.C.S.; Chang, X.; Manenti, A.; Vedovi, F.; Resevica, G.; Walton, S.M.; et al. Intranasal administration of a virus-like particles-based vaccine induces neutralizing antibodies against SARS-CoV-2 and variants of concern. Allergy 2022, 77, 2446–2458. [Google Scholar] [CrossRef]
- Varanda, C.M.R.; Machado, M.; Martel, P.; Nolasco, G.; Clara, M.I.E.; Félix, M.R. Genetic diversity of the coat protein of olive mild mosaic virus (OMMV) and tobacco necrosis virus D (TNV-D) isolates and its structural implications. PLoS ONE 2014, 9, e110941. [Google Scholar] [CrossRef]
- Smith, T.J.; Chase, E.; Schmidt, T.; Perry, K.L. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J. Virol. 2000, 74, 7578–7586. [Google Scholar] [CrossRef]
- Percival-Alwyn, J.L.; England, E.; Kemp, B.; Rapley, L.; Davis, N.H.E.; McCarthy, G.R.; Majithiya, J.B.; Corkill, D.J.; Welsted, S.; Minton, K.; et al. Generation of potent mouse monoclonal antibodies to self-proteins using T-cell epitope “tags”. MAbs 2015, 7, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Skelton, D.; Satake, N.; Kohn, D.B. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther. 2001, 8, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.J.; Osterrieder, N.; Metzger, S.M.; Buckles, E.; Doody, A.M.; DeLisa, M.P.; Putnam, D. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. USA 2010, 107, 3099–3104. [Google Scholar] [CrossRef]
- Mairuhu, A.T.; Wagenaar, J.; Brandjes, D.P.; van Gorp, E.C. Dengue: An arthropod-borne disease of global importance. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 425–433. [Google Scholar] [CrossRef]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martínez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8 (Suppl. S12), S7–S16. [Google Scholar] [CrossRef]
- Khetarpal, N.; Khanna, I. Dengue fever: Causes, complications, and vaccine strategies. J. Immunol. Res. 2016, 2016, 6803098. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Hermida, L.; Bernardo, L.; Ramirez, R.; Guillén, G. Domain III of the envelope protein as a dengue vaccine target. Expert Rev. Vaccines 2010, 9, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Fahimi, H.; Mohammadipour, M.; Haddad Kashani, H.; Parvini, F.; Sadeghizadeh, M. Dengue viruses and promising envelope protein domain III-based vaccines. Appl. Microbiol. Biotechnol. 2018, 102, 2977–2996. [Google Scholar] [CrossRef] [PubMed]
- Seesen, M.; Jearanaiwitayakul, T.; Limthongkul, J.; Midoeng, P.; Sunintaboon, P.; Ubol, S. A bivalent form of nanoparticle-based dengue vaccine stimulated responses that potently eliminate both DENV-2 particles and DENV-2-infected cells. Vaccine 2023, 41, 1638–1648. [Google Scholar] [CrossRef]
- Gromowski, G.D.; Barrett, A.D. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 2007, 366, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Sukupolvi-Petty, S.; Austin, S.K.; Purtha, W.E.; Oliphant, T.; Nybakken, G.E.; Schlesinger, J.J.; Roehrig, J.T.; Gromowski, G.D.; Barrett, A.D.; Fremont, D.H.; et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 2007, 81, 12816–12826. [Google Scholar] [CrossRef]
- Gromowski, G.D.; Barrett, N.D.; Barrett, A.D.T. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J. Virol. 2008, 82, 8828–8837. [Google Scholar] [CrossRef]
- Chen, H.W.; Liu, S.J.; Li, Y.S.; Liu, H.H.; Tsai, J.P.; Chiang, C.Y.; Chen, M.Y.; Hwang, C.S.; Huang, C.C.; Hu, H.M.; et al. A consensus envelope protein domain III can induce neutralizing antibody responses against serotype 2 of dengue virus in non-human primates. Arch. Virol. 2013, 158, 1523–1531. [Google Scholar] [CrossRef]
- Rajpoot, R.K.; Shukla, R.; Arora, U.; Swaminathan, S.; Khanna, N. Dengue envelope-based ‘four-in-one’ virus-like particles produced using Pichia pastoris induce enhancement-lacking, domain III-directed tetravalent neutralising antibodies in mice. Sci. Rep. 2018, 8, 8643. [Google Scholar] [CrossRef]
- Shukla, R.; Rajpoot, R.K.; Arora, U.; Poddar, A.; Swaminathan, S.; Khanna, N. Pichia pastoris-Expressed Bivalent Virus-Like Particulate Vaccine Induces Domain III-Focused Bivalent Neutralizing Antibodies without Antibody-Dependent Enhancement in Vivo. Front. Microbiol. 2018, 8, 2644. [Google Scholar] [CrossRef]
- Ramasamy, V.; Arora, U.; Shukla, R.; Poddar, A.; Shanmugam, R.K.; White, L.J.; Mattocks, M.M.; Raut, R.; Perween, A.; Tyagi, P.; et al. A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice. PLoS Negl. Trop. Dis. 2018, 12, e0006191. [Google Scholar] [CrossRef]
- Huang, H.-J.; Yang, M.; Chen, H.-W.; Wang, S.; Chang, C.-P.; Ho, T.-S.; Kao, Y.-S.; Tien, S.-M.; Lin, H.-H.; Chang, P.-C.; et al. A novel chimeric dengue vaccine candidate composed of consensus envelope protein domain III fused to C-terminal-modified NS1 protein. Vaccine 2022, 40, 2299–2310. [Google Scholar] [CrossRef]
- Spohn, G.; Jennings, G.T.; Martina, B.E.; Keller, I.; Beck, M.; Pumpens, P.; Osterhaus, A.D.M.E.; Bachmann, M.F. A VLP-based vaccine targeting domain III of the West Nile virus E protein protects from lethal infection in mice. Virol. J. 2010, 7, 146. [Google Scholar] [CrossRef]
- Maeda, Y.; Ueda, H.; Kazami, J.; Kawano, G.; Suzuki, E.; Nagamune, T. Engineering of functional chimeric protein G–Vargula luciferase. Anal. Biochem. 1997, 249, 147–152. [Google Scholar] [CrossRef]
- Kalnciema, I.; Balke, I.; Skrastina, D.; Ose, V.; Zeltins, A. Potato virus M-like nanoparticles: Construction and characterization. Mol. Biotechnol. 2015, 57, 982–992. [Google Scholar] [CrossRef]
- Qi, X.; Sun, Y.; Xiong, S. A single freeze-thawing cycle for highly efficient solubilization of inclusion body proteins and its refolding into bioactive form. Microb. Cell Fact. 2015, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Linke, T.; Aspelund, M.T.; Thompson, C.; Xi, G.; Fulton, A.; Wendeler, M.; Pabst, T.M.; Wang, X.; Wang, W.K.; Ram, K.; et al. Development and scale-up of a commercial fed batch refolding process for an anti-CD22 two chain immunotoxin. Biotechnol. Prog. 2014, 30, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.R.; Stanbro, H.; Merril, C.R. Endotoxins in commercial vaccines. Appl. Environ. Microbiol. 1978, 36, 445–449. [Google Scholar] [CrossRef]
- Malyala, P.; Singh, M. Endotoxin limits in formulations for preclinical research. J. Pharm. Sci. 2008, 97, 2041–2044. [Google Scholar] [CrossRef]
- Brito, L.A.; Singh, M. Acceptable levels of endotoxin in vaccine formulations during preclinical research. J. Pharm. Sci. 2011, 100, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Nair, S.; Savithri, H.S. Natively unfolded nucleic acid binding P8 domain of SeMV polyprotein 2a affects the novel ATPase activity of the preceding P10 domain. FEBS Lett. 2010, 584, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Hønsvall, B.K.; Robertson, L.J. Washed Away; How Not to Lose Your RNA during Isolation. J. Biomol. Tech. 2017, 28, 75–79. [Google Scholar] [CrossRef]
- Braun, M.; Jandus, C.; Maurer, P.; Hammann-Haenni, A.; Schwarz, K.; Bachmann, M.F.; Speiser, D.E.; Romero, P. Virus-like particles induce robust human T-helper cell responses. Eur. J. Immunol. 2012, 42, 330–340. [Google Scholar] [CrossRef]
- Farlow, M.R.; Andreasen, N.; Riviere, M.E.; Vostiar, I.; Vitaliti, A.; Sovago, J.; Caputo, A.; Winblad, B.; Graf, A. Long-term treatment with active Aβ immunotherapy with CAD106 in mild Alzheimer’s disease. Alzheimers Res. Ther. 2015, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Vandenberghe, R.; Riviere, M.E.; Caputo, A.; Sovago, J.; Maguire, R.P.; Farlow, M.; Marotta, G.; Sanchez-Valle, R.; Scheltens, P.; Ryan, J.M.; et al. Active Aβ immunotherapy CAD106 in Alzheimer’s disease: A phase 2b study. Alzheimers Dement. 2017, 3, 10–22. [Google Scholar] [CrossRef]
- Classen, D.C.; Morningstar, J.M.; Shanley, J.D. Detection of antibody to murine cytomegalovirus by enzyme-linked immunosorbent and indirect immunofluorescence assays. J. Clin. Microbiol. 1987, 25, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Lardeux, F.; Torrico, G.; Aliaga, C. Calculation of the ELISA’s cut-off based on the change-point analysis method for detection of Trypanosoma cruzi infection in Bolivian dogs in the absence of controls. Mem. Inst. Oswaldo Cruz. 2016, 111, 501–504. [Google Scholar] [CrossRef]
- Fick de Souza, V.A.U.; Fernandes, S.; Araújo, E.S.; Tateno, A.F.; Oliveira, O.M.N.P.F.; Oliveira, R.d.R.; Pannuti, C.S. Use of an Immunoglobulin G Avidity Test to Discriminate between Primary and Secondary Dengue Virus Infections. J. Clin. Microbiol. 2004, 42, 1782–1784. [Google Scholar] [CrossRef]
- Olsson, J.; Johansson, J.; Honkala, E.; Blomqvist, B.; Kok, E.; Weidung, B.; Lövheim, H.; Elgh, F. Urea dilution of serum for reproducible anti-HSV1 IgG avidity index. BMC Infect. Dis. 2019, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Correa, V.A.; Rodrigues, T.S.; Portilho, A.I.; Lima, G.T.; De Gaspari, E. Modified ELISA for antibody avidity evaluation: The need for standardization. Biomed. J. 2021, 44, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Nurmi, V.; Hedman, L.; Perdomo, M.F.; Weseslindtner, L.; Hedman, K. Comparison of approaches for IgG avidity calculation and a new highly sensitive and specific method with broad dynamic range. Int. J. Infect. Dis. 2021, 110, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Balke, I.; Silamikelis, I.; Radovica-Spalvina, I.; Zeltina, V.; Resevica, G.; Fridmanis, D.; Zeltins, A. Ryegrass mottle virus complete genome determination and development of infectious cDNA by combining two methods-3′ RACE and RNA-Seq. PLoS ONE 2023, 18, e0287278. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Medina, T.; Kirkwood, J.M.; Zakharia, Y.; Gonzalez, R.; Davar, D.; Chmielowski, B.; Campbell, K.M.; Bao, R.; Kelley, H.; et al. Overcoming PD-1 Blockade Resistance with CpG-A Toll-Like Receptor 9 Agonist Vidutolimod in Patients with Metastatic Melanoma. Cancer Discov. 2021, 11, 2998–3007. [Google Scholar] [CrossRef] [PubMed]
- Mellid-Carballal, R.; Gutierrez-Gutierrez, S.; Rivas, C.; Garcia-Fuentes, M. Viral protein-based nanoparticles (part 2): Pharmaceutical applications. Eur. J. Pharm. Sci. 2023, 189, 106558. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K. Construction of spherical virus-inspired peptide nanoassemblies. Polym. J. 2012, 44, 469–474. [Google Scholar] [CrossRef]
- Janković, P.; Šantek, I.; Pina, A.S.; Kalafatovic, D. Exploiting peptide self-assembly for the development of minimalistic viral mimetics. Front. Chem. 2021, 9, 723473. [Google Scholar] [CrossRef]
- Rötzschke, O.; Falk, K.; Strominger, J.L. Superactivation of an immune response triggered by oligomerized T cell epitopes. Proc. Natl. Acad. Sci. USA 1997, 94, 14642–14647. [Google Scholar] [CrossRef]
- Fossum, E.; Grødeland, G.; Terhorst, D.; Tveita, A.A.; Vikse, E.; Mjaaland, S.; Henri, S.; Malissen, B.; Bogen, B. Vaccine molecules targeting XCR1 on cross-presenting DCs induce protective CD8+ T-cell responses against influenza virus. Eur. J. Immunol. 2015, 45, 624–635. [Google Scholar] [CrossRef]
- Grødeland, G.; Fossum, E.; Bogen, B. Polarizing T and B cell responses by APC-targeted subunit vaccines. Front. Immunol. 2015, 6, 367. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Klein, F.; Pietzsch, J.; Seaman, M.S.; Nussenzweig, M.C.; Ravetch, J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 2014, 158, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Visciano, M.L.; Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Effects of adjuvants on IgG subclasses elicited by virus-like particles. J. Transl. Med. 2012, 10, 4. [Google Scholar] [CrossRef]
- Samal, S.; Shrivastava, T.; Sonkusre, P.; Rizvi, Z.A.; Kumar, R.; Ahmed, S.; Vishwakarma, P.; Yadav, N.; Bansal, M.; Chauhan, K.; et al. Tetramerizing TGCN4 domain facilitates production of influenza A H1N1 M2e higher order soluble oligomers that show enhanced immunogenicity in vivo. J. Biol. Chem. 2020, 295, 14352–14366. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Heilman, D.; Liu, F.; Giehl, T.; Joshi, S.; Huang, X.; Chou, T.; Goguen, J.; Lu, S. A DNA vaccine producing LcrV antigen in oligomers is effective in protecting mice from lethal mucosal challenge of plague. Vaccine 2004, 22, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Dalgediene, I.; Lasickiene, R.; Budvytyte, R.; Valincius, G.; Morkuniene, R.; Borutaite, V.; Zvirbliene, A. Immunogenic properties of amyloid beta oligomers. J. Biomed. Sci. 2013, 20, 10. [Google Scholar] [CrossRef]
- Perlmutter, R.M.; Hansburg, D.; Briles, D.E.; Nicolotti, R.A.; Davie, J.M. Subclass restriction of murine anti-carbohydrate antibodies. J. Immunol. 1978, 121, 566–572. [Google Scholar] [CrossRef]
- Slack, J.; Der-Balian, G.P.; Nahm, M.; Davie, J.M. Subclass restriction of murine antibodies. II. The IgG plaque-forming cell response to thymus-independent type 1 and type 2 antigens in normal mice and mice expressing an X-linked immunodeficiency. J. Exp. Med. 1980, 151, 853–862. [Google Scholar] [CrossRef]
- Coutelier, J.P.; van der Logt, J.T.; Heessen, F.W.; Warnier, G.; Van Snick, J. IgG2a restriction of murine antibodies elicited by viral infections. J. Exp. Med. 1987, 165, 64–69. [Google Scholar] [CrossRef]
- Coutelier, J.P.; van der Logt, J.T.; Heessen, F.W. IgG subclass distribution of primary and secondary immune responses concomitant with viral infection. J. Immunol. 1991, 147, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004, 427, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, R.J.; Zhang, W.; Rossmann, M.G.; Pletnev, S.V.; Corver, J.; Lenches, E.; Jones, C.T.; Mukhopadhyay, S.; Chipman, P.R.; Strauss, E.G.; et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell 2002, 108, 717–725. [Google Scholar] [CrossRef]
- Matsuura, K.; Shiomi, Y.; Mizuta, T.; Inaba, H. Horseradish peroxidase-decorated artificial viral capsid constructed from beta-annulus peptide via interaction between his-tag and ni-nta. Processes 2020, 8, 1455. [Google Scholar] [CrossRef]
- Chacón-Díaz, C.; Muñoz-Rodríguez, M.; Barquero-Calvo, E.; Guzmán-Verri, C.; Chaves-Olarte, E.; Grilló, M.J.; Moreno, E. The use of green fluorescent protein as a marker for brucella vaccines. Vaccine 2011, 29, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Zinkernagel, R.M. The influence of virus structure on antibody responses and virus serotype formation. Immunol. Today 1996, 17, 553–558. [Google Scholar] [CrossRef]
- Poteet, E.; Lewis, P.; Chen, C.; Ho, S.O.; Do, T.; Chiang, S.; Labranche, C.; Montefiori, D.; Fujii, G.; Yao, Q. Toll-like receptor 3 adjuvant in combination with virus-like particles elicit a humoral response against HIV. Vaccine 2016, 34, 5886–5894. [Google Scholar] [CrossRef]
- Gomes, A.C.; Mohsen, M.O.; Mueller, J.E.; Leoratti, F.M.S.; Cabral-Miranda, G.; Bachmann, M.F. Early transcriptional signature in dendritic cells and the induction of protective T cell responses upon immunization with VLPs containing TLR ligands-a role for CCL2. Front. Immunol. 2019, 10, 1679. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, S.P.; Kozlowski, P.A.; Nakaya, H.I.; Burger, M.C.; Russo, P.; Pham, M.; Kovalenkov, Y.; Silveira, E.L.V.; Havenar-Daughton, C.; Burton, S.L.; et al. Adjuvanting a simian immunodeficiency virus vaccine with toll-like receptor ligands encapsulated in nanoparticles induces persistent antibody responses and enhanced protection in TRIM5α restrictive macaques. J. Virol. 2017, 91, e01844-16. [Google Scholar] [CrossRef]
- Mitragotri, S.; Anderson, D.G.; Chen, X.; Chow, E.K.; Ho, D.; Kabanov, A.V.; Karp, J.M.; Kataoka, K.; Mirkin, C.A.; Petrosko, S.H.; et al. Accelerating the translation of nanomaterials in biomedicine. ACS Nano 2015, 9, 6644–6654. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Hasegawa, T.; Hinata, H.; Ito, F.; Inagawa, H.; Kochi, C.; Soma, G.-I.; Makino, K.; Terada, H. Optimum conditions for efficient phagocytosis of rifampicin-loaded PLGA microspheres by alveolar macrophages. J. Control. Release 2007, 119, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Lico, C.; Santi, L.; Twyman, R.M.; Pezzotti, M.; Avesani, L. The use of plants for the production of therapeutic human peptides. Plant Cell Rep. 2012, 31, 439–451. [Google Scholar] [PubMed]
- Plotkin, S.A. Immunologic correlates of protection induced by vaccination. Pediatr. Infect. Dis. J. 2001, 20, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Vaccines: Correlates of vaccine-induced immunity. Clin. Infect. Dis. 2008, 47, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Abbad, A.; Tcheou, J.; Mendu, D.R.; Firpo-Betancourt, A.; Gleason, C.; Srivastava, K.; Cordon-Cardo, C.; Simon, V.; Krammer, F.; et al. Binding and Avidity Signatures of Polyclonal Sera from Individuals with Different Exposure Histories to Severe Acute Respiratory Syndrome Coronavirus 2 Infection, Vaccination, and Omicron Breakthrough Infections. J. Infect. Dis. 2023, 228, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mu, H.; Pan, X.; Guo, W. Studying the effects of booster shots and antibody responses to the SARS-CoV-2 vaccination over time in health personnel. Front. Cell. Infect. Microbiol. 2023, 13, 1138631. [Google Scholar] [CrossRef] [PubMed]
- Puschnik, A.; Lau, L.; Cromwell, E.A.; Balmaseda, A.; Zompi, S.; Harris, E. Correlation between dengue-specific neutralizing antibodies and serum avidity in primary and secondary dengue virus 3 natural infections in humans. PLoS Negl. Trop. Dis. 2013, 7, e2274. [Google Scholar] [CrossRef]
- Saron, W.A.A.; Rathore, A.P.S.; Ting, L.; Ooi, E.E.; Low, J.; Abraham, S.N.; St John, A.L. Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci. Adv. 2018, 4, eaar4297. [Google Scholar] [CrossRef]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction between Virus-like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Armero-Gimenez, J.; Wilbers, R.; Schots, A.; Williams, C.; Finnern, R. Rapid screening and scaled manufacture of immunogenic virus-like particles in a tobacco BY-2 cell-free protein synthesis system. Front. Immunol. 2023, 14, 1088852. [Google Scholar] [CrossRef] [PubMed]
- Zupančič, E.; Curato, C.; Paisana, M.; Rodrigues, C.; Porat, Z.; Viana, A.S.; Afonso, C.A.M.; Pinto, J.; Gaspar, R.; Moreira, J.N.; et al. Rational design of nanoparticles towards targeting antigen-presenting cells and improved T cell priming. J. Control. Release 2017, 258, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Liu, J.; Yang, J.; Hu, Z.; Rao, X. Dengue virus-like particles: Construction and application. Appl. Microbiol. Biotechnol. 2012, 94, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, C.; Jiang, S.; Du, L. Recent Advances in the Development of Virus-Like Particle-Based Flavivirus Vaccines. Vaccines 2020, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Castilho, L.R.; Mattos, N.R.; Abreu, W.S.; Gutarra, M.L.E. Virus-like Particles (VLPs) as Important Tools for Flavivirus Vaccine Development. Biologics 2022, 2, 226–242. [Google Scholar] [CrossRef]
- Khetarpal, N.; Poddar, A.; Nemani, S.K.; Dhar, N.; Patil, A.; Negi, P.; Perween, A.; Viswanathan, R.; Lünsdorf, H.; Tyagi, P.; et al. Dengue-specific subviral nanoparticles: Design, creation and characterization. J. Nanobiotechnol. 2013, 11, 15–241. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Hefferon, K. Application of plant viruses in biotechnology, medicine, and human health. Viruses 2021, 13, 1697. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S.; Peabody, J.; Bradfute, S.B.; Chackerian, B. RNA phage VLP-based vaccine platforms. Pharmaceuticals 2021, 14, 764. [Google Scholar] [CrossRef]
- Warner, N.L.; Frietze, K.M. Development of Bacteriophage Virus-Like Particle Vaccines Displaying Conserved Epitopes of Dengue Virus Non-Structural Protein 1. Vaccines 2021, 9, 726. [Google Scholar] [CrossRef]
- Didierlaurent, A.M.; Morel, S.; Lockman, L.; Giannini, S.L.; Bisteau, M.; Carlsen, H.; Kielland, A.; Vosters, O.; Vanderheyde, N.; Schiavetti, F.; et al. AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J. Immunol. 2009, 183, 6186–6197. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.H.; Cha, S.B.; Lee, S.-H.; Bae, H.S.; Ham, C.S.; Lee, M.-G.; Kim, D.-H.; Han, S.H. A novel defined TLR3 agonist as an effective vaccine adjuvant. Front. Immunol. 2023, 14, 1075291. [Google Scholar] [CrossRef] [PubMed]
- Bessa, J.; Jegerlehner, A.; Hinton, H.J.; Pumpens, P.; Saudan, P.; Schneider, P.; Bachmann, M.F. Alveolar macrophages and lung dendritic cells sense RNA and drive mucosal IgA responses. J. Immunol. 2009, 183, 3788–3799. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Krenger, P.; Krueger, C.C.; Zha, L.; Han, J.; Yermanos, A.; Roongta, S.; Mohsen, M.O.; Oxenius, A.; Vogel, M.; et al. TLR7 Signaling Shapes and Maintains Antibody Diversity upon Virus-like Particle Immunization. Front. Immunol. 2022, 12, 827256. [Google Scholar] [CrossRef] [PubMed]
- Clausi, A.; Cummiskey, J.; Merkley, S.; Carpenter, J.F.; Jones Braun, L.; Randolph, T.W. Influence of particle size and antigen binding on effectiveness of aluminum salt adjuvants in a model lysozyme vaccine. J. Pharm. Sci. 2008, 97, 5252–5262. [Google Scholar] [CrossRef] [PubMed]
- Coria, L.M.; Saposnik, L.M.; Castro, C.P.; Castro, E.F.; Bruno, L.A.; Stone, W.B.; Pérez, P.S.; Darriba, M.L.; Chemes, L.B.; Alcain, J.; et al. A Novel Bacterial Protease Inhibitor Adjuvant in RBD-Based COVID-19 Vaccine Formulations Containing Alum Increases Neutralizing Antibodies, Specific Germinal Center B Cells and Confers Protection against SARS-CoV-2 Infection in Mice. Front. Immunol. 2022, 13, 844837. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, R.; Hogan, M.J.; Loré, K.; Pardi, N. Innate immune mechanisms of mRNA vaccines. Immunity 2022, 55, 1993–2005. [Google Scholar] [CrossRef] [PubMed]
- Muslimov, A.; Tereshchenko, V.; Shevyrev, D.; Rogova, A.; Lepik, K.; Reshetnikov, V.; Ivanov, R. The Dual Role of the Innate Immune System in the Effectiveness of mRNA Therapeutics. Int. J. Mol. Sci. 2023, 24, 14820. [Google Scholar] [CrossRef]
- Diebold, S.S.; Massacrier, C.; Akira, S.; Paturel, C.; Morel, Y.; Reis e Sousa, C. Nucleic acid agonists for toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur. J. Immunol. 2006, 36, 3256–3267. [Google Scholar] [CrossRef]
- Zhang, Z.; Ohto, U.; Shimizu, T. Toward a structural understanding of nucleic acid-sensing toll-like receptors in the innate immune system. FEBS Lett. 2017, 591, 3167–3181. [Google Scholar] [CrossRef]
- Sakaniwa, K.; Fujimura, A.; Shibata, T.; Shigematsu, H.; Ekimoto, T.; Yamamoto, M.; Ikeguchi, M.; Miyake, K.; Ohto, U.; Shimizu, T. TLR3 forms a laterally aligned multimeric complex along double-stranded RNA for efficient signal transduction. Nat. Commun. 2023, 14, 164. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Mir, F.F.; Jhunjhunwala, S.; Kaczmarek, J.C.; Hurtado, J.E.; Yang, J.H.; Webber, M.J.; Kowalski, P.S.; Heartlein, M.W.; DeRosa, F.; et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 2016, 109, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-based Vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Leppek, K.; Byeon, G.W.; Kladwang, W.; Wayment-Steele, H.K.; Kerr, C.H.; Xu, A.F.; Kim, D.S.; Topkar, V.V.; Choe, C.; Rothschild, D.; et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat. Commun. 2022, 13, 1536. [Google Scholar] [CrossRef]
- Chheda, U.; Pradeepan, S.; Esposito, E.; Strezsak, S.; Fernandez-Delgado, O.; Kranz, J. Factors Affecting Stability of RNA—Temperature, Length, Concentration, pH, and Buffering Species. J. Pharm. Sci. 2024, 113, 377–385. [Google Scholar] [CrossRef]
- Kyuregyan, K.K.; Jansons, J.; Isaguliants, M. Is there a future for traditional immunogens when we have mRNA? Microorganisms 2023, 11, 1004. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Zeltins, A.; Kalnins, G.; Balke, I.; Fischer, N.; Rostaher, A.; Tars, K.; Favrot, C. Vaccination against IL-31 for the treatment of atopic dermatitis in dogs. J. Allergy Clin. Immunol. 2018, 142, 279–281.e1. [Google Scholar] [CrossRef]
- Fettelschoss-Gabriel, A.; Fettelschoss, V.; Thoms, F.; Giese, C.; Daniel, M.; Olomski, F.; Kamarachev, J.; Birkmann, K.; Buhler, M.; Kummer, M. Treating insect-bite hypersensitivity in horses with active vaccination against IL-5. J. Allergy Clin. Immunol. 2018, 142, 1194–1205.e3. [Google Scholar] [CrossRef]
- von Loga, I.S.; El-Turabi, A.; Jostins, L.; Miotla-Zarebska, J.; Mackay-Alderson, J.; Zeltins, A.; Parisi, I.; Bachmann, M.F.; Vincent, T.L. Active immunisation targeting nerve growth factor attenuates chronic pain behaviour in murine osteoarthritis. Ann. Rheum. Dis. 2019, 78, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Lu, S. Heterologous prime-boost vaccination. Curr. Opin. Immunol. 2009, 21, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Deming, M.E.; Lyke, K.E. A ‘mix and match’ approach to SARS-CoV-2 vaccination. Nat. Med. 2021, 27, 1510–1511. [Google Scholar] [CrossRef] [PubMed]
- Jara, A.; Undurraga, E.A.; Zubizarreta, J.R.; González, C.; Pizarro, A.; Acevedo, J.; Leo, K.; Paredes, F.; Bralic, T.; Vergara, V.; et al. Effectiveness of homologous and heterologous booster doses for an inactivated SARS-CoV-2 vaccine: A large-scale prospective cohort study. Lancet Glob. Health 2022, 10, e798–e806. [Google Scholar] [CrossRef]
- Vogt, A.-C.S.; Jörg, L.; Martina, B.; Krenger, P.S.; Chang, X.; Zeltins, A.; Vogel, M.; Mohsen, M.O.; Bachmann, M.F. Virus-Like Particles Are Efficient Tools for Boosting mRNA-Induced Antibodies. Front. Immunol. 2022, 13, 864718. [Google Scholar] [CrossRef]




| Oligonucleotide Name | Sequence |
|---|---|
| CmN-NcoF | 5′ ATACCATGGACAAATCTGAATCAACCAGT 3′ |
| CmN-BamR | 5′ TCTGGATCCCCGGTTGGGTGGTTAATAGTTGGACGA 3′ |
| His-tag-C-eGFP-Bsp1407I-F | 5′ AGCTGTACAAGGGTGGCGGATCCCATCATCATCATCATCACCATT 3′ |
| His-tag-C-eGFP-SacI-R | 5′ AGCGAGCTCTAGGGCCGCTTTAATGGTGATGATGATGATGATGGG 3′ |
| pET-dir | 5′ GGGGAATTGTGAGCGGATAACA 3′ |
| pET-rev | 5′ TATTGCTCAGCGGTGGCAGC 3’ |
| M13seq-F | 5’ GCCAGGGTTTTCCCAGTCACGA 3’ |
| M13seq-R | 5’ GAGCGGATAACAATTTCACACAGG 3’ |
| PADRE-eGFP-BamHI-F | 5′ ACCACCCAACCGGGGATCCCGCGAAATTTGTGGCCGCGTGGACCCTC 3’ |
| PADRE-eGFP-AgeI-R | 5′ TCACCATGGTGGCCACCGGTGGCGCGGCCGCCTTGAGGGTCCACGCGGCCAC 3’ |
| nCMV-Vect_R | 5′ TGCTCGAGAATTCAAGCTTGCTTTACAATAGCGGTGGCGCGGCCGCCT 3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobczak, J.M.; Barkovska, I.; Balke, I.; Rothen, D.A.; Mohsen, M.O.; Skrastina, D.; Ogrina, A.; Martina, B.; Jansons, J.; Bogans, J.; et al. Identifying Key Drivers of Efficient B Cell Responses: On the Role of T Help, Antigen-Organization, and Toll-like Receptor Stimulation for Generating a Neutralizing Anti-Dengue Virus Response. Vaccines 2024, 12, 661. https://doi.org/10.3390/vaccines12060661
Sobczak JM, Barkovska I, Balke I, Rothen DA, Mohsen MO, Skrastina D, Ogrina A, Martina B, Jansons J, Bogans J, et al. Identifying Key Drivers of Efficient B Cell Responses: On the Role of T Help, Antigen-Organization, and Toll-like Receptor Stimulation for Generating a Neutralizing Anti-Dengue Virus Response. Vaccines. 2024; 12(6):661. https://doi.org/10.3390/vaccines12060661
Chicago/Turabian StyleSobczak, Jan M., Irena Barkovska, Ina Balke, Dominik A. Rothen, Mona O. Mohsen, Dace Skrastina, Anete Ogrina, Byron Martina, Juris Jansons, Janis Bogans, and et al. 2024. "Identifying Key Drivers of Efficient B Cell Responses: On the Role of T Help, Antigen-Organization, and Toll-like Receptor Stimulation for Generating a Neutralizing Anti-Dengue Virus Response" Vaccines 12, no. 6: 661. https://doi.org/10.3390/vaccines12060661
APA StyleSobczak, J. M., Barkovska, I., Balke, I., Rothen, D. A., Mohsen, M. O., Skrastina, D., Ogrina, A., Martina, B., Jansons, J., Bogans, J., Vogel, M., Bachmann, M. F., & Zeltins, A. (2024). Identifying Key Drivers of Efficient B Cell Responses: On the Role of T Help, Antigen-Organization, and Toll-like Receptor Stimulation for Generating a Neutralizing Anti-Dengue Virus Response. Vaccines, 12(6), 661. https://doi.org/10.3390/vaccines12060661










