Gene Augmentation and Editing to Improve TCR Engineered T Cell Therapy against Solid Tumors
Abstract
1. Advantages and Disadvantages of Using rTCR-T Cells for the Treatment of Solid Tumors: From the Bench to the Bedside
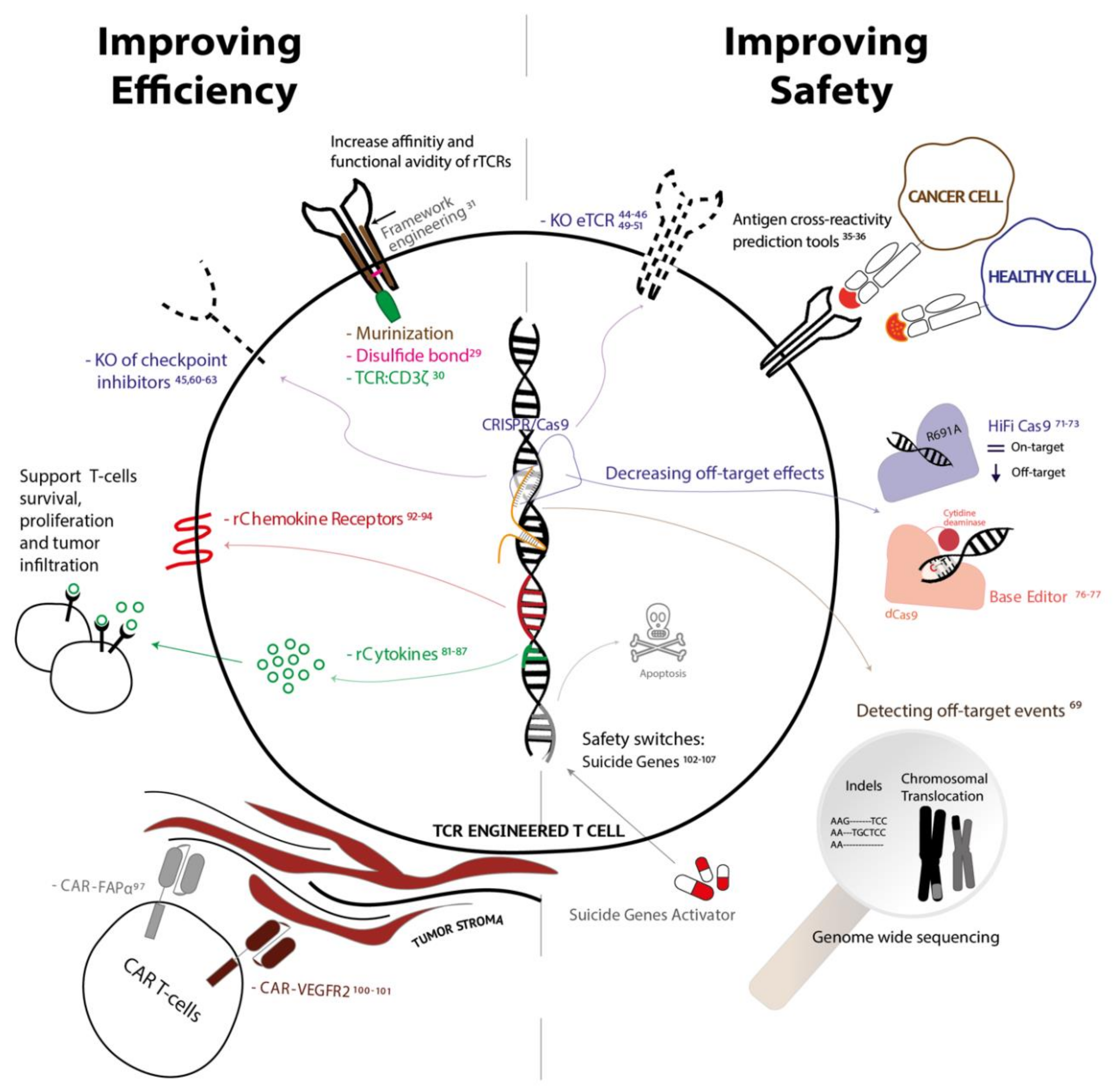
2. Increasing Affinity and Functional Avidity of rTCRs While Maintaining a Safe Profile
3. Genetic Elimination of Endogenous TCR to Improve Efficacy and Safety
4. Disrupting Inhibitory Pathways to Prevent Exhaustion
5. Risks of Using Gene-Editing Techniques
6. Incorporating Cytokines to Enhance T Cell Proliferation
7. Introduction of Chemokines Receptors to Promote Migration and Infiltration
8. Targeting the Tumor Surrounding Stroma
9. Incorporation of Suicide Genes to Safeguard Off-Target Toxicities
10. Conclusions
Funding
Conflicts of Interest
References
- Yang, Y. Cancer immunotherapy: Harnessing the immune system to battle cancer Find the latest version: Cancer immunotherapy: Harnessing the immune system to battle cancer. J. Clin. Investig. 2015, 125, 3335–3337. [Google Scholar] [CrossRef] [PubMed]
- Hargadon, K.M.; Johnson, C.E.; Williams, C.J. Immune checkpoint blockade therapy for cancer: An overview of FDA-approved immune checkpoint inhibitors. Int. Immunopharmacol. 2018, 62, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Vétizou, M.; Daillère, R.; Roberti, M.P.; Yamazaki, T.; Routy, B.; Lepage, P.; Boneca, I.G.; Chamaillard, M.; Kroemer, G.; et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016, 44, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Dudley, M.E.; Rosenberg, S.A. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat. Rev. Cancer 2003, 3, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Dudley, M.E.; Wunderlich, J.R.; Robbins, P.F.; Yang, J.C.; Hwu, P.; Schwartzentruber, D.J.; Topalian, S.L.; Sherry, R.; Restifo, N.P.; Hubicki, A.M.; et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002, 298, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.; Zhou, W.L.; Huang, Y.; Liang, X.; Jiang, L.; Yang, X.; Sun, J.; Li, Z.; Han, W.D.; et al. Genetically engineered t cells for cancer immunotherapy. Signal. Transduct. Target. Ther. 2019, 4. [Google Scholar] [CrossRef]
- Martinez, M.; Moon, E.K. CAR T cells for solid tumors: New strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 2019, 10, 1–21. [Google Scholar] [CrossRef]
- Bagley, S.J.; O’Rourke, D.M. Clinical investigation of CAR T cells for solid tumors: Lessons learned and future directions. Pharmacol. Ther. 2020, 205. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor iimune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Chen, N.; Li, X.; Chintala, N.K.; Tano, Z.E.; Adusumilli, P.S. Driving CARs on the uneven road of antigen heterogeneity in solid tumors. Curr. Opin. Immunol. 2018, 51, 103–110. [Google Scholar] [CrossRef]
- Garber, K. Driving T-cell immunotherapy to solid tumors. Nat. Biotechnol. 2018, 36, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.; Liu, C.; Zhang, Y. T-cell receptor-engineered T cells for cancer treatment: Current status and future directions. Protein Cell 2018, 9, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Debets, R.; Donnadieu, E.; Chouaib, S.; Coukos, G. TCR-engineered T cells to treat tumors: Seeing but not touching? Semin. Immunol. 2016, 28, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Fesnak, A.D.; June, C.H.; Levine, B.L. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 2016, 16, 566–581. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Jiang, X.; Zhou, X.; Weng, J. Targeting cancers through TCR-peptide/MHC interactions. J. Hematol. Oncol. 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Parkhurst, M.R.; Yang, J.C.; Langan, R.C.; Dudley, M.E.; Nathan, D.-A.N.; Feldman, S.A.; Davis, J.L.; Morgan, R.A.; Merino, M.J.; Sherry, R.M.; et al. T Cells Targeting Carcinoembryonic Antigen Can Mediate Regression of Metastatic Colorectal Cancer but Induce Severe Transient Colitis. Mol. Ther. 2011, 19, 620–626. [Google Scholar] [CrossRef]
- Morgan, R.A.; Chinnasamy, N.; Abate-daga, D.D.; Gros, A.; Robbins, F.; Zheng, Z.; Feldman, S.A.; Yang, J.C.; Sherry, R.M.; Phan, Q.; et al. Cancer regression and neurologic toxicity following anti-MAGE- A3 TCR gene therapy Richard. J. Immunother. 2014, 36, 133–151. [Google Scholar] [CrossRef]
- Linette, G.P.; Stadtmauer, E.A.; Maus, M.V.; Rapoport, A.P.; Levine, B.L.; Emery, L.; Litzky, L.; Bagg, A.; Carreno, B.M.; Cimino, P.J.; et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013, 122, 863–871. [Google Scholar] [CrossRef]
- Klebanoff, C.A.; Khong, H.T.; Antony, P.A.; Palmer, D.C.; Restifo, N.P. Sinks, suppressors and antigen presenters: How lymphodepletion enhances T cell-mediated tumor immunotherapy. Trends Immunol. 2005, 26, 111–117. [Google Scholar] [CrossRef]
- Seliger, B. Combinatorial Approaches with Checkpoint Inhibitors to Enhance Anti-tumor Immunity. Front. Immunol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Inderberg-Suso, E.M.; Trachsel, S.; Lislerud, K.; Rasmussen, A.M.; Gaudernack, G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. Oncoimmunology 2012, 1, 670–686. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Gartner, J.J.; Horovitz-Fried, M.; Shamalov, K.; Trebska-McGowan, K.; Bliskovsky, V.V.; Parkhurst, M.R.; Ankri, C.; Prickett, T.D.; Crystal, J.S.; et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Investig. 2015, 125, 3981–3991. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, C.; Heemskerk, B.; Kvistborg, P.; Kluin, R.J.C.; Bolotin, D.A.; Chen, X.; Bresser, K.; Nieuwland, M.; Schotte, R.; Michels, S.; et al. Technical Reports High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med. 2013, 19, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.K.; Sami, M.; Scott, D.R.; Rizkallah, P.J.; Borbulevych, O.Y.; Todorov, P.T.; Moysey, R.K.; Jakobsen, B.K.; Boulter, J.M.; Baker, B.M.; et al. Increased peptide contacts govern high affinity binding of a modified TCR whilst maintaining a native pMHC docking mode. Front. Immunol. 2013, 4, 1–8. [Google Scholar] [CrossRef]
- Malecek, K.; Grigoryan, A.; Zhong, S.; Gu, W.J.; Johnson, L.A.; Rosenberg, S.A.; Cardozo, T.; Krogsgaard, M. Specific Increase in Potency via Structure-Based Design of a TCR. J. Immunol. 2014, 193, 2587–2599. [Google Scholar] [CrossRef]
- Spear, T.T.; Wang, Y.; Foley, K.C.; Murray, D.C.; Scurti, G.M.; Simms, P.E.; Garrett-Mayer, E.; Hellman, L.M.; Baker, B.M.; Nishimura, M.I. Critical biological parameters modulate affinity as a determinant of function in T-cell receptor gene-modified T-cells. Cancer Immunol. Immunother. 2017, 66, 1411–1424. [Google Scholar] [CrossRef]
- Costa, C.; Hypolite, G.; Bernadin, O.; Lévy, C.; Cosset, F.-L.; Asnafi, V.; Macintyre, E.; Verhoeyen, E.; Tesio, M. Baboon envelope pseudotyped lentiviral vectors: A highly efficient new tool to genetically manipulate T-cell acute lymphoblastic leukaemia-initiating cells. Leukemia 2017, 31, 977–980. [Google Scholar] [CrossRef]
- Cavalieri, S.; Cazzaniga, S.; Geuna, M.; Magnani, Z.; Bordignon, C.; Naldini, L.; Bonini, C. Human T lymphocytes transduced by lentiviral vectors in the absence of TCR activation maintain an intact immune competence. Blood 2003, 102, 497–505. [Google Scholar] [CrossRef]
- Circosta, P.; Granziero, L.; Follenzi, A.; Vigna, E.; Stella, S.; Vallario, A.; Elia, A.R.; Gammaitoni, L.; Vitaggio, K.; Orso, F.; et al. T Cell Receptor (TCR) Gene Transfer with Lentiviral Vectors Allows Efficient Redirection of Tumor Specificity in Naive and Memory T Cells Without Prior Stimulation of Endogenous TCR. Hum. Gene Ther. 2009, 20, 1576–1588. [Google Scholar] [CrossRef]
- Kuball, J.; Dossett, M.L.; Wolfl, M.; Ho, W.Y.; Voss, R.H.; Fowler, C.; Greenberg, P.D. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood 2007, 109, 2331–2338. [Google Scholar] [CrossRef]
- Sebestyén, Z.; Schooten, E.; Sals, T.; Zaldivar, I.; José, E.S.; Alarcón, B.; Bobisse, S.; Szöllosi, J.; Gratama, J.W.; Ralph, A.; et al. Human TCR That Incorporate CD3 ζ Induce Highly Preferred Pairing between TCR α and β Chains following Gene Transfer. J. Immunol. 2020. [Google Scholar] [CrossRef]
- Thomas, S.; Mohammed, F.; Reijmers, R.M.; Woolston, A.; Stauss, T.; Kennedy, A.; Stirling, D.; Holler, A.; Green, L.; Jones, D.; et al. Framework engineering to produce dominant T cell receptors with enhanced antigen-specific function. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.C.; Guittard, G.C.; Franco, Z.; Crompton, J.G.; Eil, R.L.; Patel, S.J.; Ji, Y.; Van Panhuys, N.; Klebanoff, C.A.; Sukumar, M.; et al. Cish actively silences TCR signaling in CD8 + T cells to maintain tumor tolerance. J. Exp. Med. 2013, 2095–2113. [Google Scholar] [CrossRef]
- Duinkerken, C.W.; Rohaan, M.W.; De Weger, V.A.; Lohuis, P.J.F.M.; Latenstein, M.N.; Theunissen, E.A.R.; Balm, A.J.M.; Dreschler, W.A.; Haanen, J.B.A.G.; Zuur, C.L. Sensorineural Hearing Loss after Adoptive Cell Immunotherapy for Melanoma Using MART-1 Specific T Cells: A Case Report and Its Pathophysiology. Otol. Neurotol. 2019, 40, e674–e678. [Google Scholar] [CrossRef]
- Morgan, R.A.; Dudley, M.E.; Wunderlich, J.R.; Hughes, M.S.; Yang, J.C.; Sherry, R.M.; Royal, R.E.; Topalian, S.L.; Kammula, U.S.; Restifo, N.P.; et al. Cancer Regression in Patients After Transfer of Genetically Engineered Lymphocytes. Science 2006, 314, 126–129. [Google Scholar] [CrossRef]
- Bijen, H.M.; van der Steen, D.M.; Hagedoorn, R.S.; Wouters, A.K.; Wooldridge, L.; Falkenburg, J.H.F.; Heemskerk, M.H.M. Preclinical Strategies to Identify Off-Target Toxicity of High-Affinity TCRs. Mol. Ther. 2018, 26, 1206–1214. [Google Scholar] [CrossRef]
- Bentzen, A.K.; Hadrup, S.R. Evolution of MHC-based technologies used for detection of antigen-responsive T cells. Cancer Immunol. Immunother. 2017, 66, 657–666. [Google Scholar] [CrossRef]
- Shao, H.; Zhang, W.; Hu, Q.; Wu, F.; Shen, H.; Huang, S. TCR mispairing in genetically modified T cells was detected by fluorescence resonance energy transfer. Mol. Biol. Rep. 2010, 37, 3951–3956. [Google Scholar] [CrossRef]
- Bendle, G.M.; Linnemann, C.; Hooijkaas, A.I.; Bies, L.; de Witte, M.A.; Jorritsma, A.; Kaiser, A.D.M.; Pouw, N.; Debets, R.; Kieback, E.; et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 2010, 16, 565–570. [Google Scholar] [CrossRef]
- Legut, M.; Dolton, G.; Mian, A.A.; Ottmann, O.G.; Sewell, A.K. CRISPR-mediated TCR replacement generates superior anticancer transgenic t cells. Blood 2018, 131, 311–322. [Google Scholar] [CrossRef]
- Provasi, E.; Genovese, P.; Lombardo, A.; Magnani, Z.; Liu, P.-Q.; Reik, A.; Chu, V.; Paschon, D.E.; Zhang, L.; Kuball, J.; et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012, 18, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cheng, C.; Sun, W.; Wang, H. Engineering T Cells Using CRISPR/Cas9 for Cancer Therapy. In RNA Interference and CRISPR Technologies; Springer: Berlin, Germany, 2020; Volume 2115, pp. 503–509. [Google Scholar]
- Ren, J.; Zhang, X.; Liu, X.; Fang, C.; Jiang, S. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget 2017, 8, 17002–17011. [Google Scholar] [CrossRef] [PubMed]
- Morton, L.T.; Reijmers, R.M.; Wouters, A.K.; Kweekel, C.; Remst, D.F.G.; Pothast, C.R.; Falkenburg, J.H.F.; Heemskerk, M.H.M. Simultaneous Deletion of Endogenous TCRαβ for TCR Gene Therapy Creates an Improved and Safe Cellular Therapeutic. Mol. Ther. 2020, 28, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Qasim, W.; Zhan, H.; Samarasinghe, S.; Adams, S.; Amrolia, P.; Stafford, S.; Butler, K.; Rivat, C.; Wright, G.; Somana, K.; et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017, 9, eaaj2013. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Cheng, C.; Cheng, A.W.; Zhang, X.; Li, N.; Xia, C.; Wei, X.; Liu, X.; Wang, H. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017, 27, 154–157. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, 1–64. [Google Scholar] [CrossRef]
- Wang, J.; DeClercq, J.J.; Hayward, S.B.; Li, P.W.L.; Shivak, D.A.; Gregory, P.D.; Lee, G.; Holmes, M.C. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 2016, 44, 1–9. [Google Scholar] [CrossRef]
- Ortinski, P.I.; O’Donovan, B.; Dong, X.; Kantor, B. Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing. Mol. Ther. Methods Clin. Dev. 2017, 5. [Google Scholar] [CrossRef]
- Hale, M.; Lee, B.; Honaker, Y.; Leung, W.H.; Grier, A.E.; Jacobs, H.M.; Sommer, K.; Sahni, J.; Jackson, S.W.; Scharenberg, A.M.; et al. Homology-Directed Recombination for Enhanced Engineering of Chimeric Antigen Receptor T Cells. Mol. Ther. Methods Clin. Dev. 2017. [Google Scholar] [CrossRef]
- Eyquem, J.; Mansilla-soto, J.; Giavridis, T.; Van Der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nat. Publ. Gr. 2017, 543, 113–117. [Google Scholar] [CrossRef]
- MacLeod, D.T.; Antony, J.; Martin, A.J.; Moser, R.J.; Hekele, A.; Wetzel, K.J.; Brown, A.E.; Triggiano, M.A.; Hux, J.A.; Pham, C.D.; et al. Integration of a CD19 CAR into the TCR Alpha Chain Locus Streamlines Production of Allogeneic Gene-Edited CAR T Cells. Mol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Louis Jeune, V.; Joergensen, J.A.; Hajjar, R.J.; Weber, T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods 2013, 24, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Roth, T.L.; Puig-saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Repromming human T cell function and specificity with non-viral genome targeting. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Schober, K.; Müller, T.R.; Busch, D.H. Orthotopic T-Cell Receptor Replacement-An “Enabler” for TCR-Based Therapies. Cells 2020, 9, 1367. [Google Scholar] [CrossRef]
- Abate-Daga, D.; Hanada, K.I.; Davis, J.L.; Yang, J.C.; Rosenberg, S.A.; Morgan, R.A. Expression profiling of TCR-engineered T cells demonstrates overexpression of multiple inhibitory receptors in persisting lymphocytes. Blood 2013, 122, 1399–1410. [Google Scholar] [CrossRef]
- Datar, I.; Sanmamed, M.F.; Wang, J.; Henick, B.S.; Choi, J.; Badri, T.; Dong, W.; Mani, N.; Toki, M.; Mejías, L.D.; et al. Expression analysis and significance of PD-1, LAG-3, and TIM-3 in human non-small cell lung cancer using spatially resolved and multiparametric single-cell analysis. Clin. Cancer Res. 2019, 25, 4663–4673. [Google Scholar] [CrossRef]
- Li, X.; Wang, R.; Fan, P.; Yao, X.; Qin, L.; Peng, Y.; Ma, M.; Asley, N.; Chang, X.; Feng, Y.; et al. A comprehensive analysis of key immune checkpoint receptors on tumor-infiltrating t cells from multiple types of cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Moon, E.K.; Ranganathan, R.; Eruslanov, E.; Kim, S.; Newick, K.; O’Brien, S.; Lo, A.; Liu, X.; Zhao, Y.; Albelda, S.M. Blockade of programmed death 1 augments the ability of human T cells engineered to target NY-ESO-1 to control tumor growth after adoptive transfer. Clin. Cancer Res. 2016, 22, 436–447. [Google Scholar] [CrossRef]
- Cherkassky, L.; Morello, A.; Villena-Vargas, J.; Feng, Y.; Dimitrov, D.S.; Jones, D.R.; Sadelain, M.; Adusumilli, P.S. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Investig. 2016, 126, 3130–3144. [Google Scholar] [CrossRef]
- Rupp, L.J.; Schumann, K.; Roybal, K.T.; Gate, R.E.; Ye, C.J.; Lim, W.A.; Marson, A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017, 7, 737. [Google Scholar] [CrossRef]
- Su, S.; Hu, B.; Shao, J.; Shen, B.; Du, J.; Du, Y.; Zhou, J.; Yu, L. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Nat. Publ. Gr. 2016, 1–14. [Google Scholar] [CrossRef]
- Menger, L.; Sledzinska, A.; Bergerhoff, K.; Varga, F.A.; Smith, J.; Poirot, L.; Pule, M.; Hererro, J.; Peggs, K.S.; Quezada, S.A. TALEN-Mediated Inactivation of PD-1 in Tumor-Reactive Lymphocytes Promotes Intratumoral T-cell Persistence and Rejection of Established Tumors. Cancer Res. 2016, 76, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.D.; Yu, X.; Castano, A.P.; Darr, H.; Henderson, D.B.; Bouffard, A.A.; Larson, R.C.; Scarfò, I.; Bailey, S.R.; Gerhard, G.M.; et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J. Immunother. Cancer 2019, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McGowan, E.; Lin, Q.; Ma, G.; Yin, H.; Chen, S.; Lin, Y. PD-1 disrupted CAR-T cells in the treatment of solid tumors: Promises and challenges. Biomed. Pharmacother. 2020, 121, 109625. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, A.P.; Stadtmauer, E.A.; Binder-Scholl, G.K.; Goloubeva, O.; Vogl, D.T.; Lacey, S.F.; Badros, A.Z.; Garfall, A.; Weiss, B.; Finklestein, J.; et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 2015, 21, 914–921. [Google Scholar] [CrossRef]
- Ghosh, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. CRISPR–Cas9 a boon or bane: The bumpy road ahead to cancer therapeutics. Cancer Cell Int. 2019, 1–10. [Google Scholar] [CrossRef]
- Weber, J.; Öllinger, R.; Friedrich, M.; Ehmer, U.; Barenboim, M.; Steiger, K.; Heid, I. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl. Acad. Sci. USA 2015, 112. [Google Scholar] [CrossRef]
- Li, J.; Hong, S.; Chen, W.; Zuo, E.; Yang, H. Advances in detecting and reducing off-target effects generated by CRISPR-mediated genome editing. J. Genet. Genom. 2019, 46, 513–521. [Google Scholar] [CrossRef]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xue, J.; Deng, T.; Zhou, X.; Yu, K.; Deng, L.; Huang, M.; Yi, X.; Liang, M.; Wang, Y.; et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020, 26, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A-T to G-C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Webber, B.R.; Lonetree, C.; Kluesner, M.G.; Johnson, M.J.; Pomeroy, E.J.; Diers, M.D.; Lahr, W.S.; Draper, G.M.; Slipek, N.J.; Smeester, B.S.; et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Dwyer, C.J.; Knochelmann, H.M.; Smith, A.S.; Wyatt, M.M.; Rivera, G.O.R.; Arhontoulis, D.C.; Bartee, E.; Li, Z.; Rubinstein, M.P.; Paulos, C.M. Fueling Cancer Immunotherapy With Common Gamma Chain Cytokines. Front. Immmunol. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- White, D.E.; Rosenberg, S.A. Trends in the Safety of High Dose Bolus Interleukin-2 Administration in Patients with Metastatic Cancer. Cancer 1998, 2, 797–805. [Google Scholar]
- Tang, A.; Harding, F. The challenges and molecular approaches surrounding interleukin-2-based therapeutics in cancer. Cytokine X 2019, 1, 100001. [Google Scholar] [CrossRef]
- Liu, K.; Rosenberg, S.A. Transduction of an IL-2 Gene into Human Melanoma-Reactive Lymphocytes Results in Their Continued Growth in the Absence of Exogenous IL-2 and Maintenance of Specific Antitumor Activity. J. Immunol. 2001. [Google Scholar] [CrossRef]
- Heemskerk, B.; Liu, K.; Dudley, M.E.; Johnson, L.A.; Kaiser, A.; Downey, S.; Zheng, Z.; Shelton, T.E.; Matsuda, K.; Robbins, P.F.; et al. Adoptive cell therapy for patients with melanoma, using tumor-infiltrating lymphocytes genetically engineered to secrete interleukin-2. Hum. Gene Ther. 2008, 19, 496–510. [Google Scholar] [CrossRef]
- Kunert, A.; Chmielewski, M.; Wijers, R.; Berrevoets, C.; Abken, H.; Debets, R. Intra-tumoral production of IL18, but not IL12, by TCR-engineered T cells is non-toxic and counteracts immune evasion of solid tumors. Oncoimmunology 2018, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Alsaieedi, A.; Holler, A.; Velica, P.; Bendle, G.; Stauss, H.J. Safety and efficacy of Tet-regulated IL-12 expression in cancer-specific T cells. Oncoimmunology 2019, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Davies, J.S.; Serna, C.; Yu, Z.; Restifo, N.P.; Rosenberg, S.A.; Morgan, R.A.; Hinrichs, C.S. Enhanced efficacy and limited systemic cytokine exposure with membrane-anchored interleukin-12 T-cell therapy in murine tumor models. J. Immunother. Cancer 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Morgan, R.A.; Beane, J.D.; Zheng, Z.; Dudley, M.E.; Kassim, S.H.; Nahvi, A.V.; Ngo, L.T.; Sherry, R.M.; Phan, G.Q.; et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res. 2015, 21, 2278–2288. [Google Scholar] [CrossRef] [PubMed]
- Koneru, M.; O’Cearbhaill, R.; Pendharkar, S.; Spriggs, D.R.; Brentjens, R.J. A phase I clinical trial of adoptive T cell therapy using IL-12 secreting MUC-16ecto directed chimeric antigen receptors for recurrent ovarian cancer. J. Transl. Med. 2015, 13, 1–11. [Google Scholar] [CrossRef]
- Gonzales, C.; Yoshihara, H.A.I.; Dilek, N.; Leignadier, J.; Irving, M.; Mieville, P.; Helm, L.; Michielin, O.; Schwitter, J. In-vivo detection and tracking of T cells in various organs in a melanoma tumor model by 19F-fluorine MRS/MRI. PLoS ONE 2016, 11, 1–18. [Google Scholar] [CrossRef]
- Boschi, F.; De Sanctis, F.; Ugel, S.; Spinelli, A.E. T-cell tracking using Cerenkov and radioluminescence imaging. J. Biophotonics 2018, 11, 1–11. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Publ. Gr. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Bronger, H.; Singer, J.; Windmu, C.; Reuning, U.; Zech, D.; Delbridge, C. CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br. J. Cancer 2016, 553–563. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Kershaw, M.H.; Wang, G.; Westwood, J.A.; Pachynski, R.K.; Tiffany, H.L.; Marincola, F.M.; Wang, E.; Young, H.A.; Murphy, P.M.; Hwu, P. Redirecting migration of T cells to chemokine secreted from tumors by genetic modification with CXCR2. Hum. Gene Ther. 2002, 13, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Idorn, M.; Skadborg, S.K.; Kellermann, L.; Halldórsdóttir, H.R.; Holmen Olofsson, G.; Met, Ö.; Thor Straten, P. Chemokine receptor engineering of T cells with CXCR2 improves homing towards subcutaneous human melanomas in xenograft mouse model. Oncoimmunology 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Tao, H.; Karachi, A.; Long, Y.; Hou, A.Y.; Na, M.; Dyson, K.A.; Grippin, A.J.; Deleyrolle, L.P.; Zhang, W.; et al. CXCR1- or CXCR2-modified CAR T cells co-opt IL- 8 for maximal antitumor efficacy in solid tumors. Nat. Commun. 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bremnes, R.M.; Dønnem, T.; Al-Saad, S.; Al-Shibli, K.; Andersen, S.; Sirera, R.; Camps, C.; Marinez, I.; Busund, L.T. The role of tumor stroma in cancer progression and prognosis: Emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 209–217. [Google Scholar] [CrossRef]
- Liu, R.; Li, H.; Liu, L.; Yu, J.; Ren, X. Fibroblast activation protein: A potential therapeutic target in cancer. Cancer Biol. Ther. 2012, 4047. [Google Scholar] [CrossRef]
- Wang, L.S.; Lo, A.; Scholler, J.; Sun, J.; Majumdar, R.S.; Kapoor, V.; Antzis, M.; Cotner, C.E.; Johnson, L.A.; Durham, A.C.; et al. Targeting Fibroblast Activation Protein in Tumor Stroma with Chimeric Antigen Receptor T Cells Can Inhibit Tumor Growth and Augment Host Immunity without Severe Toxicity. Cancer Immunol. Res. 2014, 2, 154–167. [Google Scholar] [CrossRef]
- Mpekris, F.; Voutouri, C.; Baish, J.W.; Duda, D.G.; Munn, L.L. Combining microenvironment normalization strategies to improve cancer immunotherapy. Proc. Natl. Acad. Sci. USA 2020, 117. [Google Scholar] [CrossRef]
- Lanitis, E.; Irving, M.; Coukos, G. Targeting the tumor vasculature to enhance T cell activity. Curr. Opin. Immunol. 2015, 33, 55–63. [Google Scholar] [CrossRef]
- Chinnasamy, D.; Yu, Z.; Theoret, M.R.; Zhao, Y.; Shrimali, R.K.; Morgan, R.A.; Feldman, S.A.; Restifo, N.P.; Rosenberg, S.A. Gene therapy using genetically modified lymphocytes targeting VEGFR-2 inhibits the growth of vascularized syngenic tumors in mice. J. Clin. Investig. 2010, 120, 3953–3968. [Google Scholar] [CrossRef]
- Chinnasamy, D.; Tran, E.; Yu, Z.; Morgan, R.A.; Restifo, N.P.; Rosenberg, S.A. Simultaneous targeting of tumor antigens and the tumor vasculature using t lymphocyte transfer synergize to induce regression of established tumors in mice. Cancer Res. 2013, 73, 3371–3380. [Google Scholar] [CrossRef]
- Casucci, M.; Di Robilant, B.N.; Falcone, L.; Camisa, B.; Norelli, M.; Genovese, P.; Gentner, B.; Gullotta, F.; Ponzoni, M.; Bernardi, M.; et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 2013, 122, 3461–3472. [Google Scholar] [CrossRef] [PubMed]
- Porcellini, S.; Asperti, C.; Corna, S.; Cicoria, E.; Valtolina, V.; Stornaiuolo, A.; Valentinis, B.; Bordignon, C.; Traversari, C. CAR T Cells Redirected to CD44v6 Control Tumor Growth in Lung and Ovary Adenocarcinoma Bearing Mice. Front. Immunol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Murty, S.; Labanieh, L.; Murty, T.; Gowrishankar, G.; Haywood, T.; Alam, I.S.; Beinat, C.; Robinson, E.; Aalipour, A.; Klysz, D.D.; et al. PET reporter gene imaging and ganciclovir-mediated ablation of chimeric antigen receptor T-cells in solid tumors. Cancer Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tiberghien, P.; Ferrand, C.; Lioure, B.; Milpied, N.; Angonin, R.; Deconinck, E.; Certoux, J.M.; Robinet, E.; Saas, P.; Petracca, B.; et al. Administration of herpes simplex-thymidine kinase-expressing donor T cells with a T-cell-depleted allogeneic marrow graft. Blood 2001, 97, 63–72. [Google Scholar] [CrossRef]
- Straathof, K.C.; Pulè, M.A.; Yotnda, P.; Dotti, G.; Vanin, E.F.; Brenner, M.K.; Heslop, H.E.; Spencer, D.M.; Rooney, C.M. An inducible caspase 9 safety switch for T-cell therapy. Blood 2005, 105, 4247–4254. [Google Scholar] [CrossRef]
- Di Stasi, A.; Tey, S.K.; Dotti, G.; Fujita, Y.; Kennedy-Nasser, A.; Martinez, C.; Straathof, K.; Liu, E.; Durett, A.G.; Grilley, B.; et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011, 365, 1673–1683. [Google Scholar] [CrossRef]
- Stavrou, M.; Philip, B.; Traynor-White, C.; Davis, C.G.; Onuoha, S.; Cordoba, S.; Thomas, S.; Pule, M. A Rapamycin-Activated Caspase 9-Based Suicide Gene. Mol. Ther. 2018, 26, 1266–1276. [Google Scholar] [CrossRef]


| Target | Trial ID Number | Cancer Type | Additional Treatment | Location | Date | Status |
|---|---|---|---|---|---|---|
| NY-ESO-1 | NCT01967823 | Melanoma Meningioma Breast Cancer (and 2 more…) | Lymphodepletion Aldesleukin | USA | 24 October 2013 | Completed |
| NCT02775292 | Adult Solid Neoplasm Childhood Solid Neoplasm Metastatic Neoplasm | Lymphodepletion Aldesleukin Nivolumab | Unites States | 3 January 2017 | Completed | |
| NCT03029273 | Lung Cancer, Non-small Cell, Recurrent | Lymphodepletion | China | 21 March 2017 | Recruiting | |
| NCT02650986 | Advanced Fallopian Tube Carcinoma Advanced Malignant Solid Neoplasm Advanced Melanoma (and 47 more…) | Lymphodepletion TGF-β blocker | USA | 30 June 2017 | Recruiting | |
| NCT01567891 | Ovarian Cancer | Lymphodepletion Aldesleukin | USA | 3 July 2017 | Completed | |
| NCT03017131 | Recurrent Fallopian Tube Carcinoma Recurrent Ovarian Carcinoma Recurrent Primary Peritoneal Carcinoma | Lymphodepletion Aldesleukin | USA | 8 December 2017 | Recruiting | |
| NCT03638206 | Multiple Myeloma Oesophagus Cancer Lung Cancer (and 13 more…) | Lymphodepletion | China | 1 March 2018 | Recruiting | |
| NCT03462316 | Bone Sarcoma | Lymphodepletion Aldesleukin | China | 21 May 2018 | Recruiting | |
| NCT03709706 | NSCLC | Pembrolizimab | USA | 31December 2018 | Recruiting | |
| NCT03691376 | Platinum-Resistant Fallopian Tube Carcinoma Platinum-Resistant Ovarian Carcinoma Platinum-Resistant Primary Peritoneal Carcinoma (and 9 more…) | Chemotherapy Aldesleukin Cellular Therapy | USA | 24 January 2019 | Recruiting | |
| NCT03941626 | Oesophagus Cancer Hepatoma Glioma Gastric Cancer | Lymphodepletion | China | 1 September 2019 | Recruiting | |
| NCT03967223 | Neoplasms | Lymphodepletion | USA | 31 December 2019 | Recruiting | |
| HPV E7 | NCT02858310 | Papillomavirus Infections Cervical Intraepithelial Neoplasia Carcinoma In Situ (and 2 more…) | Lymphodepletion | USA | 27 January 2017 | Recruiting |
| NCT03912831 | Human Papillomavirus (HPV) 16+ Relapsed/Refractory Cancer | Lymphodepletion | USA | 8 June 2019 | Recruiting | |
| NCT03937791 | Squamous Intraepithelial Lesions of Vulva Neoplasms, Squamous Cell Vulvar HSIL | N/A | USA | 9 October 2019 | Recruiting | |
| NCT04411134 | Cervical Intraepithelial Neoplasia | N/A | USA | 5 June 2020 | Not yet recruiting | |
| NCT04044950 | Papillomavirus Infections Oropharyngeal Neoplasms | N/A | USA | 5 June 2020 | Not yet recruiting | |
| NCT04015336 | Papillomavirus Infections Oropharyngeal Neoplasms | Lymphodepletion Aldesleukin | USA | 5 June 2020 | Recruiting | |
| HPV E6 | NCT02280811 | Vaginal Cancer Cervical Cancer Anal Cancer (and 2 more…) | Lymphodepletion Aldesleukin | USA | 14 October 2014 | Completed |
| NCT03197025 | Human Papillomavirus HPV-16 High Grade Squamous Intraepithelial Lesion | Aldesleukin | USA | 9 January 2018 | Completed | |
| NCT03578406 | Cervical Cancer Head and Neck Squamous Cell Carcinoma | PD-1 Antagonist | China | 1 September 2018 | Recruiting | |
| NCT04139057 | Head and Neck Squamous Cell Carcinoma | PD-1 Antagonist | China | 1 March 2019 | Recruiting | |
| MAGE family | NCT02111850 | Cervical Cancer Renal Cancer Urothelial Cancer (and 2 more…) | USA | 7 February 2014 | Recruiting | |
| NCT03139370 | Solid Tumor | USA | 8 May 2017 | Recruiting | ||
| NCT03247309 | Solid Tumor Cancer Head and Neck Squamous Cell Carcinoma Non-small Cell Lung Cancer | USA | 19 December 2018 | Recruiting | ||
| NCT03441100 | Solid Tumor, Adult Cancer Hepatocellular Carcinoma (and 4 more…) | USA Germany | 2 May 2019 | Recruiting | ||
| Neoantigens | NCT03412877 | Glioblastoma Non-Small Cell Lung Cancer Ovarian Cancer (and 2 more…) | USA | 6 September 2018 | Recruiting | |
| NCT03970382 | Solid Tumor | USA | 3 July 2019 | Recruiting | ||
| NCT04102436 | Glioblastoma Non-Small Cell Lung Cancer Breast Cancer (and 2 more…) | USA | 5 June 2020 | Recruiting | ||
| MART1 F5 | NCT00509288 | Melanoma Skin Cancer | USA | June 2007 | Completed | |
| NCT00910650 | Metastatic Melanoma | USA | 13 October 2009 | Completed | ||
| HBV antigen | NCT02719782 | Recurrent Hepatocellular Carcinoma | China | 2 July 2015 | Recruiting | |
| NCT02686372 | Hepatocellular Carcinoma | China | December 2015 | Recruiting | ||
| EBV antigen | NCT03648697 | Nasopharyngeal Carcinoma | China | 10 October 2018 | Not yet recruiting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Presti, V.; Buitenwerf, F.; van Til, N.P.; Nierkens, S. Gene Augmentation and Editing to Improve TCR Engineered T Cell Therapy against Solid Tumors. Vaccines 2020, 8, 733. https://doi.org/10.3390/vaccines8040733
Lo Presti V, Buitenwerf F, van Til NP, Nierkens S. Gene Augmentation and Editing to Improve TCR Engineered T Cell Therapy against Solid Tumors. Vaccines. 2020; 8(4):733. https://doi.org/10.3390/vaccines8040733
Chicago/Turabian StyleLo Presti, Vania, Frank Buitenwerf, Niek P. van Til, and Stefan Nierkens. 2020. "Gene Augmentation and Editing to Improve TCR Engineered T Cell Therapy against Solid Tumors" Vaccines 8, no. 4: 733. https://doi.org/10.3390/vaccines8040733
APA StyleLo Presti, V., Buitenwerf, F., van Til, N. P., & Nierkens, S. (2020). Gene Augmentation and Editing to Improve TCR Engineered T Cell Therapy against Solid Tumors. Vaccines, 8(4), 733. https://doi.org/10.3390/vaccines8040733







