Asian Flush Gene Variant Enhances Cellular Immunogenicity of COVID-19 Vaccine: Prospective Observation in the Japanese General Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Design of This Study and Participants
2.2. Questionnaire
2.3. Genotyping
2.4. Separation of Peripheral Blood Mononuclear Cells (PBMCs)
2.5. ELISPOT Assay
2.6. Cross-Reactivity with SARS-CoV-2 Delta and Omicron Variants
2.7. Statistical Analysis
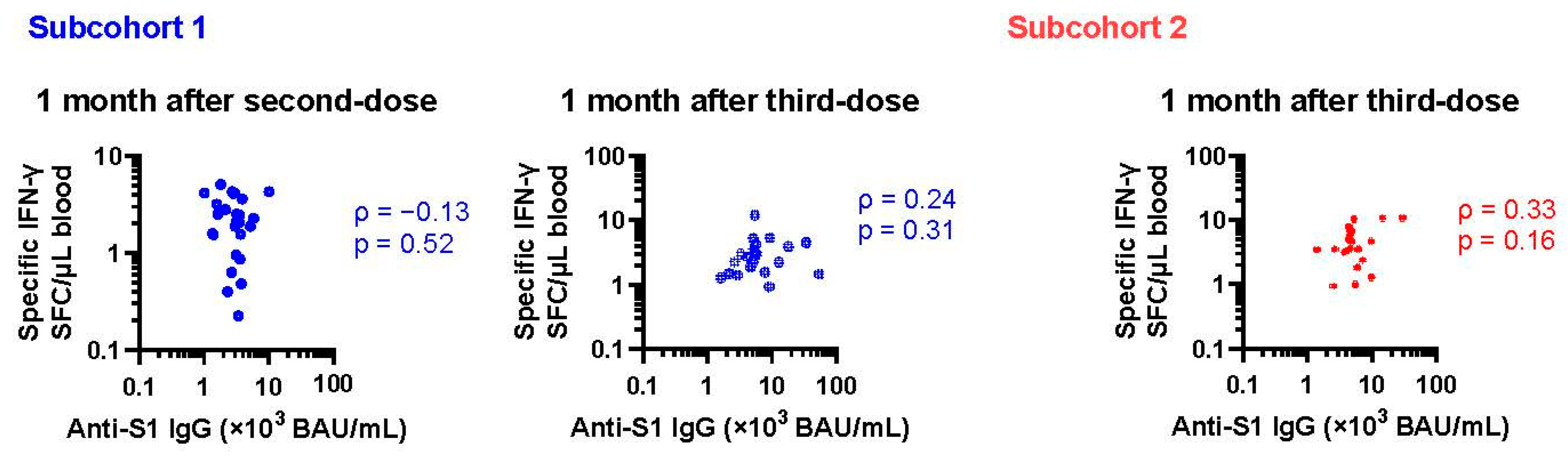
3. Results
3.1. Baseline Characters of the Participants
3.2. Changes in Cellular Immune Response after Vaccination
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Farres, J.; Wang, X.; Takahashi, K.; Cunningham, S.J.; Wang, T.T.; Weiner, H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J. Biol. Chem. 1994, 269, 13854–13860. [Google Scholar] [CrossRef]
- Wang, M.F.; Han, C.L.; Yin, S.J. Substrate specificity of human and yeast aldehyde dehydrogenases. Chem. Biol. Interact. 2009, 178, 36–39. [Google Scholar] [CrossRef]
- Peng, G.S.; Chen, Y.C.; Tsao, T.P.; Wang, M.F.; Yin, S.J. Pharmacokinetic and pharmacodynamic basis for partial protection against alcoholism in Asians, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet. Genom. 2007, 17, 845–855. [Google Scholar] [CrossRef]
- Chen, C.H.; Ferreira, J.C.; Gross, E.R.; Mochly-Rosen, D. Targeting aldehyde dehydrogenase 2: New therapeutic opportunities. Physiol. Rev. 2014, 94, 1–34. [Google Scholar] [CrossRef]
- Moh, I.; Simon, D.; Gross, E.R. The Alcohol Flush Response. Graph. Med. Rev. 2024, 4, e807. [Google Scholar] [CrossRef]
- Brooks, P.J.; Enoch, M.A.; Goldman, D.; Li, T.K.; Yokoyama, A. The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009, 6, e50. [Google Scholar] [CrossRef]
- Hishimoto, A.; Fukutake, M.; Mouri, K.; Nagasaki, Y.; Asano, M.; Ueno, Y.; Nishiguchi, N.; Shirakawa, O. Alcohol and aldehyde dehydrogenase polymorphisms and risk for suicide: A preliminary observation in the Japanese male population. Genes. Brain Behav. 2010, 9, 498–502. [Google Scholar] [CrossRef]
- Matsumoto, A. The bidirectional effect of defective ALDH2 polymorphism and disease prevention. Adv. Exp. Med. Biol. 2019, 1193, 69–87. [Google Scholar]
- Matsumoto, A.; Nakashima, C.; Kimura, S.; Sueoka, E.; Aragane, N. ALDH2 polymorphism rs671 is a predictor of PD-1/PD-L1 inhibitor efficacy against thoracic malignancies. BMC Cancer 2021, 21, 584. [Google Scholar] [CrossRef]
- Matsumoto, A.; Thompson, D.C.; Chen, Y.; Kitagawa, K.; Vasiliou, V. Roles of defective ALDH2 polymorphism on liver protection and cancer development. Environ. Health Prev. Med. 2016, 21, 395–402. [Google Scholar] [CrossRef]
- Mizuno, Y.; Harada, E.; Morita, S.; Kinoshita, K.; Hayashida, M.; Shono, M.; Morikawa, Y.; Murohara, T.; Nakayama, M.; Yoshimura, M.; et al. East asian variant of aldehyde dehydrogenase 2 is associated with coronary spastic angina: Possible roles of reactive aldehydes and implications of alcohol flushing syndrome. Circulation 2015, 131, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.L.; Tan, C.H.; Lu, Y.C.; Wu, R.M. Aldehyde dehydrogenase 2 is associated with cognitive functions in patients with Parkinson’s disease. Sci. Rep. 2016, 6, 30424. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Terashima, Y.; Toda, E.; Akao, C.; Miyake, Y.; Matsuo, K.; Shimanoe, C.; Nishida, Y.; Hara, M.; Tanaka, K.; et al. Reduced T cell immunity in variant ALDH2 allele carriers. In Proceedings of the 91th Annual Meeting of the Japanese Society for Hygiene, Toyama, Japan, 6–8 March 2021. [Google Scholar]
- Matsumoto, A.; Hara, M.; Ashenagar, M.S.; Tokiya, M.; Sawada, T.; Iwasaka, C.; Furukawa, T.; Kitagawa, K.; Miyake, Y.; Hirota, Y. Variant Allele of ALDH2, rs671, Associates with Attenuated Post-Vaccination Response in Anti-SARS-CoV-2 Spike Protein IgG: A Prospective Study in the Japanese General Population. Vaccines 2022, 10, 1035. [Google Scholar] [CrossRef]
- Takashima, S.; Tokiya, M.; Izui, K.; Miyamoto, H.; Matsumoto, A. Asian flush is a potential protective factor against COVID-19: A web-based retrospective survey in Japan. Environ. Health Prev. Med. 2024, 29, 14. [Google Scholar] [CrossRef] [PubMed]
- Rydyznski Moderbacher, C.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 2020, 183, 996–1012.e1019. [Google Scholar] [CrossRef]
- Devassikutty, F.M.; Jain, A.; Edavazhippurath, A.; Joseph, M.C.; Peedikayil, M.M.T.; Scaria, V.; Sandhya, P.; Govindaraj, G.M. X-Linked Agammaglobulinemia and COVID-19: Two Case Reports and Review of Literature. Pediatr. Allergy Immunol. Pulmonol. 2021, 34, 115–118. [Google Scholar] [CrossRef]
- Burgos-Barragan, G.; Wit, N.; Meiser, J.; Dingler, F.A.; Pietzke, M.; Mulderrig, L.; Pontel, L.B.; Rosado, I.V.; Brewer, T.F.; Cordell, R.L.; et al. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature 2017, 548, 549–554. [Google Scholar] [CrossRef]
- Wagner, M.A.; Jorns, M.S. Monomeric sarcosine oxidase: 2. Kinetic studies with sarcosine, alternate substrates, and a substrate analogue. Biochemistry 2000, 39, 8825–8829. [Google Scholar] [CrossRef]
- Ai, L.; Tan, T.; Tang, Y.; Yang, J.; Cui, D.; Wang, R.; Wang, A.; Fei, X.; Di, Y.; Wang, X.; et al. Endogenous formaldehyde is a memory-related molecule in mice and humans. Commun. Biol. 2019, 2, 446. [Google Scholar] [CrossRef] [PubMed]
- Shoeb, M.; Ansari, N.H.; Srivastava, S.K.; Ramana, K.V. 4-Hydroxynonenal in the pathogenesis and progression of human diseases. Curr. Med. Chem. 2014, 21, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, Y.; Ohashi, S.; Amanuma, Y.; Nakai, Y.; Tsurumaki, M.; Kikuchi, O.; Miyamoto, S.; Oyama, T.; Kawamoto, T.; Chiba, T.; et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am. J. Cancer Res. 2014, 4, 279–284. [Google Scholar]
- Mulderrig, L.; Garaycoechea, J.I.; Tuong, Z.K.; Millington, C.L.; Dingler, F.A.; Ferdinand, J.R.; Gaul, L.; Tadross, J.A.; Arends, M.J.; O’Rahilly, S.; et al. Aldehyde-driven transcriptional stress triggers an anorexic DNA damage response. Nature 2021, 600, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Traverso, N.; Menini, S.; Maineri, E.P.; Patriarca, S.; Odetti, P.; Cottalasso, D.; Marinari, U.M.; Pronzato, M.A. Malondialdehyde, a lipoperoxidation-derived aldehyde, can bring about secondary oxidative damage to proteins. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, B890–B895. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Ren, J. ALDH2 Polymorphism and Ethanol Consumption: A Genetic-Environmental Interaction in Carcinogenesis. Adv. Exp. Med. Biol. 2019, 1193, 229–236. [Google Scholar]
- Mali, V.R.; Ning, R.; Chen, J.; Yang, X.P.; Xu, J.; Palaniyandi, S.S. Impairment of aldehyde dehydrogenase-2 by 4-hydroxy-2-nonenal adduct formation and cardiomyocyte hypertrophy in mice fed a high-fat diet and injected with low-dose streptozotocin. Exp. Biol. Med. 2014, 239, 610–618. [Google Scholar] [CrossRef]
- Zhang, H.; Forman, H.J. 4-hydroxynonenal-mediated signaling and aging. Free Radic. Biol. Med. 2017, 111, 219–225. [Google Scholar] [CrossRef]
- Inoue, S.; Niikura, M.; Mineo, S.; Kobayashi, F. Roles of IFN-γ and γδ T Cells in Protective Immunity Against Blood-Stage Malaria. Front. Immunol. 2013, 4, 258. [Google Scholar] [CrossRef]
- Cao, J.; Liao, S.; Zeng, F.; Liao, Q.; Luo, G.; Zhou, Y. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis. 2023, 14, 407. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Darwin, K.H.; Stanley, S.A. The aldehyde hypothesis: Metabolic intermediates as antimicrobial effectors. Open Biol. 2022, 12, 220010. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.B.; Espich, S.; Thuong, N.T.T.; Chang, X.; Dorajoo, R.; Khor, C.C.; Heng, C.K.; Yuan, J.M.; Fox, D.; Anaya-Sanchez, A.; et al. Disruption of aldehyde dehydrogenase 2 protects against bacterial infection. bioRxiv 2023. [Google Scholar] [CrossRef]
- Oota, H.; Pakstis, A.J.; Bonne-Tamir, B.; Goldman, D.; Grigorenko, E.; Kajuna, S.L.; Karoma, N.J.; Kungulilo, S.; Lu, R.B.; Odunsi, K.; et al. The evolution and population genetics of the ALDH2 locus: Random genetic drift, selection, and low levels of recombination. Ann. Hum. Genet. 2004, 68, 93–109. [Google Scholar] [CrossRef]
- Deiana, G.; Sun, R.; Huang, J.; Napolioni, V.; Ciccocioppo, R. Infection burden and ALDH2 rs671, East Asian genetic diversity: A reply. Alcohol. Clin. Exp. Res. 2024. Early View. [Google Scholar] [CrossRef]
- Deiana, G.; Sun, R.; Huang, J.; Napolioni, V.; Ciccocioppo, R. Contribution of infectious diseases to the selection of ADH1B and ALDH2 gene variants in Asian populations. Alcohol Clin. Exp. Res. 2024, 48, 855–866. [Google Scholar] [CrossRef]
- Moon, K.H.; Hood, B.L.; Kim, B.J.; Hardwick, J.P.; Conrads, T.P.; Veenstra, T.D.; Song, B.J. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology 2006, 44, 1218–1230. [Google Scholar] [CrossRef]
- Lee, S.L.; Lee, Y.P.; Wu, M.L.; Chi, Y.C.; Liu, C.M.; Lai, C.L.; Yin, S.J. Inhibition of human alcohol and aldehyde dehydrogenases by aspirin and salicylate: Assessment of the effects on first-pass metabolism of ethanol. Biochem. Pharmacol. 2015, 95, 71–79. [Google Scholar] [CrossRef]
- Lee, Y.P.; Liao, J.T.; Cheng, Y.W.; Wu, T.L.; Lee, S.L.; Liu, J.K.; Yin, S.J. Inhibition of human alcohol and aldehyde dehydrogenases by acetaminophen: Assessment of the effects on first-pass metabolism of ethanol. Alcohol 2013, 47, 559–565. [Google Scholar] [CrossRef]
- Landin, J.S.; Cohen, S.D.; Khairallah, E.A. Identification of a 54-kDa mitochondrial acetaminophen-binding protein as aldehyde dehydrogenase. Toxicol. Appl. Pharmacol. 1996, 141, 299–307. [Google Scholar] [CrossRef]
- Liu, M.; Yang, Z.; Wu, Q.; Yang, Y.; Zhao, D.; Cheng, Q.; Li, Y.; Liu, G.; Zhao, C.; Pan, J.; et al. IL-4-secreting CD40L(+) MAIT cells support antibody production in the peripheral blood of Heonch-Schönlein purpura patients. Inflamm. Res. 2024, 73, 35–46. [Google Scholar] [CrossRef]


| Subcohort 1 University Employees and Students | Subcohort 2 University Students | |||||
|---|---|---|---|---|---|---|
| n | 26 | 19 | ||||
| Type of Vaccine | mRNA-1273 × 2 BNT162b2 × 1 | BNT162b2 × 3 | ||||
| ALDH2 rs671 | GG | GA | AA | GG | GA | AA |
| Males, n | 3 | 7 | 1 | 10 | 4 | 5 |
| Females, n | 7 | 6 | 2 | 0 | 0 | 0 |
| Age, years | ||||||
| Median | 39 | 47 | 21 | 23 | 23 | 22 |
| (IQR) | (22–56) | (21–55) | (21–42) | (22–23) | (23–24) | (22–23) |
| Body height, cm | ||||||
| Median | 162 | 166 | 157 | 171 | 173 | 172 |
| (IQR) | (155–166) | (163–170) | (151–163) | (167–175) | (167–178) | (170–173) |
| Cigarette smoke, yes | 0 | 1 | 0 | 0 | 0 | 0 |
| Ethanol intake * | ||||||
| <1 g/d | 6 (60%) | 9 (69%) | 3 (100%) | 2 (20%) | 2 (50%) | 4 (80%) |
| ≥1, <20 g/d | 4 (40%) | 2 (15%) | 0 (0%) | 8 (80%) | 2 (50%) | 1 (20%) |
| ≥20 g/d | 0 (0%) | 2 (15%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Exercise habit | ||||||
| No habit | 3 (30%) | 4 (31%) | 1 (33%) | 1 (10%) | 1 (25%) | 2 (40%) |
| <1 d/w | 2 (20%) | 2 (15%) | 0 (0%) | 4 (40%) | 1 (25%) | 1 (20%) |
| 1 to 3 d/w | 2 (20%) | 4 (31%) | 1 (33%) | 2 (20%) | 2 (50%) | 2 (40%) |
| ≥3 d/w | 3 (30%) | 3 (23%) | 1 (33%) | 3 (30%) | 0 (0%) | 0 (0%) |
| Perceived stress | ||||||
| 0 (no) | 1 (10%) | 5 (38%) | 1 (33%) | 5 (50%) | 1 (25%) | 1 (20%) |
| 1 | 3 (30%) | 1 (8%) | 0 (0%) | 1 (10%) | 1 (25%) | 0 (0%) |
| 2 | 2 (20%) | 2 (15%) | 0 (0%) | 1 (10%) | 2 (50%) | 3 (60%) |
| 3 | 4 (40%) | 4 (31%) | 0 (0%) | 3 (30%) | 0 (0%) | 1 (20%) |
| 4 (yes) | 0 (0%) | 1 (8%) | 2 (67%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Steroid use, yes | 0 (0%) | 0 (0%) | 0 (0%) | 2 (20%) | 0 (0%) | 0 (0%) |
| Allergic disease, yes | 2 (20%) | 6 (46%) | 1 (33%) | 5 (50%) | 1 (25%) | 1 (20%) |
| AIC = 486 | AIC = 498 | |||
|---|---|---|---|---|
| 183 Datapoints (n = 45) | 183 Datapoints (n = 45) | |||
| Fixed Effect | β | p-Value | β | p-Value |
| BNT162b2 (reference) | ||||
| mRNA-1273 | −0.58 | 0.067 | −0.64 | 0.065 |
| Age (per 10 years) | 0.08 | 0.108 | 0.09 | 0.137 |
| Female sex | 0.10 | 0.536 | 0.24 | 0.358 |
| Height (per 10 cm) | 0.04 | 0.802 | ||
| Tobacco smoking, yes | −0.52 | 0.294 | ||
| Ethanol intake (per category) | 0.10 | 0.500 | ||
| Exercise (per category) | 0.07 | 0.337 | ||
| Perceived stress (per category) | 0.02 | 0.767 | ||
| Steroid use, yes | 0.17 | 0.759 | ||
| Allergic diseases | −0.22 | 0.185 | ||
| Number of rs671 variant allele | 0.27 | 0.010 | 0.30 | 0.007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogahawaththa, S.; Hara, M.; Furukawa, T.; Iwasaka, C.; Sawada, T.; Yamada, G.; Tokiya, M.; Kitagawa, K.; Miyake, Y.; Kido, M.A.; et al. Asian Flush Gene Variant Enhances Cellular Immunogenicity of COVID-19 Vaccine: Prospective Observation in the Japanese General Population. Vaccines 2024, 12, 1015. https://doi.org/10.3390/vaccines12091015
Bogahawaththa S, Hara M, Furukawa T, Iwasaka C, Sawada T, Yamada G, Tokiya M, Kitagawa K, Miyake Y, Kido MA, et al. Asian Flush Gene Variant Enhances Cellular Immunogenicity of COVID-19 Vaccine: Prospective Observation in the Japanese General Population. Vaccines. 2024; 12(9):1015. https://doi.org/10.3390/vaccines12091015
Chicago/Turabian StyleBogahawaththa, Sudarma, Megumi Hara, Takuma Furukawa, Chiharu Iwasaka, Takeshi Sawada, Goki Yamada, Mikiko Tokiya, Kyoko Kitagawa, Yasunobu Miyake, Mizuho Aoki Kido, and et al. 2024. "Asian Flush Gene Variant Enhances Cellular Immunogenicity of COVID-19 Vaccine: Prospective Observation in the Japanese General Population" Vaccines 12, no. 9: 1015. https://doi.org/10.3390/vaccines12091015
APA StyleBogahawaththa, S., Hara, M., Furukawa, T., Iwasaka, C., Sawada, T., Yamada, G., Tokiya, M., Kitagawa, K., Miyake, Y., Kido, M. A., Hirota, Y., & Matsumoto, A. (2024). Asian Flush Gene Variant Enhances Cellular Immunogenicity of COVID-19 Vaccine: Prospective Observation in the Japanese General Population. Vaccines, 12(9), 1015. https://doi.org/10.3390/vaccines12091015






