Enhanced Immune Responses with Serum Proteomic Analysis of Hu Sheep to Foot-and-Mouth Disease Vaccine Emulsified in a Vegetable Oil Adjuvant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Vaccine Preparation
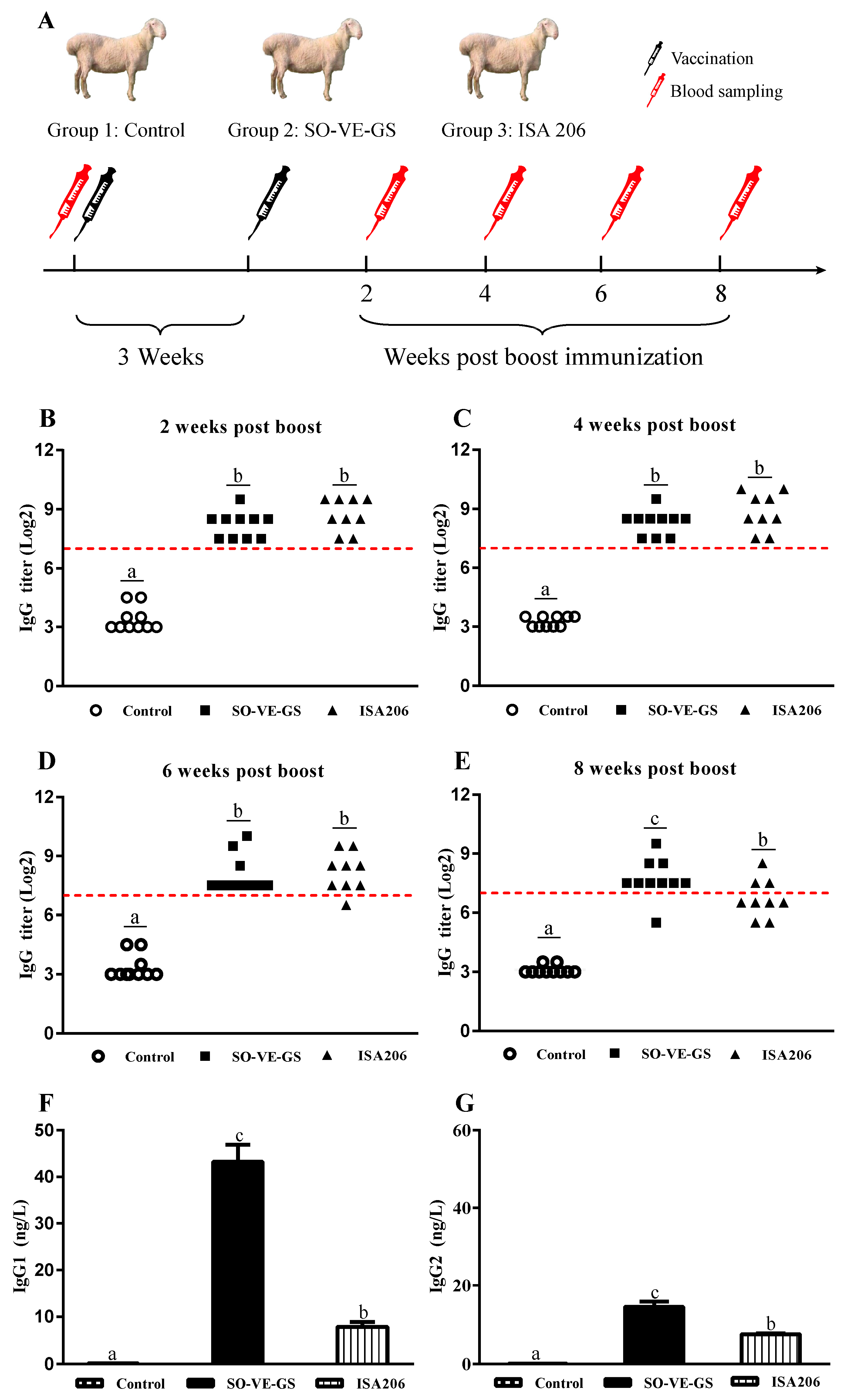
2.3. Immunization and Sampling
2.4. Analysis of FMDV-Specific Antibody and Isotypes
2.5. Serum Neutralizing Antibody Test
2.6. Determination of Serum Cytokine Levels
2.7. Proteomic Analysis
2.8. Differential Expression Proteins (DEPs) Analysis
2.9. Statistical Analysis
3. Results
3.1. FMDV-Specific Antibody Response
3.2. Serum Neutralizing Antibody Assay
3.3. Cytokines Assay
3.4. Protein Identification and Quantitation
3.5. Differential Expression Proteins (DEPs) Analysis
3.6. Gene Ontology (GO) Enrichment Analysis of DEPs
3.7. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of DEPs
3.8. Interaction Network of DEPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maree, F.F.; Nsamba, P.; Mutowembwa, P.; Rotherham, L.S.; Esterhuysen, J.; Scott, K. Intra-serotype SAT2 chimeric foot-and-mouth disease vaccine protects cattle against FMDV challenge. Vaccine 2015, 33, 2909–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Yu, X.; Zheng, Q.; Hou, L.; Du, L.; Zhang, Y.; Qiao, X.; Hou, J.; Huang, K. The immunopotentiator CVC1302 enhances immune efficacy and protective ability of foot-and-mouth disease virus vaccine in pigs. Vaccine 2018, 36, 7929–7935. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.; El-Sayed, A.; Castañeda Vazquez, H. Foot-and-mouth disease vaccines: recent updates and future perspectives. Arch. Virol. 2019, 164, 1501–1513. [Google Scholar] [CrossRef]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Li, H.; Qi, X.; Ma, Y.; Yang, B.; Zhang, S.; Chang, H.; Yin, X.; Li, Z. Immunogenicity and protective efficacy of a novel foot-and-mouth disease virus empty-capsid-like particle with improved acid stability. Vaccine 2019, 37, 2016–2025. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.-K.; Jo, H.-E.; Choi, J.-H.; You, S.-H.; Shin, S.H.; Jo, H.; Lee, M.-J.; Kim, S.-M.; Kim, B.; Park, J.-H. Chimeric vaccine strain of type O foot-and-mouth disease elicits a strong immune response in pigs against ME-SA and SEA topotypes. Vet. Microbiol. 2019, 229, 124–129. [Google Scholar] [CrossRef]
- Mahapatra, M.; Parida, S. Foot and mouth disease vaccine strain selection: current approaches and future perspectives. Expert Rev. Vaccines 2018, 17, 577–591. [Google Scholar] [CrossRef]
- OIE. Manual of Diagnositic Tests and Vaccines for Terrestrial Animals (Terrestrial Manual); OIE: Paris, France, 2019. [Google Scholar]
- Feng, H.; Fan, J.; Qiu, H.; Wang, Z.; Yan, Z.; Yuan, L.; Guan, L.; Du, X.; Song, Z.; Han, X.; et al. Chuanminshen violaceum polysaccharides improve the immune responses of foot-and-mouth disease vaccine in mice. Int. J. Biol. Macromol. 2015, 78, 405–416. [Google Scholar] [CrossRef]
- Cai, C.; Li, H.; Edwards, J.; Hawkins, C.; Robertson, I.D. Meta-analysis on the efficacy of routine vaccination against foot and mouth disease (FMD) in China. Prev. Vet. Med. 2014, 115, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Depa, P.M.; Dimri, U.; Sharma, M.C.; Tiwari, R. Update on epidemiology and control of foot and mouth disease-a menace to international trade and global animal enterprise. Vet. World 2012, 5, 694–704. [Google Scholar] [CrossRef]
- Terhuja, M.; Saravanan, P.; Tamilselvan, R.P. Comparative efficacy of virus like particle (VLP) vaccine of foot-and-mouth-disease virus (FMDV) type O adjuvanted with poly I:C or CpG in guinea pigs. Biologicals 2015, 43, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.M.; Cox, S.J.; Aggarwal, N.; Mackay, D.J.; Davies, P.R.; Hamblin, P.A.; Dani, P.; Barnett, P.V.; Paton, D.J. Detection of antibody to the foot-and-mouth disease virus (FMDV) non-structural polyprotein 3ABC in sheep by ELISA. J. Virol. Methods 2005, 125, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Alexandersen, S.; Zhang, Z.; Donaldson, A.; Garland, A.J. The pathogenesis and diagnosis of Foot-and-Mouth Disease. J. Comp. Pathol. 2003, 129, 1–36. [Google Scholar] [CrossRef]
- Arzt, J.; Juleff, N.; Zhang, Z.; Rodriguez, L.L. The Pathogenesis of Foot-and-Mouth Disease I: Viral Pathways in Cattle. Transbound. Emerg. Dis. 2011, 58, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Pacheco, J.M.; Singanallur, N.B.; de Carvalho Ferreira, H.C.; Vosloo, W.; Rodriguez, L.L.; Arzt, J. Clinical and virological dynamics of a serotype O 2010 South East Asia lineage foot-and-mouth disease virus in sheep using natural and simulated natural inoculation and exposure systems. Vet. Microbiol. 2015, 178, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Horsington, J.; Zhang, Z.; Bittner, H.; Hole, K.; Singanallur, N.B.; Alexandersen, S.; Vosloo, W. Early protection in sheep against intratypic heterologous challenge with serotype O foot-and-mouth disease virus using high-potency, emergency vaccine. Vaccine 2015, 33, 422–429. [Google Scholar] [CrossRef]
- Cao, Y.; Lu, Z.; Liu, Z. Foot-and-mouth disease vaccines: progress and problems. Expert Rev. Vaccines 2016, 15, 783–789. [Google Scholar] [CrossRef]
- de Bravo Rueda, C.; de Jong, M.C.; Eblé, P.L.; Dekker, A. Estimation of the transmission of foot-and-mouth disease virus from infected sheep to cattle. Vet. Res. 2014, 45, 58. [Google Scholar] [CrossRef] [Green Version]
- de Bravo Rueda, C.; Dekker, A.; Eblé, P.L.; de Jong, M.C.M. Identification of factors associated with increased excretion of foot-and-mouth disease virus. Prev. Vet. Med. 2014, 113, 23–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFadden, A.M.J.; Tsedenkhuu, P.; Bold, B.; Purevsuren, B.; Bold, D.; Morris, R. Epidemiology of the 2010 Outbreak of Foot-and-Mouth Disease in Mongolia. Transbound. Emerg. Dis. 2015, 62, e45–e51. [Google Scholar] [CrossRef]
- Ferguson, N.M. The Foot-and-Mouth Epidemic in Great Britain: Pattern of Spread and Impact of Interventions. Science 2001, 292, 1155–1160. [Google Scholar] [CrossRef] [Green Version]
- Diaz-San Segundo, F.; Medina, G.N.; Stenfeldt, C.; Arzt, J.; de los Santos, T. Foot-and-mouth disease vaccines. Vet. Microbiol. 2017, 206, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Adjuvants for foot-and-mouth disease virus vaccines: Recent progress. Expert Rev. Vaccines 2014, 13, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Guo, Y.; Fan, Y.; Nie, H.; Wang, R.; Wang, F. Metabolic profiling of stages of healthy pregnancy in Hu sheep using nuclear magnetic resonance (NMR). Theriogenology 2017, 92, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, P.; He, X.; Lu, T.; Guan, W.; Ma, Y. Attempt at conserving the genetic resources of Hu sheep by fibroblast line cryopreservation. J. Appl. Anim. Res. 2014, 42, 352–355. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Sun, W.; Yin, J.; Ni, R.; Su, R.; Wang, Q.; Gao, W.; Bao, J.; Yu, J.; Wang, L.; et al. An integrated analysis of microRNA and mRNA expression profiles to identify RNA expression signatures in lambskin hair follicles in Hu sheep. PLoS ONE 2016, 11, 1–16. [Google Scholar] [CrossRef]
- Ding, Y.-Z.; Chen, H.-T.; Zhang, J.; Zhou, J.-H.; Ma, L.-N.; Zhang, L.; Gu, Y.; Liu, Y.-S. An overview of control strategy and diagnostic technology for foot-and-mouth disease in China. Virol. J. 2013, 10, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Patil, P.K.; Bayry, J.; Ramakrishna, C.; Hugar, B.; Misra, L.D.; Prabhudas, K.; Natarajan, C. Immune Responses of Sheep to Quadrivalent Double Emulsion Foot-and-Mouth Disease Vaccines: Rate of Development of Immunity and Variations among Other Ruminants. J. Clin. Microbiol. 2002, 40, 4367–4371. [Google Scholar] [CrossRef] [Green Version]
- Madhanmohan, M.; Nagendrakumar, S.B.; Srinivasan, V.A. Protection against direct in-contact challenge following foot-and-mouth disease vaccination in sheep and goats: the effect on virus excretion and carrier status. Vet. Res. Commun. 2010, 34, 285–299. [Google Scholar] [CrossRef]
- Cox, S.J.; Barnett, P.V.; Dani, P.; Salt, J.S. Emergency vaccination of sheep against foot-and-mouth disease: protection against disease and reduction in contact transmission. Vaccine 1999, 17, 1858–1868. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.; Maqbool, B.; Yuan, L.; He, S.; Zhang, C.; Xu, W.; Hu, S. Early IgG Response to Foot and Mouth Disease Vaccine Formulated with a Vegetable Oil Adjuvant. Vaccines 2019, 7, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, S.; Didierlaurent, A.; Bourguignon, P.; Delhaye, S.; Baras, B.; Jacob, V.; Planty, C.; Elouahabi, A.; Harvengt, P.; Carlsen, H. Adjuvant System AS03 containing α-tocopherol modulates innate immune response and leads to improved adaptive immunity. Vaccine 2011, 29, 2461–2473. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Akalkotkar, A.; Bivona, J.J.; Lee, J.Y.; Park, Y.K.; Yu, M.; Colpitts, S.L.; Vajdy, M. Vitamin A or E and a catechin synergize as vaccine adjuvant to enhance immune responses in mice by induction of early interleukin-15 but not interleukin-1β responses. Immunology 2016, 148, 352–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, I.; Borggren, M.; Nielsen, J.; Christensen, D.; Williams, J.; Fomsgaard, A. Increased humoral immunity by DNA vaccination using an α-tocopherol-based adjuvant. Hum. Vaccin. Immunother. 2017, 13, 1823–1830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Feng, X.; Jin, Y.; Ma, J.; Cai, H.; Zhang, X. Immunoprotective mechanisms in swine within the “grey zone” in antibody response after immunization with foot-and-mouth disease vaccine. Virus Res. 2016, 220, 39–46. [Google Scholar] [CrossRef]
- Hendrix, C.M. Manual of Standards for Diagnostic Tests and Vaccines. J. Parasitol. 1997, 83, 605. [Google Scholar] [CrossRef]
- Reed, L.; Muench, H. A simple method of estimating fifty percent endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Qu, Y.; Zhou, B.; Yang, W.; Han, B.; Yu-Rice, Y.; Gao, B.; Johnson, J.; Svendsen, C.N.; Freeman, M.R.; Giuliano, A.E.; et al. Transcriptome and proteome characterization of surface ectoderm cells differentiated from human iPSCs. Sci. Rep. 2016, 6, 32007. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Wu, Q.; Zhou, B.; Choi, M.Y.; Ding, B.; Yang, W.; Dong, M. Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex. Cell Rep. 2019, 28, 2517–2526. [Google Scholar] [CrossRef]
- Yang, J.; Wang, G.-Q.; Zhou, Q.; Lu, W.; Ma, J.-Q.; Huang, J.-H. Transcriptomic and proteomic response of Manihot esculenta to Tetranychus urticae infestation at different densities. Exp. Appl. Acarol. 2019, 78, 273–293. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Chi, X.; Ma, X.; Xu, W.; Shi, F.; Hu, S. Anti-inflammatory mechanism of ginsenoside Rg1: Proteomic analysis of milk from goats with mastitis induced with lipopolysaccharide. Int. Immunopharmacol. 2019, 71, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-X.; Yan, R.; Shi, H.-Y.; Shi, D.; Fang, D.-Q.; Jiang, H.-Y.; Wu, W.-R.; Guo, F.-F.; Jiang, X.-W.; Gu, S.-L.; et al. Integrated transcriptomic and proteomic analysis of the bile stress response in probiotic Lactobacillus salivarius LI01. J. Proteom. 2017, 150, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Wang, Z.; Chen, X.; Yu, J.; Li, Z.; Nie, Q. Proteomic analysis of chicken skeletal muscle during embryonic development. Front. Physiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dar, P.; Kalaivanan, R.; Sied, N.; Mamo, B.; Kishore, S.; Suryanarayana, V.V.S.; Kondabattula, G. Montanide ISATM 201 adjuvanted FMD vaccine induces improved immune responses and protection in cattle. Vaccine 2013, 31, 3327–3332. [Google Scholar] [CrossRef]
- Xiao, C.; Rajput, Z.I.; Hu, S. Improvement of a commercial foot-and-mouth disease vaccine by supplement of Quil A. Vaccine 2007, 25, 4795–4800. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, C.; She, D.; Li, P.; Sun, P.; Bai, X.; Chen, Y.; Xie, B.; Liu, Z. The comparison of the efficacy of swine FMD vaccine emulsified with oil adjuvant of ISA 201 VG or ISA 206 VG. J. Biosci. Med. 2013, 1, 22–25. [Google Scholar] [CrossRef] [Green Version]
- Parida, S. Vaccination against foot-and-mouth disease virus: Strategies and effectiveness. Expert Rev. Vaccines 2009, 8, 347–365. [Google Scholar] [CrossRef]
- Doel, T.R. Natural and Vaccine Induced Immunity to FMD. In Foot-and-Mouth Disease Virus; Springer: Berlin/Heidelberg, Germany, 2005; pp. 103–131. [Google Scholar]
- Selim, A.M.A.; Abouzeid, N.Z.; Aggour, A.M.; Sobhy, N.M. Comparative study for immune efficacy of two different adjuvants bivalent FMD vaccines in sheep. J. Am. Sci. 2010, 6, 1292–1298. [Google Scholar]
- Zhang, Y.; Dominguez-Medina, C.; Cumley, N.J.; Heath, J.N.; Essex, S.J.; Bobat, S.; Schager, A.; Goodall, M.; Kracker, S.; Buckley, C.D.; et al. IgG1 Is Required for Optimal Protection after Immunization with the Purified Porin OmpD from Salmonella Typhimurium. J. Immunol. 2017, 199, 4103–4109. [Google Scholar] [CrossRef]
- Mailybayeva, A.; Yespembetov, B.; Ryskeldinova, S.; Zinina, N.; Sansyzbay, A.; Renukaradhya, G.J.; Petrovsky, N.; Tabynov, K. Improved influenza viral vector based Brucella abortus vaccine induces robust B and T-cell responses and protection against Brucella melitensis infection in pregnant sheep and goats. PLoS ONE 2017, 12, e0186484. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Pérez, J.M.; Robles-Pérez, D.; Rojo-Vázquez, F.A.; Martínez-Valladares, M. Immunological features of LPS from Ochrobactrum intermedium on sheep experimentally infected with Fasciola hepatica. Res. Vet. Sci. 2014, 97, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.S.; Altmann, K.; Cross, M.L.; Husband, A.J. Induction of T helper 1- and T helper 2-type immune responses duringHaemonchus contortus infection in sheep. Immunology 2000, 99, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Schallig, H.D.F.H. Immunological responses of sheep to Haemonchus contortus. Parasitology 2000, 120, 63–72. [Google Scholar] [CrossRef]
- Zhang, Q.; Lou, Y.; Yang, J.; Wang, J.; Feng, J.; Zhao, Y.; Wang, L.; Huang, X.; Fu, Q.; Ye, M.; et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 2019, 68, 2019–2031. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Zhao, H.; Qin, X. Ovarian proteomic study reveals the possible molecular mechanism for hyperprolificacy of Small Tail Han sheep. Sci. Rep. 2016, 6, 27606. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shao, C.; Xu, W.; Zhou, Q.; Wang, N.; Chen, S. Proteome profiling reveals immune responses in Japanese flounder ( Paralichthys olivaceus ) infected with Edwardsiella tarda by iTRAQ analysis. Fish Shellfish Immunol. 2017, 66, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Li, Y.; Yu, K.; Huang, B.; Ma, X.; Liu, C.; Guo, X.; Song, M.; Wu, J. ITRAQ-Based Quantitative Proteomics Reveals the Proteome Profiles of Primary Duck Embryo Fibroblast Cells Infected with Duck Tembusu Virus. Biomed. Res. Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Li, L.; Wu, J.; Cao, X.; Zhou, J.; Yin, S.; Yang, S.; Feng, Q.; Du, P.; Liu, Y.; Shang, Y.; et al. Proteomic analysis of murine bone marrow derived dendritic cells in response to peste des petits ruminants virus. Res. Vet. Sci. 2019, 125, 195–204. [Google Scholar] [CrossRef]
- Jia, J.; Liu, X.; Li, L.; Lei, C.; Dong, Y.; Wu, G.; Hu, G. Transcriptional and translational relationship in environmental stress: RNAseq and ITRAQ proteomic analysis between sexually reproducing and parthenogenetic females in Moina micrura. Front. Physiol. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Lo, U.; Selvaraj, V.; Plane, J.M.; Chechneva, O.V.; Otsu, K.; Deng, W. p38α (MAPK14) critically regulates the immunological response and the production of specific cytokines and chemokines in astrocytes. Sci. Rep. 2015, 4, 7405. [Google Scholar] [CrossRef] [Green Version]
- Deak, M. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activationofCREB. EMBO J. 1998, 17, 4426–4441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Yu, J.; Wang, H.; Liu, B.; Yue, X. p38 MAPK is involved in the immune response to pathogenic Vibrio in the clam Meretrix petechialis. Fish Shellfish Immunol. 2019, 95, 456–463. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yao, W.; Liu, P.; Li, J.; Wang, Q. Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp Fenneropenaeus chinensis in response to viral and bacterial infections. Gene 2018, 642, 381–388. [Google Scholar] [CrossRef]
- Ma, F.; Chang, X.; Wang, G.; Zhou, H.; Ma, Z.; Lin, H.; Fan, H. Streptococcus Suis Serotype 2 Stimulates Neutrophil Extracellular Traps Formation via Activation of p38 MAPK and ERK1/2. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Lin, J.T.; Martin, S.L.; Xia, L.; Gorham, J.D. TGF-β1 Uses Distinct Mechanisms to Inhibit IFN-γ Expression in CD4 + T Cells at Priming and at Recall: Differential Involvement of Stat4 and T-bet. J. Immunol. 2005, 174, 5950–5958. [Google Scholar] [CrossRef] [Green Version]






| Group | Animal | SN Antibody Titer | Log10 SN Titer |
|---|---|---|---|
| Control | 1-1 | <1:16 | <1.20 |
| 1-2 | <1:16 | <1.20 | |
| 1-3 | <1:16 | <1.20 | |
| 1-4 | <1:16 | <1.20 | |
| 1-5 | < 1:16 | <1.20 | |
| 1-6 | < 1:16 | <1.20 | |
| 1-7 | <1:16 | <1.20 | |
| 1-8 | <1:16 | <1.20 | |
| 1-9 | <1:16 | <1.20 | |
| 1-10 | <1:16 | <1.20 | |
| SO-VE-GS | 2-1 | 1:190 | 2.27 |
| 2-2 | 1:256 | 2.40 | |
| 2-3 | 1:177 | 2.24 | |
| 2-4 | 1:273 | 2.43 | |
| 2-5 | 1:97 | 1.98 | |
| 2-6 | 1:215 | 2.33 | |
| 2-7 | 1:328 | 2.51 | |
| 2-8 | 1:49 | 1.69 | |
| 2-9 | 1:156 | 2.19 | |
| 2-10 | 1:153 | 1.72 | |
| ISA 206 | 3-1 | <1:16 | <1.20 |
| 3-2 | <1:16 | <1.20 | |
| 3-3 | <1:16 | <1.20 | |
| 3-4 | <1:16 | <1.20 | |
| 3-5 | <1:16 | <1.20 | |
| 3-6 | <1:16 | <1.20 | |
| 3-7 | <1:16 | <1.20 | |
| 3-8 | <1:16 | <1.20 | |
| 3-9 | <1:16 | <1.20 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Wang, Y.; Guan, R.; Lu, M.; Yuan, L.; Xu, W.; Hu, S. Enhanced Immune Responses with Serum Proteomic Analysis of Hu Sheep to Foot-and-Mouth Disease Vaccine Emulsified in a Vegetable Oil Adjuvant. Vaccines 2020, 8, 180. https://doi.org/10.3390/vaccines8020180
Cui X, Wang Y, Guan R, Lu M, Yuan L, Xu W, Hu S. Enhanced Immune Responses with Serum Proteomic Analysis of Hu Sheep to Foot-and-Mouth Disease Vaccine Emulsified in a Vegetable Oil Adjuvant. Vaccines. 2020; 8(2):180. https://doi.org/10.3390/vaccines8020180
Chicago/Turabian StyleCui, Xuemei, Yong Wang, Ran Guan, Meiqian Lu, Lijia Yuan, Wei Xu, and Songhua Hu. 2020. "Enhanced Immune Responses with Serum Proteomic Analysis of Hu Sheep to Foot-and-Mouth Disease Vaccine Emulsified in a Vegetable Oil Adjuvant" Vaccines 8, no. 2: 180. https://doi.org/10.3390/vaccines8020180





