Exploiting Pan Influenza A and Pan Influenza B Pseudotype Libraries for Efficient Vaccine Antigen Selection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Production and Transformation
2.2. Propagation and Maintenance of Cell Cultures
2.3. Production of Influenza HA Pseudotypes (PV)
2.4. Influenza Pseudotype Titration
2.5. Reference Antisera and Bat Surveillence Sera
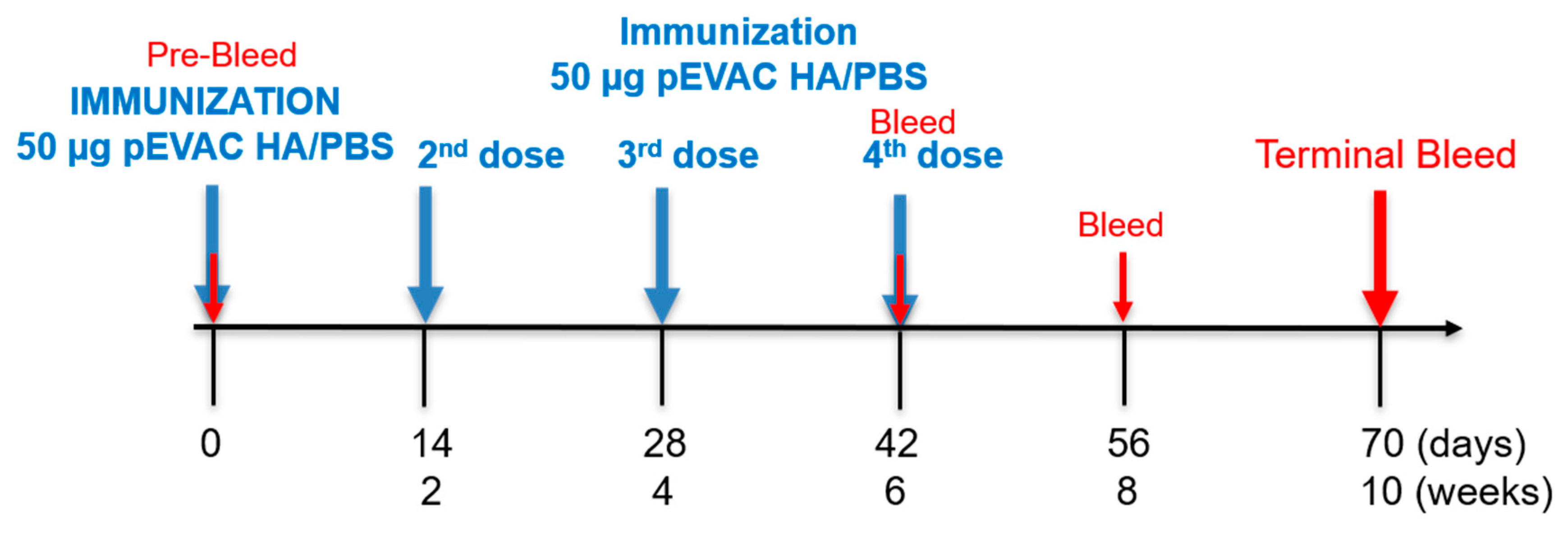
2.6. Mouse Immunogenicity Studies
2.7. Pseudotype Microneutralization (pMN) Assay
2.8. Statistical Analysis
2.9. Bioinformatic Analysis
3. Results
3.1. Production of the IAV and IBV Pseudotype Library
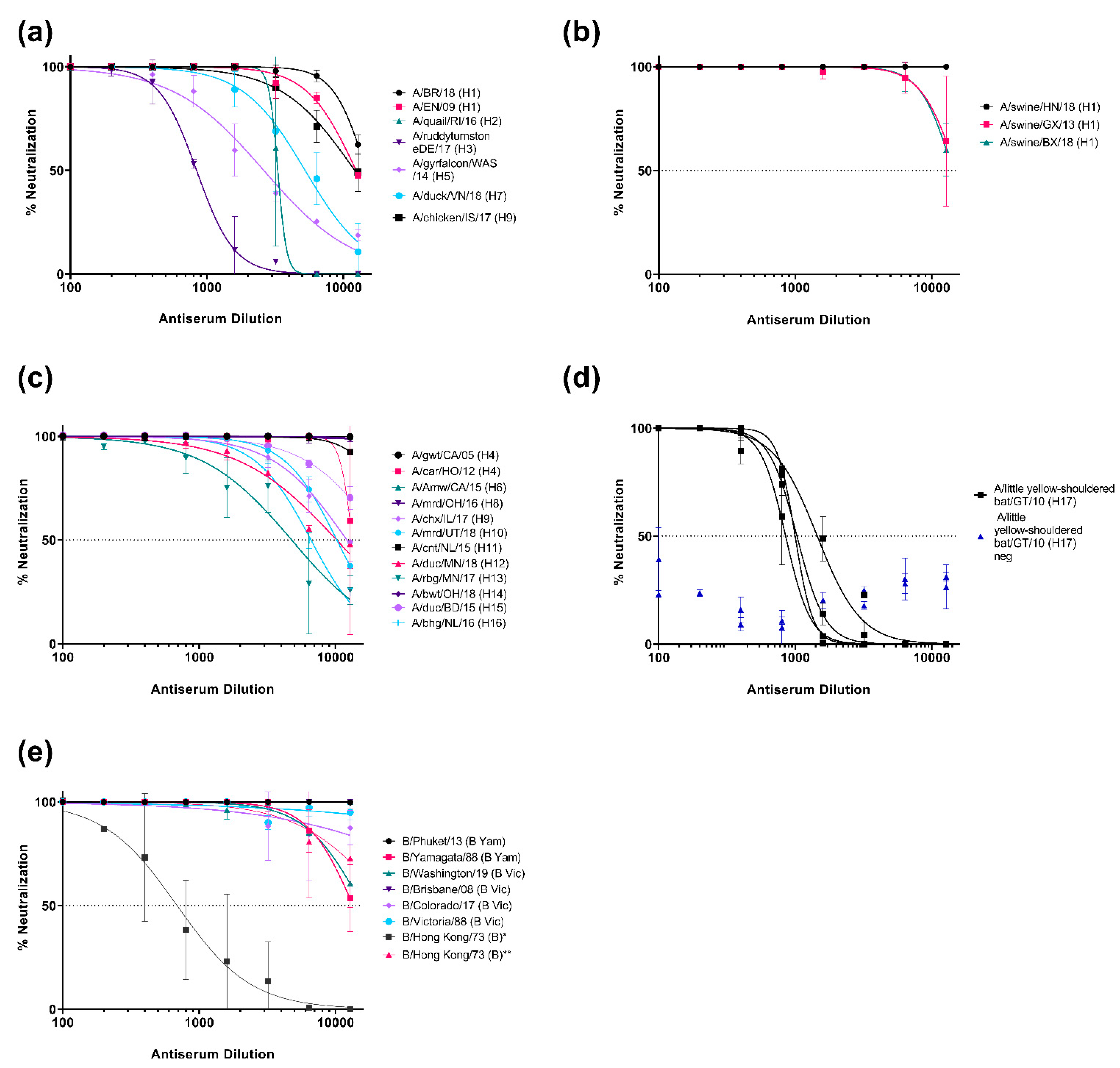
3.2. Neutralization of Pseudotypes by Reference Antisera
3.3. Mouse Immunogenicity Studies
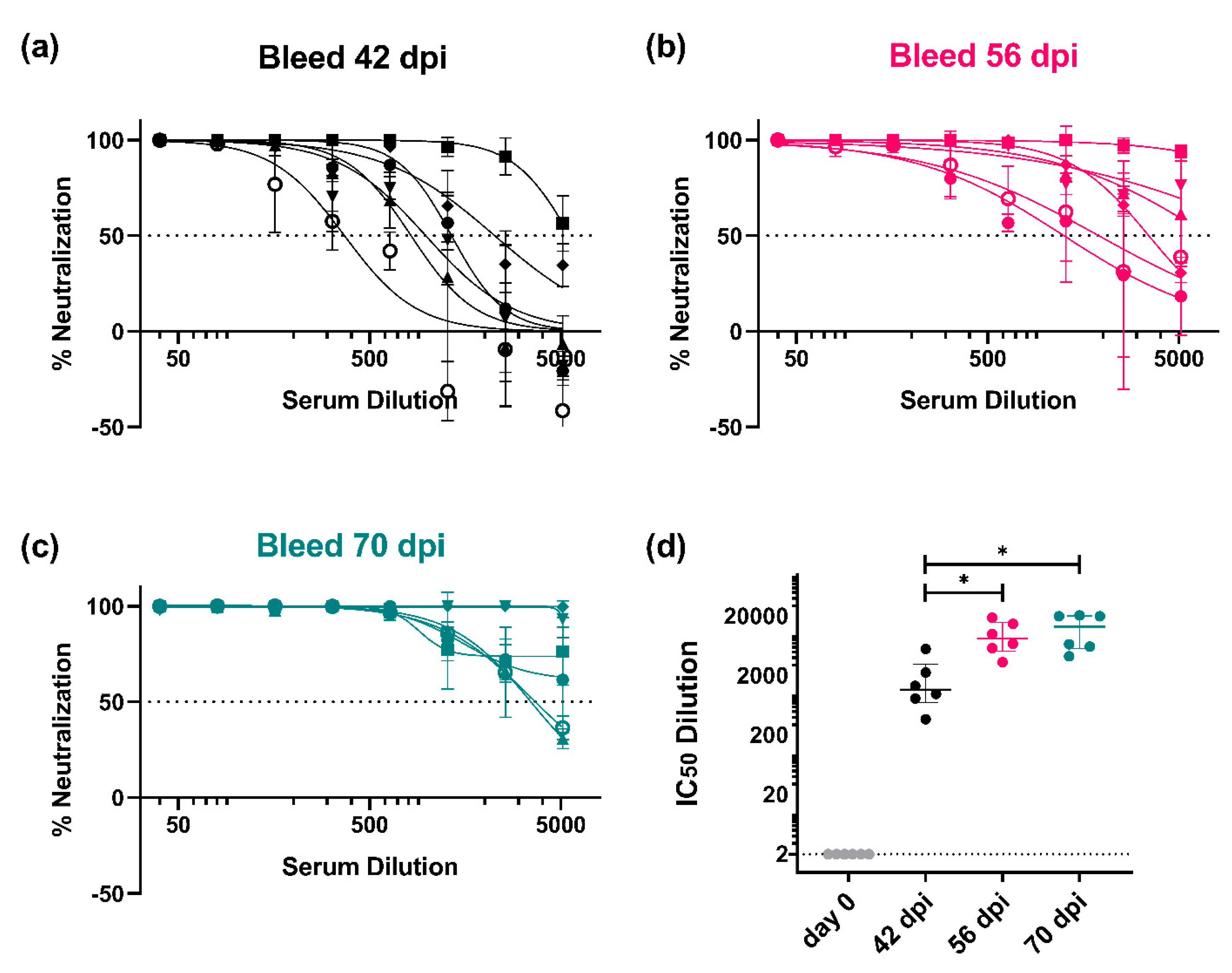
3.3.1. Monitoring of the Immune Response throughout the Vaccination Protocol
3.3.2. Strain-Specific and Subtype-Specific Immune Responses Post-Vaccination
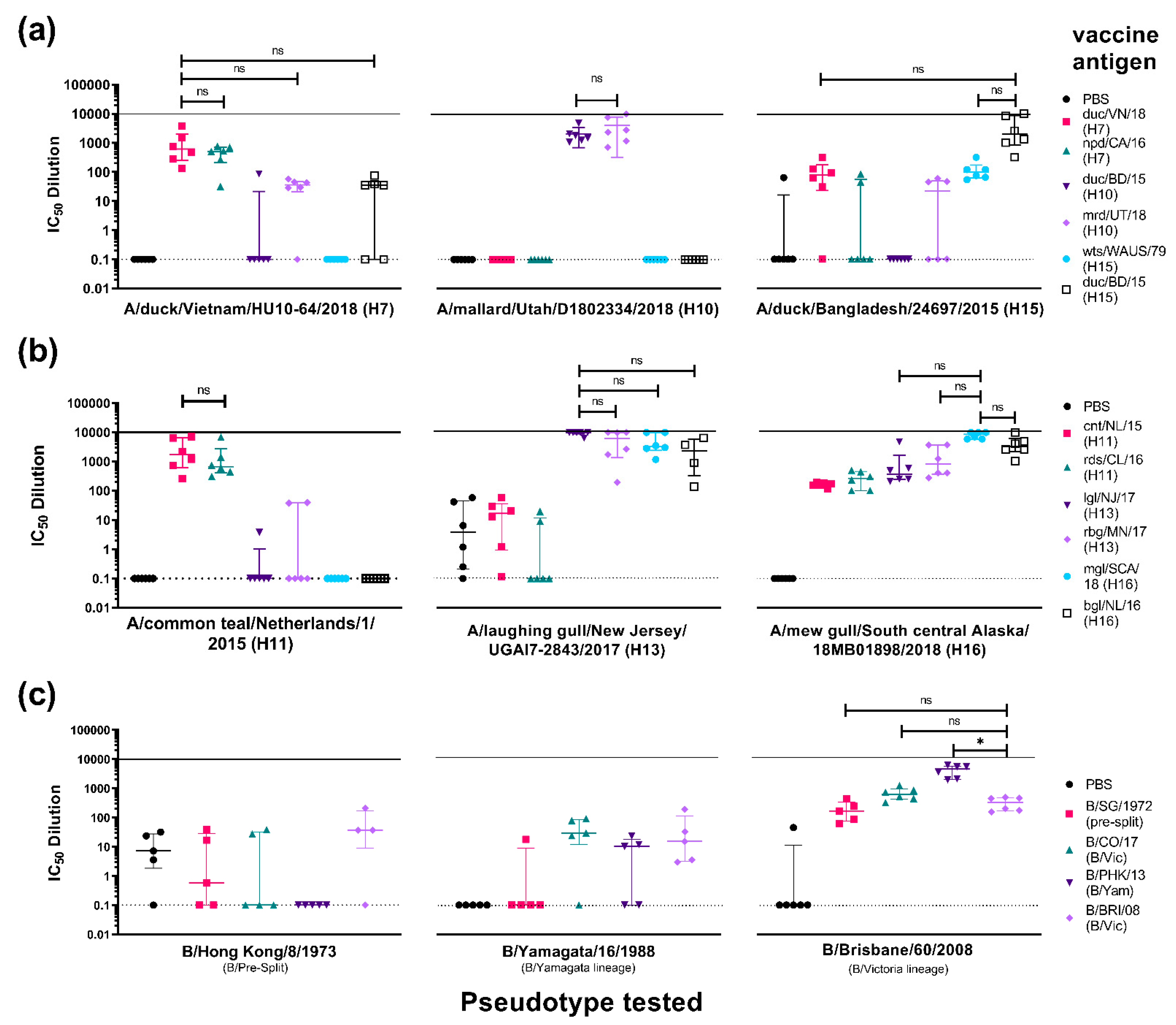
3.3.3. Cross-Subtype Immune Responses Post-Vaccination
3.4. In Vitro Neutralization of HA Pseudotypes by HA-Stem Directed Monoclonal Antibodies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26, D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Wolff, T.; Veit, M. Influenza B, C and D Viruses (Orthomyxoviridae). Encycl. Virol. 2021, 561–574. [Google Scholar] [CrossRef]
- Cauldwell, A.V.; Long, J.S.; Moncorgé, O.; Barclay, W.S. Viral determinants of influenza A virus host range. J. Gen. Virol. 2014, 95, 1193–1210. [Google Scholar] [CrossRef]
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog 2013, 9, e1003657. [Google Scholar] [CrossRef]
- World Health Organization. Influenza Virus Infection in Humans. Available online: http://www.who.int/influenza/human_animal_interface/virology_laboratories_and_vaccines/influenza_virus_infections_humans_feb14.pdf?ua=1 (accessed on 5 April 2021).
- World Health Organization. Influenza (Seasonal). Available online: http://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 5 April 2021).
- Hobbelen, P.H.F.; Elbers, A.R.W.; Werkman, M.; Koch, G.; Velkers, F.C.; Stegeman, A.; Hagenaars, T.J. Estimating the introduction time of highly pathogenic avian influenza into poultry flocks. Sci. Rep. 2020, 10, 12388. [Google Scholar] [CrossRef]
- Russell, C.J.; Hu, M.; Okda, F.A. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Morick, D.; de Mutsert, G.; Osinga, N.; Bestebroer, T.; van der Vliet, S.; Smits, S.L.; Kuiken, T.; Rimmelzwaan, G.F.; Fouchier, R.A.M.; et al. Recurring influenza B virus infections in seals. Emerg. Infect. Dis. 2013, 19, 511–512. [Google Scholar] [CrossRef]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A.M. Influenza B virus in seals. Science (N. Y.) 2000, 288, 1051–1053. [Google Scholar] [CrossRef]
- Ferrara, F.; Del Rosario, J.M.M.; da Costa, K.A.S.; Kinsley, R.; Scott, S.; Fereidouni, S.; Thompson, C.; Kellam, P.; Gilbert, S.; Carnell, G.; et al. Development of lentiviral vectors pseudotyped with influenza B hemagglutinins: Application in vaccine immunogenicity, mAb potency, and sero-surveillance studies. Front. Immunol. 2021, 12, 1904. [Google Scholar] [CrossRef]
- de Vries, R.P.; de Vries, E.; Bosch, B.J.; de Groot, R.J.; Rottier, P.J.M.; de Haan, C.A.M. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology 2010, 403, 17–25. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann. Rev. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef]
- Gamblin, S.J.; Skehel, J.J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 2010, 285, 28403–28409. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gamblin, S.J.; Haire, L.F.; Stevens, D.J.; Xiao, B.; Ha, Y.; Skehel, J.J. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 2004, 325, 287–296. [Google Scholar] [CrossRef]
- Rota, P.A.; Wallis, T.R.; Harmon, M.W.; Rota, J.S.; Kendal, A.P.; Nerome, K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990, 175, 59–68. [Google Scholar] [CrossRef]
- Benton, D.J.; Nans, A.; Calder, L.J.; Turner, J.; Neu, U.; Lin, Y.P.; Ketelaars, E.; Kallewaard, N.L.; Corti, D.; Lanzavecchia, A.; et al. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 2018, 115, 10112–10117. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 10432. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.C.-C.; Hsiao, T.-C.; Ho, M.-S.; Li, W.-H. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 6283–6288. [Google Scholar] [CrossRef] [PubMed]
- Bizebard, T.; Gigant, B.t.; Rigolet, P.; Rasmussen, B.; Diat, O.; Böseckei, P.; Wharton, S.A.; Skehel, J.J.; Knossow, M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 1995, 376, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Couzens, L.; Burke, D.F.; Wan, H.; Wilson, P.; Memoli, M.J.; Xu, X.; Harvey, R.; Wrammert, J.; Ahmed, R.; et al. Antigenic Drift of the Influenza A(H1N1)pdm09 virus neuraminidase results in reduced effectiveness of A/California/7/2009 (H1N1pdm09)-specific antibodies. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Kasson, P.M.; Pande, V.S. Combining mutual information with structural analysis to screen for functionally important residues in influenza hemagglutinin. Pac. Symp. Biocomput. 2009, 492–503. [Google Scholar] [CrossRef]
- Popova, L.; Smith, K.; West, A.H.; Wilson, P.C.; James, J.A.; Thompson, L.F.; Air, G.M. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE 2012, 7, e41895. [Google Scholar] [CrossRef]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef]
- Wilson, J.R.; Guo, Z.; Tzeng, W.-P.; Garten, R.J.; Xiyan, X.; Blanchard, E.G.; Blanchfield, K.; Stevens, J.; Katz, J.M.; York, I.A. Diverse antigenic site targeting of influenza hemagglutinin in the murine antibody recall response to A(H1N1)pdm09 virus. Virology 2015, 485, 252–262. [Google Scholar] [CrossRef]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist Med. 2002, 76, 105–115. [Google Scholar] [CrossRef]
- Lee, V.J.; Chen, M.I.; Chan, S.P.; Wong, C.S.; Cutter, J.; Goh, K.T.; Tambyah, P.A. Influenza pandemics in Singapore, a tropical, globally connected city. Emerg. Infect. Dis. 2007, 13, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. Influenza: The once and future pandemic. Public Health Rep. 2010, 125, 15–26. [Google Scholar] [CrossRef]
- Viboud, C.; Simonsen, L.; Fuentes, R.; Flores, J.; Miller, M.A.; Chowell, G. Global mortality impact of the 1957–1959 influenza pandemic. J. Infect. Dis. 2016, 213, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Dalby, A.R.; Iqbal, M. The European and Japanese outbreaks of H5N8 derive from a single source population providing evidence for the dispersal along the long distance bird migratory flyways. PeerJ 2015, 3, e934. [Google Scholar] [CrossRef] [PubMed]
- Isoda, N.; Twabela, A.T.; Bazarragchaa, E.; Ogasawara, K.; Hayashi, H.; Wang, Z.-J.; Kobayashi, D.; Watanabe, Y.; Saito, K.; Kida, H.; et al. Re-invasion of H5N8 high pathogenicity avian influenza virus clade 2.3.4.4b in Hokkaido, Japan, 2020. Viruses 2020, 12, 1439. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.S.; Banyard, A.C.; Whittard, E.; Karibayev, T.; Al Kafagi, T.; Chvala, I.; Byrne, A.; Meruyert, S.; King, J.; Harder, T.; et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021, 10, 148–151. [Google Scholar] [CrossRef]
- Vigeveno, R.M.; Poen, M.J.; Parker, E.; Holwerda, M.; de Haan, K.; van Montfort, T.; Lewis, N.S.; Russell, C.A.; Fouchier, R.A.M.; de Jong, M.D.; et al. Outbreak Severity of Highly Pathogenic Avian Influenza A(H5N8) viruses is inversely correlated to polymerase complex activity and interferon induction. J. Virol. 2020, 94, e00375-20. [Google Scholar] [CrossRef]
- Shi, J.; Deng, G.; Kong, H.; Gu, C.; Ma, S.; Yin, X.; Zeng, X.; Cui, P.; Chen, Y.; Yang, H.; et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017, 27, 1409–1421. [Google Scholar] [CrossRef]
- Subbarao, K. Avian influenza H7N9 viruses: A rare second warning. Cell Res. 2018, 28, 1–2. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, E.; Bao, C.; Xiang, N.; Wu, J.; Wu, S.; Shi, J.; Wang, X.; Zheng, Y.; Zhang, Y.; et al. Clusters of human infection and Human-to-Human Transmission of Avian Influenza A(H7N9) virus, 2013–2017. Emerg. Infect. Dis. 2018, 24. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef]
- Beran, J.; Peeters, M.; Dewé, W.; Raupachová, J.; Hobzová, L.; Devaster, J.-M. Immunogenicity and safety of quadrivalent versus trivalent inactivated influenza vaccine: A randomized, controlled trial in adults. BMC Infect. Dis. 2013, 13, 224. [Google Scholar] [CrossRef] [PubMed]
- Pépin, S.; Donazzolo, Y.; Jambrecina, A.; Salamand, C.; Saville, M. Safety and immunogenicity of a quadrivalent inactivated influenza vaccine in adults. Vaccine 2013, 31, 5572–5578. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.D.; Schultz-Cherry, S. Development of a universal influenza vaccine. J. Immunol. 2019, 202, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, K.; Matsuoka, Y. The prospects and challenges of universal vaccines for influenza. Trends Microbiol. 2013, 21, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Chatziprodromidou, I.P.; Arvanitidou, M.; Guitian, J.; Apostolou, T.; Vantarakis, G.; Vantarakis, A. Global avian influenza outbreaks 2010–2016: A systematic review of their distribution, avian species and virus subtype. Syst Rev. 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Del Rosario, J.M.M.; Smith, M.; Zaki, K.; Risley, P.; Temperton, N.; Engelhardt, O.G.; Collins, M.; Takeuchi, Y.; Hufton, S.E. Protection from influenza by intramuscular gene vector delivery of a broadly neutralizing nanobody does not depend on antibody dependent cellular cytotoxicity. Front. Immunol. 2020, 11, 627. [Google Scholar] [CrossRef]
- Kanjilal, S.; Mina, M.J. Passive immunity for the treatment of influenza: Quality not quantity. Lancet Respir. Med. 2019, 7, 922–923. [Google Scholar] [CrossRef]
- Manenti, A.; Maciola, A.K.; Trombetta, C.M.; Kistner, O.; Casa, E.; Hyseni, I.; Razzano, I.; Torelli, A.; Montomoli, E. Influenza anti-stalk antibodies: Development of a new method for the evaluation of the immune responses to universal vaccine. Vaccines 2020, 8, 43. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Bernstein, H.H.; Bradley, J.S.; Englund, J.A.; File, T.M.; Fry, A.M.; Gravenstein, S.; Hayden, F.G.; Harper, S.A.; Hirshon, J.M.; et al. Clinical practice guidelines by the infectious diseases society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa. Clin. Infect. Dis. 2019, 68, e1–e47. [Google Scholar] [CrossRef]
- WHO Global Influenza Surveillance Network. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. Available online: http://www.who.int/influenza/gisrs_laboratory/manual_diagnosis_surveillance_influenza/en/index.html (accessed on 5 April 2021).
- World Health Organization. Evaluation of Influenza Vaccine Effectiveness: A Guide to the Design and Interpretation of Observational Studies; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Alladi, C.S.H.; Jagadesh, A.; Prabhu, S.G.; Arunkumar, G. Hemagglutination Inhibition Antibody Response Following Influenza A(H1N1)pdm09 virus natural infection: A cross-sectional study from Thirthahalli, Karnataka, India. Viral Immunol. 2019, 32, 230–233. [Google Scholar] [CrossRef]
- Cox, R.J. Correlates of protection to influenza virus, where do we go from here? Hum. Vaccines Immunother. 2013, 9, 405–408. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (Lond.) 1972, 70, 767–777. [Google Scholar] [CrossRef]
- Agor, J.K.; Özaltın, O.Y. Models for predicting the evolution of influenza to inform vaccine strain selection. Hum. Vaccines Immunother. 2018, 14, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Carnell, G.W.; Trombetta, C.M.; Ferrara, F.; Montomoli, E.; Temperton, N.J. Correlation of influenza B haemagglutination inhibiton, single-radial haemolysis and pseudotype-based microneutralisation assays for immunogenicity testing of seasonal vaccines. Vaccines 2021, 9, 100. [Google Scholar] [CrossRef]
- Wallerström, S.; Lagerqvist, N.; Temperton, N.J.; Cassmer, M.; Moreno, A.; Karlsson, M.; Leijon, M.; Lundkvist, Å.; Falk, K.I. Detection of antibodies against H5 and H7 strains in birds: Evaluation of influenza pseudovirus particle neutralization tests. Infect. Ecol. Epidemiol. 2014, 4, 23011. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carnell, G.W.; Ferrara, F.; Grehan, K.; Thompson, C.P.; Temperton, N.J. Pseudotype-based neutralization assays for influenza: A systematic analysis. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Temperton, N.J.; Hoschler, K.; Major, D.; Nicolson, C.; Manvell, R.; Hien, V.M.; Ha, D.Q.; De Jong, M.; Zambon, M.; Takeuchi, Y.; et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Other Respir. Viruses 2007, 1, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Toon, K.; Bentley, E.M.; Mattiuzzo, G. More than just gene therapy vectors: Lentiviral vector pseudotypes for serological investigation. Viruses 2021, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L.; Blömer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Zufferey, R.; Nagy, D.; Mandel, R.J.; Naldini, L.; Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997, 15, 871–875. [Google Scholar] [CrossRef]
- World Health Organization. Global Influenza Strategy 2019–2030; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Cox, R.J.; Mykkeltvedt, E.; Robertson, J.; Haaheim, L.R. Non-lethal viral challenge of influenza haemagglutinin and nucleoprotein DNA vaccinated mice results in reduced viral replication. Scand. J. Immunol. 2002, 55, 14–23. [Google Scholar] [CrossRef]
- Raab, D.; Graf, M.; Notka, F.; Schödl, T.; Wagner, R. The GeneOptimizer Algorithm: Using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst. Synth. Biol. 2010, 4, 215–225. [Google Scholar] [CrossRef]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef]
- Böttcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.-D.; Garten, W.; Matrosovich, M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Blazejewska, P.; Soilleux, E.; Allen, P.; Danisch, S.; Steffen, I.; Choi, S.-Y.; Park, Y.; Schneider, H.; et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010, 84, 10016–10025. [Google Scholar] [CrossRef]
- Ferrara, F.; Temperton, N. Pseudotype neutralization assays: From laboratory bench to data analysis. Methods Protoc. 2018, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Zhang, Y.; Aevermann, B.D.; Anderson, T.K.; Burke, D.F.; Dauphin, G.; Gu, Z.; He, S.; Kumar, S.; Larsen, C.N.; Lee, A.J.; et al. Influenza Research Database: An integrated bioinformatics resource for influenza virus research. Nucleic Acids Res. 2016, 45, D466–D474. [Google Scholar] [CrossRef] [PubMed]
- Bosch, F.X.; Garten, W.; Klenk, H.D.; Rott, R. Proteolytic cleavage of influenza virus hemagglutinins: Primary structure of the connecting peptide between HA, and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology 1981, 113, 725–735. [Google Scholar] [CrossRef]
- Böttcher-Friebertshäuser, E.; Garten, W.; Matrosovich, M.; Klenk, H.D. The hemagglutinin: A determinant of pathogenicity. In Influenza Pathogenesis and Control; Compans, R.W., Oldstone, M.B.A., Eds.; Springer: New York, NY, USA, 2014; Volume 1. [Google Scholar]
- Garten, W.; Klenk, H.D. Understanding influenza virus pathogenicity. Trends Microbiol. 1999, 7, 99–100. [Google Scholar] [CrossRef]
- Ferrara, F.; Molesti, E.; Böttcher-Friebertshäuser, E.; Cattoli, G.; Corti, D.; Scott, S.D.; Temperton, N.J. The human Transmembrane Protease Serine 2 is necessary for the production of Group 2 influenza A virus pseudotypes. J. Mol. Genet. Med. 2012, 7, 309–314. [Google Scholar] [CrossRef]
- World Health Organization. Avian Influenza: Assessing the Pandemic Threat; World Health Organization: Geneva, Switzerland, 2005. [Google Scholar]
- World Health Organization; World Organisation for Animal Health; Food and Agriculture Organization; H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir. Viruses 2014, 8, 384–388. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov 2015, 14, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Sautto, G.A.; Kirchenbaum, G.A.; Ecker, J.W.; Bebin-Blackwell, A.G.; Pierce, S.R.; Ross, T.M. Elicitation of broadly protective antibodies following infection with influenza viruses expressing H1N1 computationally optimized broadly reactive hemagglutinin antigens. Immunohorizons 2018, 2, 226–237. [Google Scholar] [CrossRef]
- Hillaire, M.L.B.; van Trierum, S.E.; Kreijtz, J.; Bodewes, R.; Geelhoed-Mieras, M.M.; Nieuwkoop, N.J.; Fouchier, R.A.M.; Kuiken, T.; Osterhaus, A.; Rimmelzwaan, G.F. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol 2011, 92, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to Group 1 and Group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef]
- Kay, M.A.; Glorioso, J.C.; Naldini, L. Viral vectors for gene therapy: The art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001, 7, 33–40. [Google Scholar] [CrossRef]
- Mather, S.T.; Wright, E.; Scott, S.D.; Temperton, N.J. Lyophilisation of influenza, rabies and Marburg lentiviral pseudotype viruses for the development and distribution of a neutralisation—Assay-based diagnostic kit. J. Virol. Methods 2014, 210, 51–58. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O’Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Wang, W.; Alvarado-Facundo, E.; Chen, Q.; Anderson, C.M.; Scott, D.; Vassell, R.; Weiss, C.D. Serum samples from middle-aged adults vaccinated annually with seasonal influenza vaccines cross-neutralize some potential pandemic influenza viruses. J. Infect. Dis. 2016, 213, 403–406. [Google Scholar] [CrossRef]
- Wang, W.; Vassell, R.; Song, H.S.; Chen, Q.; Keller, P.W.; Verma, S.; Alvarado-Facundo, E.; Wan, H.; Schmeisser, F.; Meseda, C.A.; et al. Generation of a protective murine monoclonal antibody against the stem of influenza hemagglutinins from group 1 viruses and identification of resistance mutations against it. PLoS ONE 2019, 14, e0222436. [Google Scholar] [CrossRef]
- Guo, L.; Wang, D.; Zhou, H.; Wu, C.; Gao, X.; Xiao, Y.; Ren, L.; Paranhos-Baccalà, G.; Shu, Y.; Jin, Q.; et al. Cross-reactivity between avian influenza A (H7N9) virus and divergent H7 subtypic- and heterosubtypic influenza A viruses. Sci. Rep. 2016, 6, 22045. [Google Scholar] [CrossRef]
- Strohmeier, S.; Amanat, F.; Krammer, F. Cross-reactive antibodies binding to the influenza virus subtype H11 hemagglutinin. Pathogens 2019, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, C.; Han, X.; Lin, S.; Ao, X.; Han, X.; Wang, J.; Ye, H. Structural insights for anti-influenza vaccine design. Comput. Struct. Biotechnol. J. 2019, 17, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Yoshida, R.; Ekiert, D.C.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 2012, 109, 17040–17045. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Uraki, R.; Ito, M.; Kiso, M.; Nakatsu, S.; Yasuhara, A.; Oishi, K.; Sasaki, T.; Ikuta, K.; Kawaoka, Y. A broadly reactive human anti-hemagglutinin stem monoclonal antibody that inhibits influenza A virus particle release. EBioMedicine 2017, 17, 182–191. [Google Scholar] [CrossRef]
- Guthmiller, J.J.; Han, J.; Li, L.; Freyn, A.W.; Liu, S.T.H.; Stovicek, O.; Stamper, C.T.; Dugan, H.L.; Tepora, M.E.; Utset, H.A.; et al. First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci. Transl. Med. 2021, 13, eabg4535. [Google Scholar] [CrossRef]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [PubMed]





| Solutions/Plasmids | Amount |
|---|---|
| OptiMEM | 100 µL |
| p8.91 | 250 ng |
| pCSFLW | 375 ng |
| HA in pEVAC | 10 ng (50 ng for HA18) |
| HA in pI.18 | 50–500 ng |
| HA in phCMV1 | 50–500 ng |
| Protease-encoding plasmid | 2.5–500 ng |
| FuGENE® HD | 3 µL per µg of total plasmid DNA |
| HA Subtype | Antiserum Strain | Source |
|---|---|---|
| H1 | A/duck/Italy/447/2005 (H1) | OIE |
| H2 | A/duck/Germany/1215/1973 (H2) | OIE |
| H3 | A/psittacine/Italy/2873/2000 (H3) | OIE |
| H4 | A/cockatoo/England/1972 (H4) | OIE |
| H5 | A/chicken/Scotland/1959 (H5) | APHA |
| H6 | A/turkey/Canada/1965 (H6) | OIE |
| H7 | A/Anhui/1/2013 (H7) | NIBSC |
| H8 | A/turkey/Ontario/6118/1968 (H8) | OIE |
| H9 | A/mallard/Italy/3817-34/2005 (H9) | OIE |
| H10 | A/ostrich/South Africa/2001 (H10) | OIE |
| H11 | A/duck/Memphis/546/1974 (H11) | OIE |
| H12 | A/duck/Alberta/60/1976 (H12) | OIE |
| H13 | A/gull/Maryland/704/1977 (H13) | OIE |
| H14 | A/mallard/Gurjev/263/1982 (H14) | OIE |
| H15 | A/shearwater/Australia/2576/1979 (H15) | OIE |
| H16 | A/gull/Denmark/68110/2002 (H16) | OIE |
| H17 | Polyclonal sera (BATS) | APHA |
| B/YAM | B/Phuket/3073/2013 | NIBSC |
| B/VIC | B/Brisbane/60/2008 | NIBSC |
| Group I IAV HA | Group II IAV HA | ||||
|---|---|---|---|---|---|
| HA Envelope | Titre (RLU/mL) | Protease | HA Envelope | Titre (RLU/mL) | Protease |
| H1 | 2.25 × 108 | T4 | H3 | 5.39 × 1010 | T2 |
| H2 | 6.62 × 107 | T2 | H4 | 6.12 × 109 | T4 |
| H5 | 1.32 × 109 | * | H7 | 5.25 × 1010 | * |
| H6 | 2.35 × 1010 | T4 | H10 | 2.68 × 1010 | T4 |
| H8 | 4.75 × 1010 | T4 | H14 | 2.92 × 1010 | T4 |
| H9 | 4.88 × 108 | T4 | H15 | 5.16 × 1010 | T4 |
| H11 | 8.78 × 109 | T4 | IBV HA | ||
| H12 | 1.21 × 1010 | T4 | B pre-split | 3.87 × 1010 | HAT |
| H13 | 1.44 × 109 | T4 | B/Vic-like | 2.89 × 1010 | HAT |
| H16 | 5.81 × 109 | T4 | B/Yam-like | 1.78 × 109 | HAT |
| H17 | 2.94 × 108 | HAT | |||
| H18 | 5.33 × 107 | T4 | |||
| Pseudotype Virus (PV) | IC50 (ng/mL) | ||
|---|---|---|---|
| Subtype | Strain | CR9114 | FI6 |
| H1 | A/England/195/2009 | 3.63 | 13.25 |
| H2 | A/quail/Rhode Island/16-018622-1/2016 | 5.06 | 26.70 |
| H3 | A/ruddy turnstone/Delaware Bay/606/2017 | 51.62 | 9.83 |
| H4 | A/Calidris ruficollis/Hokkaido/12EY0172/2012 | 1.68 | 8.36 |
| H5 | A/gyrfalcon/Washington/41088-6/2014 | 10.74 | 60.15 |
| H6 | A/American wigeon/California/HS007A/2015 | 0.68 | 2.91 |
| H7 | A/Shanghai/02/2013 | 11.88 | 17.39 |
| H8 | A/mallard duck/Ohio/16OS0672/2016 | 0.71 | 0.23 |
| H9 | A/chicken/Israel/291417/2017 | 0.39 | 6.41 |
| H10 | A/mallard/Utah/D1802334/2018 | 1.26 | 0.57 |
| H11 | A/red shoveler/Chile/C14653/2016 | 120.90 | 0.02 |
| H12 | A/duck/Mongolia/850/2018 | 15.06 | 0.51 |
| H13 | A/laughing gull/New Jersey/UGAI17-2843/2017 | 31.97 | 52.05 |
| H14 | A/blue-winged Teal/Ohio/18OS1695/2018 | 0.58 | 0.06 |
| H15 | A/wedge-tailed shearwater/Western Australia/2576/1979 | 10.73 | 14.12 |
| H16 | A/black-headed gull/Netherlands/1/2016 | 45.41 | 55.50 |
| H17 | A/little yellow-shouldered bat/Guatemala/60/2010 | 0.54 | 0.34 |
| H18 | A/flat-faced bat/Peru/033/2010 | 0.26 | 3.14 |
| B | B/Hong Kong/8/1973 | - | n.d. |
| B/Vic | B/Victoria/1/1987 | - | n.d. |
| B/Yam | B/Yamagata/16/1988 | - | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Rosario, J.M.M.; da Costa, K.A.S.; Asbach, B.; Ferrara, F.; Ferrari, M.; Wells, D.A.; Mann, G.S.; Ameh, V.O.; Sabeta, C.T.; Banyard, A.C.; et al. Exploiting Pan Influenza A and Pan Influenza B Pseudotype Libraries for Efficient Vaccine Antigen Selection. Vaccines 2021, 9, 741. https://doi.org/10.3390/vaccines9070741
Del Rosario JMM, da Costa KAS, Asbach B, Ferrara F, Ferrari M, Wells DA, Mann GS, Ameh VO, Sabeta CT, Banyard AC, et al. Exploiting Pan Influenza A and Pan Influenza B Pseudotype Libraries for Efficient Vaccine Antigen Selection. Vaccines. 2021; 9(7):741. https://doi.org/10.3390/vaccines9070741
Chicago/Turabian StyleDel Rosario, Joanne Marie M., Kelly A. S. da Costa, Benedikt Asbach, Francesca Ferrara, Matteo Ferrari, David A. Wells, Gurdip Singh Mann, Veronica O. Ameh, Claude T. Sabeta, Ashley C. Banyard, and et al. 2021. "Exploiting Pan Influenza A and Pan Influenza B Pseudotype Libraries for Efficient Vaccine Antigen Selection" Vaccines 9, no. 7: 741. https://doi.org/10.3390/vaccines9070741
APA StyleDel Rosario, J. M. M., da Costa, K. A. S., Asbach, B., Ferrara, F., Ferrari, M., Wells, D. A., Mann, G. S., Ameh, V. O., Sabeta, C. T., Banyard, A. C., Kinsley, R., Scott, S. D., Wagner, R., Heeney, J. L., Carnell, G. W., & Temperton, N. J. (2021). Exploiting Pan Influenza A and Pan Influenza B Pseudotype Libraries for Efficient Vaccine Antigen Selection. Vaccines, 9(7), 741. https://doi.org/10.3390/vaccines9070741










