Structural Assessment of Chlamydia trachomatis Major Outer Membrane Protein (MOMP)-Derived Vaccine Antigens and Immunological Profiling in Mice with Different Genetic Backgrounds
Abstract
:1. Introduction
2. Materials and Methods
3. Results
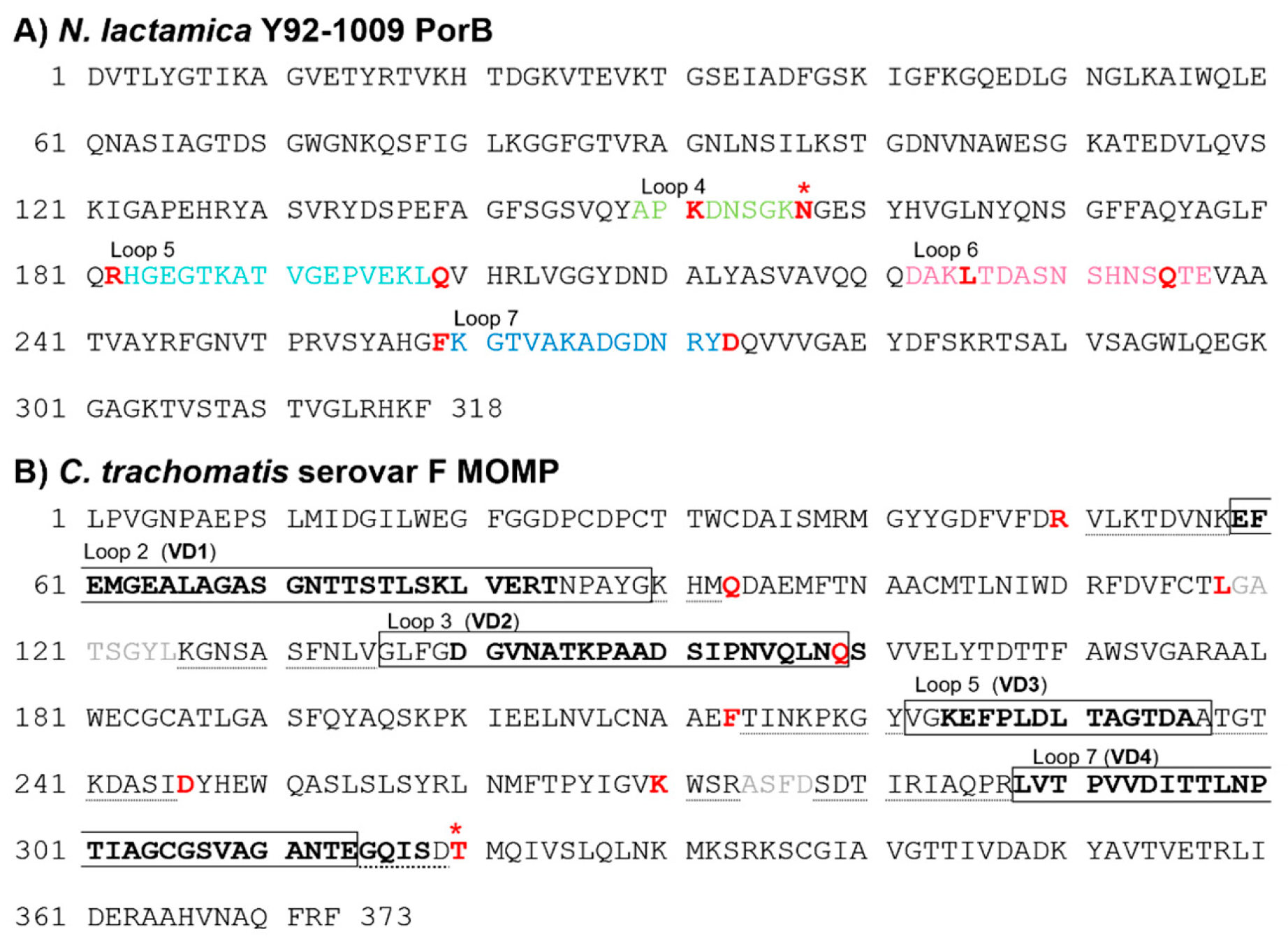
3.1. Sequence Analysis of C. trachomatis Serovar F MOMP and N. lactamica Y92-1009 PorB
3.2. PorB/VDs Sequence Analysis and Structure Predictions
3.3. Immunogenicity of PorB/VD Antigens in Mice with Different Genetic Backgrounds
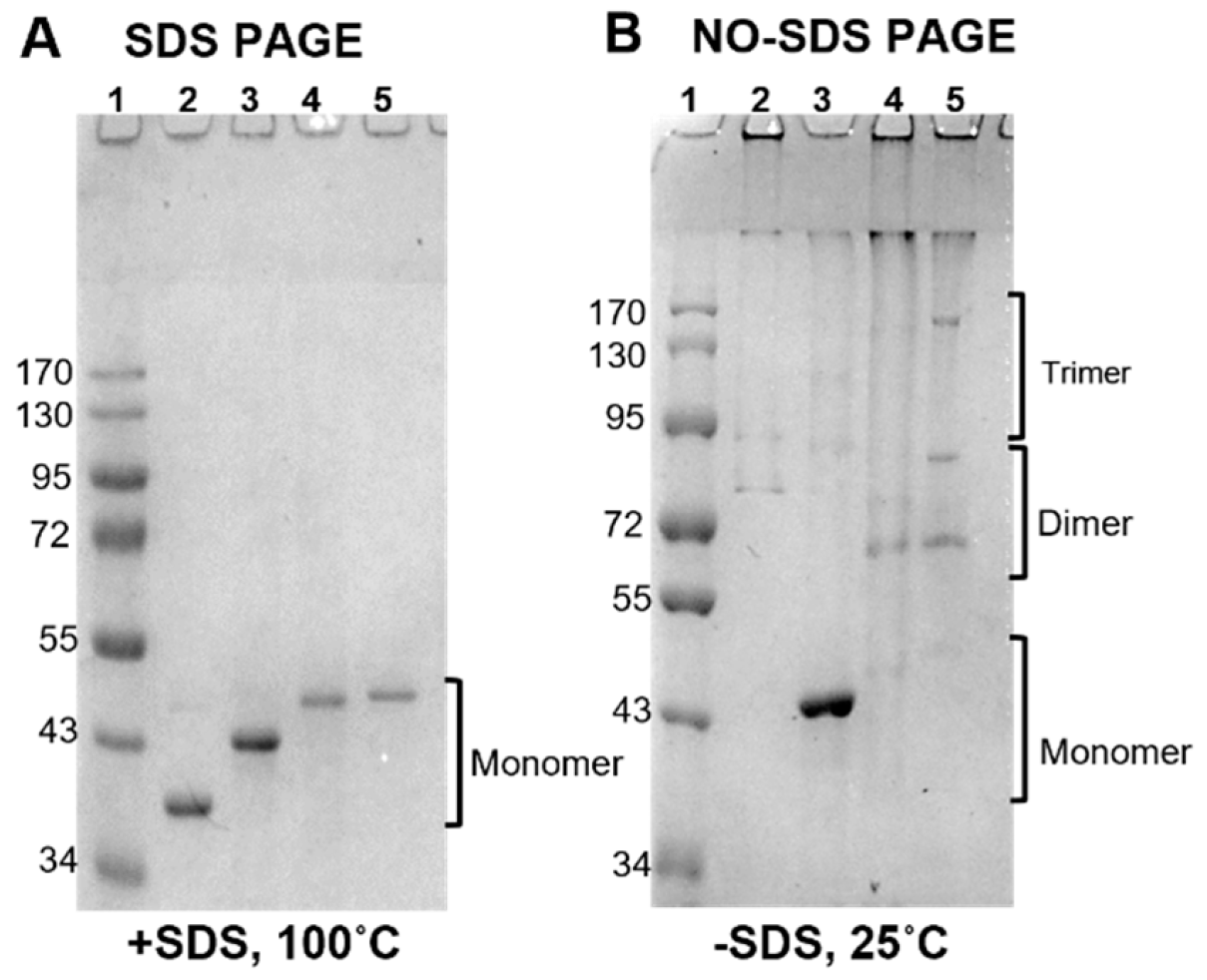
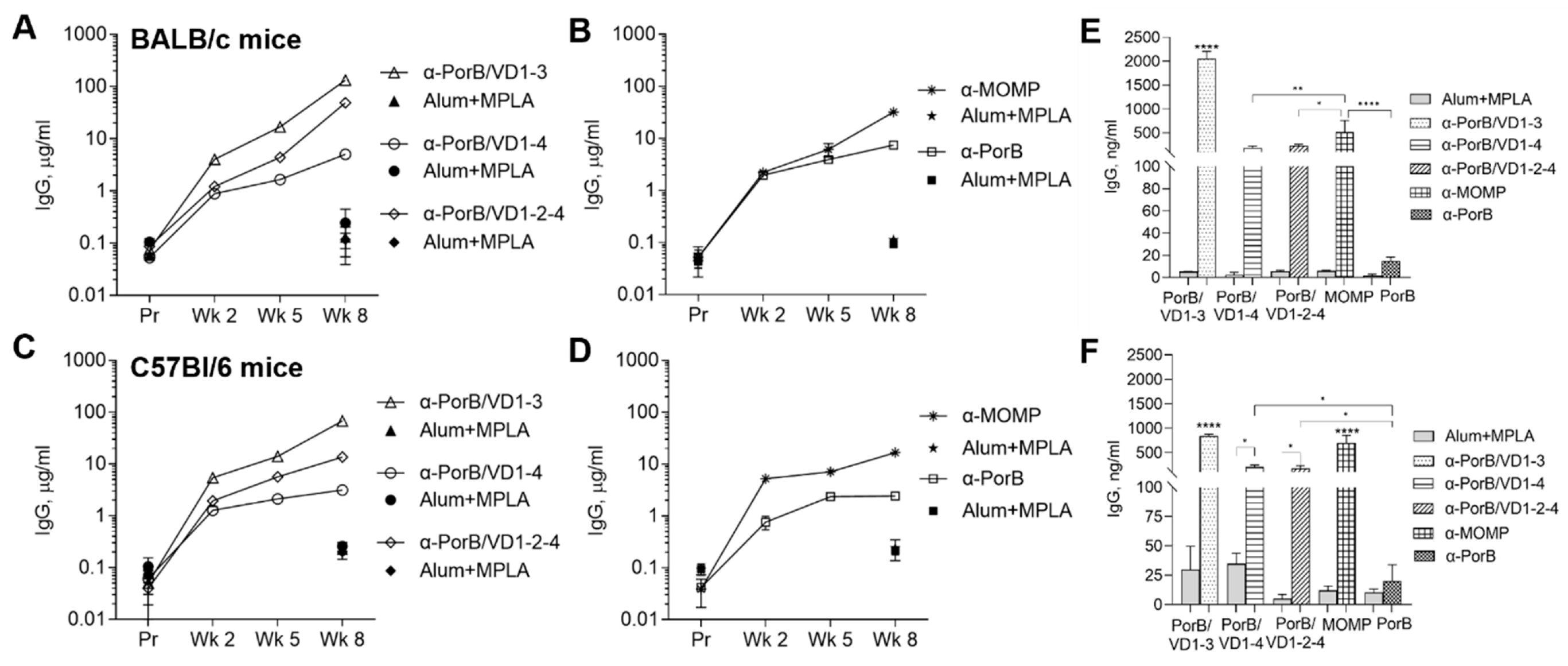
3.3.1. Antibody Responses to Purified Antigens
3.3.2. Antigen and Antibody Cross-Reactivity
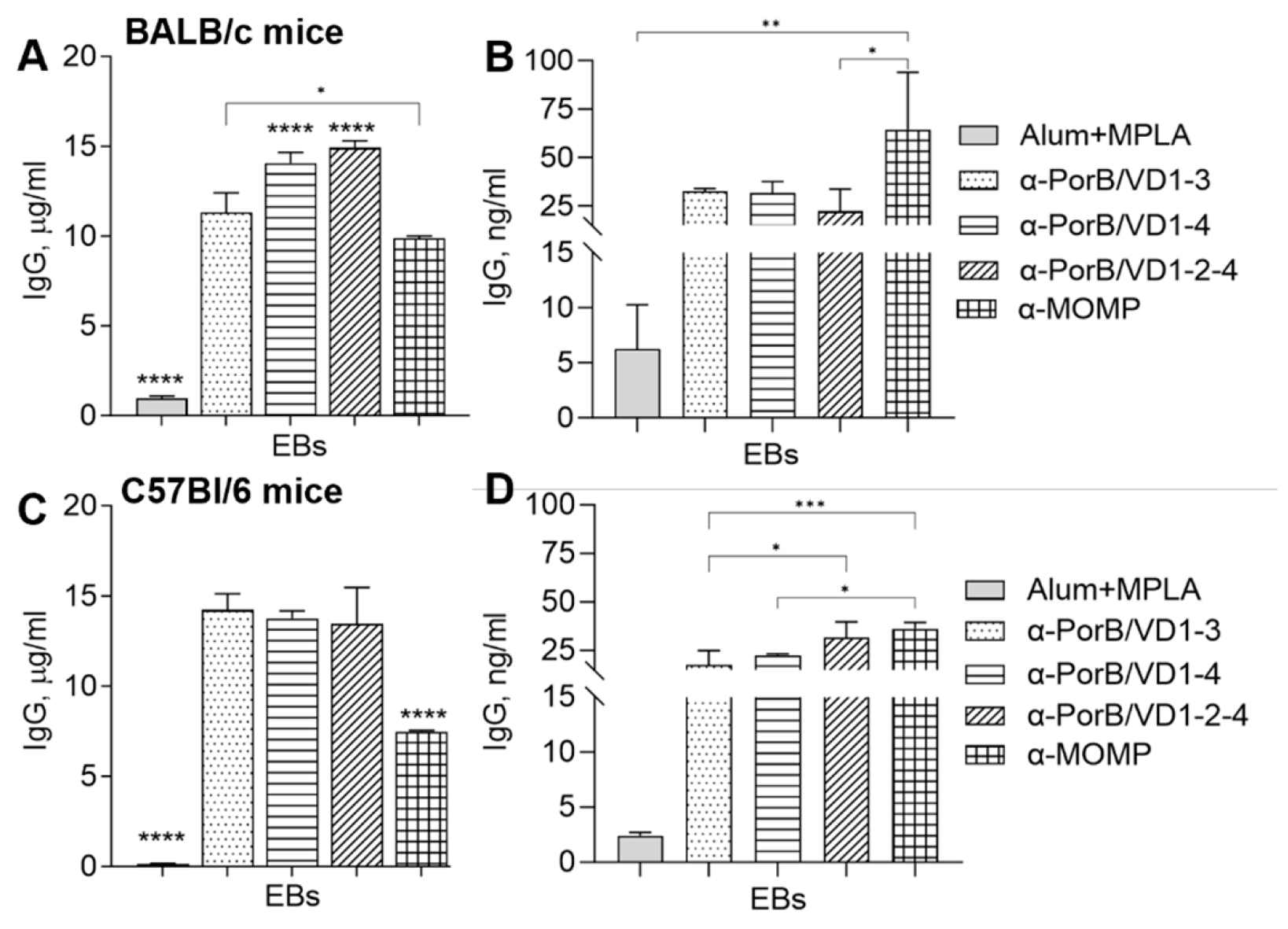
3.3.3. Antibody Cross-Reactivity with C. trachomatis Whole Organisms
3.3.4. Neutralization Titers
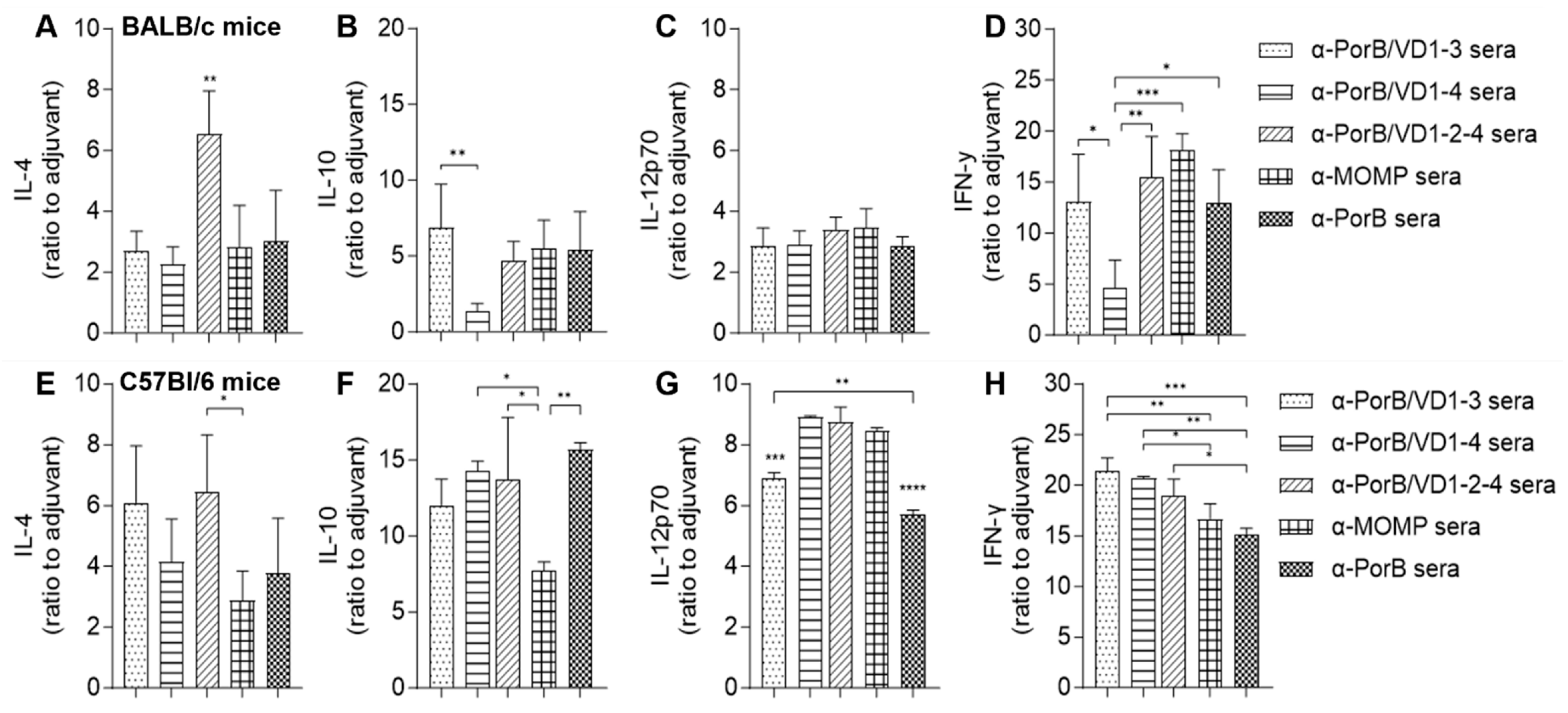
3.3.5. Serum Cytokines
3.3.6. Splenocytes Proliferation and Cytokine Response
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Centers for Disease Control and Prevention. Sexually Transmitted Disease Surveillance 2018; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2019. [CrossRef]
- Newman, L.; Rowley, J.; Vander Hoorn, S.; Wijesooriya, N.S.; Unemo, M.; Low, N.; Stevens, G.; Gottlieb, S.; Kiarie, J.; Temmerman, M. Global Estimates of the Prevalence and Incidence of Four Curable Sexually Transmitted Infections in 2012 Based on Systematic Review and Global Reporting. PLoS ONE 2015, 10, e0143304. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.M.; Cheng, X.; Markoff, B.A.; Fielder, T.J.; de la Maza, L.M. Functional and structural mapping of Chlamydia trachomatis species-specific major outer membrane protein epitopes by use of neutralizing monoclonal antibodies. Infect. Immun. 1991, 59, 4147–4153. [Google Scholar] [CrossRef]
- Schachter, J. Chlamydial infections (first of three parts). N. Engl. J. Med. 1978, 298, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Stamm, W.E. Chlamydia trachomatis infections: Progress and problems. J. Infect. Dis. 1999, 179 (Suppl. S2), S380–S383. [Google Scholar] [CrossRef] [PubMed]
- Darville, T. Pelvic Inflammatory Disease Due to Neisseria gonorrhoeae and Chlamydia trachomatis: Immune Evasion Mechanisms and Pathogenic Disease Pathways. J. Infect. Dis. 2021, 224, S39–S46. [Google Scholar] [CrossRef] [PubMed]
- Gotz, H.; Lindback, J.; Ripa, T.; Arneborn, M.; Ramsted, K.; Ekdahl, K. Is the increase in notifications of Chlamydia trachomatis infections in Sweden the result of changes in prevalence, sampling frequency or diagnostic methods? Scand. J. Infect. Dis. 2002, 34, 28–34. [Google Scholar] [CrossRef]
- Brunham, R.C.; Pourbohloul, B.; Mak, S.; White, R.; Rekart, M.L. The unexpected impact of a Chlamydia trachomatis infection control program on susceptibility to reinfection. J. Infect. Dis. 2005, 192, 1836–1844. [Google Scholar] [CrossRef]
- Ness, R.B.; Soper, D.E.; Holley, R.L.; Peipert, J.; Randall, H.; Sweet, R.L.; Sondheimer, S.J.; Hendrix, S.L.; Amortegui, A.; Trucco, G.; et al. Effectiveness of inpatient and outpatient treatment strategies for women with pelvic inflammatory disease: Results from the Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) Randomized Trial. Am. J. Obstet. Gynecol. 2002, 186, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Brunham, R.C.; de la Maza, L.M.; Darville, T.; Deal, C. National Institute of Allergy and Infectious Diseases workshop report: “Chlamydia vaccines: The way forward”. Vaccine 2019, 37, 7346–7354. [Google Scholar] [CrossRef]
- Rodrigues, R.; Marques, L.; Vieira-Baptista, P.; Sousa, C.; Vale, N. Therapeutic Options for Chlamydia trachomatis Infection: Present and Future. Antibiotics 2022, 11, 1634. [Google Scholar] [CrossRef]
- Wang, S.P.; Grayston, J.T. Pannus with experimental trachoma and inclusion conjunctivitis agent infection of Taiwan monkeys. Am. J. Ophthalmol. 1967, 63, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.R. Trachoma: A Blinding Scourge from the Bronze Age to the Twenty-First Century; Centre for Eye Research Australia: East Melbourne, Australia, 2008; p. 282. [Google Scholar]
- Vasilevsky, S.; Stojanov, M.; Greub, G.; Baud, D. Chlamydial polymorphic membrane proteins: Regulation, function and potential vaccine candidates. Virulence 2016, 7, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.K.; Li, W.; Guentzel, M.N.; Zhong, G.; Arulanandam, B.P. Vaccination with the defined chlamydial secreted protein CPAF induces robust protection against female infertility following repeated genital chlamydial challenge. Vaccine 2011, 29, 2519–2522. [Google Scholar] [CrossRef]
- Pal, S.; Peterson, E.M.; de la Maza, L.M. Vaccination with the Chlamydia trachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteria. Infect. Immun. 2005, 73, 8153–8160. [Google Scholar] [CrossRef]
- Farris, C.M.; Morrison, R.P. Vaccination against Chlamydia genital infection utilizing the murine C. muridarum model. Infect. Immun. 2011, 79, 986–996. [Google Scholar] [CrossRef]
- Kari, L.; Whitmire, W.M.; Crane, D.D.; Reveneau, N.; Carlson, J.H.; Goheen, M.M.; Peterson, E.M.; Pal, S.; de la Maza, L.M.; Caldwell, H.D. Chlamydia trachomatis native major outer membrane protein induces partial protection in nonhuman primates: Implication for a trachoma transmission-blocking vaccine. J. Immunol. 2009, 182, 8063–8070. [Google Scholar] [CrossRef]
- Simpson, S.J.; Higgins, D.P.; Timms, P.; Mella, V.S.A.; Crowther, M.S.; Fernandez, C.M.; McArthur, C.; Phillips, S.; Krockenberger, M.B. Efficacy of a synthetic peptide Chlamydia pecorum major outer membrane protein vaccine in a wild koala (Phascolarctos cinereus) population. Sci. Rep. 2023, 13, 15087. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Barnhart, K.M.; Wei, Q.; Abai, A.M.; Peterson, E.M.; de la Maza, L.M. Vaccination of mice with DNA plasmids coding for the Chlamydia trachomatis major outer membrane protein elicits an immune response but fails to protect against a genital challenge. Vaccine 1999, 17, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, Y.; Xiong, Y.; Du, W.; Cen, D.; Zhang, C.; Song, Y.; Zhu, S.; Xue, X.; Zhang, L. DNA plasmid vaccine carrying Chlamydia trachomatis (Ct) major outer membrane and human papillomavirus 16L2 proteins for anti-Ct infection. Oncotarget 2017, 8, 33241–33251. [Google Scholar] [CrossRef]
- Rodriguez-Maranon, M.J.; Bush, R.M.; Peterson, E.M.; Schirmer, T.; de la Maza, L.M. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci. 2002, 11, 1854–1861. [Google Scholar] [CrossRef]
- Sun, G.; Pal, S.; Sarcon, A.K.; Kim, S.; Sugawara, E.; Nikaido, H.; Cocco, M.J.; Peterson, E.M.; de la Maza, L.M. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 2007, 189, 6222–6235. [Google Scholar] [CrossRef] [PubMed]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef]
- Stephens, R.S.; Wagar, E.A.; Schoolnik, G.K. High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 1988, 167, 817–831. [Google Scholar] [CrossRef]
- Ortiz, L.; Angevine, M.; Kim, S.K.; Watkins, D.; DeMars, R. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect. Immun. 2000, 68, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Nunes, A.; Nogueira, P.J.; Borrego, M.J.; Gomes, J.P. Adaptive evolution of the Chlamydia trachomatis dominant antigen reveals distinct evolutionary scenarios for B- and T-cell epitopes: Worldwide survey. PLoS ONE 2010, 5, e13171. [Google Scholar] [CrossRef]
- Olsen, A.W.; Lorenzen, E.K.; Rosenkrands, I.; Follmann, F.; Andersen, P. Protective Effect of Vaccine Promoted Neutralizing Antibodies against the Intracellular Pathogen Chlamydia trachomatis. Front. Immunol. 2017, 8, 1652. [Google Scholar] [CrossRef]
- Naglak, E.K.; Morrison, S.G.; Morrison, R.P. IFNgamma is Required for Optimal Antibody-Mediated Immunity against Genital Chlamydia Infection. Infect. Immun. 2016, 84, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Gondek, D.C.; Olive, A.J.; Stary, G.; Starnbach, M.N. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J. Immunol. 2012, 189, 2441–2449. [Google Scholar] [CrossRef]
- Pal, S.; Peterson, E.M.; Rappuoli, R.; Ratti, G.; de la Maza, L.M. Immunization with the Chlamydia trachomatis major outer membrane protein, using adjuvants developed for human vaccines, can induce partial protection in a mouse model against a genital challenge. Vaccine 2006, 24, 766–775. [Google Scholar] [CrossRef]
- Pal, S.; Tifrea, D.F.; Follmann, F.; Andersen, P.; de la Maza, L.M. The cationic liposomal adjuvants CAF01 and CAF09 formulated with the major outer membrane protein elicit robust protection in mice against a Chlamydia muridarum respiratory challenge. Vaccine 2017, 35, 1705–1711. [Google Scholar] [CrossRef]
- Pal, S.; Slepenkin, A.; Felgner, J.; Huw Davies, D.; Felgner, P.; de la Maza, L.M. Evaluation of Four Adjuvant Combinations, IVAX-1, IVAX-2, CpG-1826+Montanide ISA 720 VG and CpG-1018+Montanide ISA 720 VG, for Safety and for Their Ability to Elicit Protective Immune Responses in Mice against a Respiratory Challenge with Chlamydia muridarum. Pathogens 2023, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Pal, S.; Tifrea, D.; Jia, Z.; de la Maza, L.M. A vaccine formulated with a combination of TLR-2 and TLR-9 adjuvants and the recombinant major outer membrane protein elicits a robust immune response and significant protection against a Chlamydia muridarum challenge. Microbes. Infect. 2014, 16, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Dixit, S.; Verma, R.; Duncan, S.A.; Coats, M.T.; Giambartolomei, G.H.; Singh, S.R.; Dennis, V.A. A nanovaccine formulation of Chlamydia recombinant MOMP encapsulated in PLGA 85:15 nanoparticles augments CD4(+) effector (CD44(high) CD62L(low)) and memory (CD44(high) CD62L(high)) T-cells in immunized mice. Nanomedicine 2020, 29, 102257. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Pal, S.; Weiland, J.; Peterson, E.M.; de la Maza, L.M. Protection against an intranasal challenge by vaccines formulated with native and recombinant preparations of the Chlamydia trachomatis major outer membrane protein. Vaccine 2009, 27, 5020–5025. [Google Scholar] [CrossRef] [PubMed]
- Ralli-Jain, P.; Tifrea, D.; Cheng, C.; Pal, S.; de la Maza, L.M. Enhancement of the protective efficacy of a Chlamydia trachomatis recombinant vaccine by combining systemic and mucosal routes for immunization. Vaccine 2010, 28, 7659–7666. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; Juel, H.B.; Bang, P.; Cheeseman, H.M.; Dohn, R.B.; Cole, T.; Kristiansen, M.P.; Korsholm, K.S.; Lewis, D.; Olsen, A.W.; et al. Safety and immunogenicity of the chlamydia vaccine candidate CTH522 adjuvanted with CAF01 liposomes or aluminium hydroxide: A first-in-human, randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect. Dis. 2019, 19, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, E.; Contreras, V.; Olsen, A.W.; Andersen, P.; Desjardins, D.; Rosenkrands, I.; Juel, H.B.; Delache, B.; Langlois, S.; Delaugerre, C.; et al. Multi-component prime-boost Chlamydia trachomatis vaccination regimes induce antibody and T cell responses and accelerate clearance of infection in a non-human primate model. Front. Immunol. 2022, 13, 1057375. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.W.; Rosenkrands, I.; Jacobsen, C.S.; Cheeseman, H.M.; Kristiansen, M.P.; Dietrich, J.; Shattock, R.J.; Follmann, F. Immune signature of Chlamydia vaccine CTH522/CAF(R)01 translates from mouse-to-human and induces durable protection in mice. Nat. Commun. 2024, 15, 1665. [Google Scholar] [CrossRef]
- Pollock, K.M.; Borges, A.H.; Cheeseman, H.M.; Rosenkrands, I.; Schmidt, K.L.; Sondergaard, R.E.; Day, S.; Evans, A.; McFarlane, L.R.; Joypooranachandran, J.; et al. An investigation of trachoma vaccine regimens by the chlamydia vaccine CTH522 administered with cationic liposomes in healthy adults (CHLM-02): A phase 1, double-blind trial. Lancet Infect. Dis. 2024. [Google Scholar] [CrossRef]
- Madico, G.; Gursky, O.; Fairman, J.; Massari, P. Structural and Immunological Characterization of Novel Recombinant MOMP-Based Chlamydial Antigens. Vaccines 2017, 6, 2. [Google Scholar] [CrossRef]
- Tifrea, D.F.; Pal, S.; Fairman, J.; Massari, P.; de la Maza, L.M. Protection against a chlamydial respiratory challenge by a chimeric vaccine formulated with the Chlamydia muridarum major outer membrane protein variable domains using the Neisseria lactamica porin B as a scaffold. NPJ Vaccines 2020, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Berg, E.A.; Feng, X.; Shen, L.; Smith, T.; Costello, C.E.; Zhang, Y.X. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 2006, 15, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Kattner, C.; Toussi, D.N.; Zaucha, J.; Wetzler, L.M.; Ruppel, N.; Zachariae, U.; Massari, P.; Tanabe, M. Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition. J. Struct. Biol. 2014, 185, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, Inc. The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, Inc.: New York, NY, USA, 2015. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef]
- Hepler, R.W.; Nahas, D.D.; Lucas, B.; Kaufhold, R.; Flynn, J.A.; Galli, J.D.; Swoyer, R.; Wagner, J.M.; Espeseth, A.S.; Joyce, J.G.; et al. Spectroscopic analysis of chlamydial major outer membrane protein in support of structure elucidation. Protein Sci. 2018, 27, 1923–1941. [Google Scholar] [CrossRef]
- Cheng, C.; Pal, S.; Bettahi, I.; Oxford, K.L.; Barry, P.A.; de la Maza, L.M. Immunogenicity of a vaccine formulated with the Chlamydia trachomatis serovar F, native major outer membrane protein in a nonhuman primate model. Vaccine 2011, 29, 3456–3464. [Google Scholar] [CrossRef]
- Peterson, E.M.; Zhong, G.M.; Carlson, E.; de la Maza, L.M. Protective role of magnesium in the neutralization by antibodies of Chlamydia trachomatis infectivity. Infect. Immun. 1988, 56, 885–891. [Google Scholar] [CrossRef]
- Liu, X.; Wetzler, L.M.; Massari, P. The PorB porin from commensal Neisseria lactamica induces Th1 and Th2 immune responses to ovalbumin in mice and is a potential immune adjuvant. Vaccine 2008, 26, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Liu, E.; Graham, J.; Chen, W.; Keten, S. B-factor prediction in proteins using a sequence-based deep learning model. Patterns 2023, 4, 100805. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-Factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 119, 1626–1665. [Google Scholar] [CrossRef]
- Massari, P.; King, C.A.; Macleod, H.; Wetzler, L.M. Improved purification of native meningococcal porin PorB and studies on its structure/function. Protein Expr. Purif. 2005, 44, 136–146. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Nourishirazi, E.; Guinet, E.; Nouri-Shirazi, M. The genetic background influences the cellular and humoral immune responses to vaccines. Clin. Exp. Immunol. 2016, 186, 190–204. [Google Scholar] [CrossRef]
- Oleszycka, E.; Lavelle, E.C. Immunomodulatory properties of the vaccine adjuvant alum. Curr. Opin. Immunol. 2014, 28, 1–5. [Google Scholar] [CrossRef]
- Casella, C.R.; Mitchell, T.C. Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell Mol. Life Sci. 2008, 65, 3231–3240. [Google Scholar] [CrossRef]
- Collar, A.L.; Linville, A.C.; Core, S.B.; Frietze, K.M. Epitope-Based Vaccines against the Chlamydia trachomatis Major Outer Membrane Protein Variable Domain 4 Elicit Protection in Mice. Vaccines 2022, 10, 875. [Google Scholar] [CrossRef] [PubMed]
- Baehr, W.; Zhang, Y.X.; Joseph, T.; Su, H.; Nano, F.E.; Everett, K.D.; Caldwell, H.D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 1988, 85, 4000–4004. [Google Scholar] [CrossRef]
- Herve, C.; Laupeze, B.; Del Giudice, G.; Didierlaurent, A.M.; Tavares Da Silva, F. The how’s and what’s of vaccine reactogenicity. NPJ Vaccines 2019, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Didierlaurent, A.M.; Morel, S.; Lockman, L.; Giannini, S.L.; Bisteau, M.; Carlsen, H.; Kielland, A.; Vosters, O.; Vanderheyde, N.; Schiavetti, F.; et al. AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J. Immunol. 2009, 183, 6186–6197. [Google Scholar] [CrossRef] [PubMed]
- Visan, L.; Sanchez, V.; Kania, M.; de Montfort, A.; de la Maza, L.M.; Ausar, S.F. Phosphate substitution in an AlOOH-TLR4 adjuvant system (SPA08) modulates the immunogenicity of Serovar E MOMP from Chlamydia trachomatis. Hum. Vaccin. Immunother. 2016, 12, 2341–2350. [Google Scholar] [CrossRef] [PubMed]
- Eyes, T.J.; Austerberry, J.I.; Dearman, R.J.; Johannissen, L.O.; Kimber, I.; Smith, N.; Thistlethwaite, A.; Derrick, J.P. Identification of B cell epitopes enhanced by protein unfolding and aggregation. Mol. Immunol. 2019, 105, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Athearn, K.; Sample, C.J.; Barefoot, B.E.; Williams, K.L.; Ramsburg, E.A. Acute reactogenicity after intramuscular immunization with recombinant vesicular stomatitis virus is linked to production of IL-1beta. PLoS ONE 2012, 7, e46516. [Google Scholar] [CrossRef]
- Burny, W.; Marchant, A.; Herve, C.; Callegaro, A.; Caubet, M.; Fissette, L.; Gheyle, L.; Legrand, C.; Ndour, C.; Tavares Da Silva, F.; et al. Inflammatory parameters associated with systemic reactogenicity following vaccination with adjuvanted hepatitis B vaccines in humans. Vaccine 2019, 37, 2004–2015. [Google Scholar] [CrossRef]





| Sequence a | Net Charge | PorB Loop Replaced | Charge Change |
|---|---|---|---|
| * 292 KWSRASFDSDTIRIAQPRLVTPVVDITTLNPTIAGCGSVAGANTEGQISDT 342 | −1 (+4/−5) | 4 | +1 → −1 |
| 72RVLKTDVNKEFEMGEALAGASGNTTSTLSKLVERTNPAYGKHMQ 115 | +2 (+7/−5) | 5 | +1 →+2 |
| 140LGATSGYLKGNSASFNLVGLFGDGVNATKPAADSIPNVQLNQ181 | 0 (+2/−2) | 6 | −1 → 0 |
| 235FTINKPKGYVGKEFPLDLTAGTDAATGTKDASID268 | −1 (+4/−5) | 7 | +1 → −1 |
| PorB | PorB/VD1-3 | PorB/VD1-4 | PorB/VD1-2-4 | |
|---|---|---|---|---|
| α-helix (%) | 16.04 | 16.25 | 16.24 | 14.32 |
| β-sheet (%) | 24.53 | 25.34 | 26.29 | 25.06 |
| Random coil (%) | 59.43 | 58.40 | 57.47 | 60.62 |
| PorB/VD1-3 | PorB/VD1-4 | PorB/VD1-2-4 | MOMP | PorB | |
|---|---|---|---|---|---|
| BALB/c mice a | 0.59 ± 0.04 | 0.96 ± 0.13 | 0.14 ± 0.02 | 1.2 ± 0.4 | 1.07 ± 0.3 |
| C57Bl/6 mice b | 2.8 ± 0.22 c | 1.9 ± 0.22 c | 0.25 ± 0.02 c | 1.13 ± 0.03 | 0.86 ± 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roe, S.K.; Zhu, T.; Slepenkin, A.; Berges, A.; Fairman, J.; de la Maza, L.M.; Massari, P. Structural Assessment of Chlamydia trachomatis Major Outer Membrane Protein (MOMP)-Derived Vaccine Antigens and Immunological Profiling in Mice with Different Genetic Backgrounds. Vaccines 2024, 12, 789. https://doi.org/10.3390/vaccines12070789
Roe SK, Zhu T, Slepenkin A, Berges A, Fairman J, de la Maza LM, Massari P. Structural Assessment of Chlamydia trachomatis Major Outer Membrane Protein (MOMP)-Derived Vaccine Antigens and Immunological Profiling in Mice with Different Genetic Backgrounds. Vaccines. 2024; 12(7):789. https://doi.org/10.3390/vaccines12070789
Chicago/Turabian StyleRoe, Shea K., Tianmou Zhu, Anatoli Slepenkin, Aym Berges, Jeff Fairman, Luis M. de la Maza, and Paola Massari. 2024. "Structural Assessment of Chlamydia trachomatis Major Outer Membrane Protein (MOMP)-Derived Vaccine Antigens and Immunological Profiling in Mice with Different Genetic Backgrounds" Vaccines 12, no. 7: 789. https://doi.org/10.3390/vaccines12070789
APA StyleRoe, S. K., Zhu, T., Slepenkin, A., Berges, A., Fairman, J., de la Maza, L. M., & Massari, P. (2024). Structural Assessment of Chlamydia trachomatis Major Outer Membrane Protein (MOMP)-Derived Vaccine Antigens and Immunological Profiling in Mice with Different Genetic Backgrounds. Vaccines, 12(7), 789. https://doi.org/10.3390/vaccines12070789





