Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer?
Abstract
1. Introduction
2. HBV Vaccines
3. HCV Vaccines
4. Influenza Virus Vaccines
5. Papillomavrius Vaccines
6. HIV Vaccines
7. SARS-CoV-2 Vaccines
8. Zika Virus Vaccines
9. Other Plant-Based Vaccines
10. Plant-Based Therapeutic Antibodies
11. Caveats of Plant-Derived Vaccines
12. Risks of Plant-Made Vaccines
13. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Aggarwal, S. What’s fueling the biotech engine-2008. Nat. Biotechnol. 2009, 27, 987–993. [Google Scholar] [CrossRef]
- Su, J.; Zhu, L.; Sherman, A.; Wang, X.; Lin, S.; Kamesh, A.; Norikane, J.H.; Streatfield, S.J.; Herzog, R.W.; Daniell, H. Low cost industrial production of coagulation factor IX bioencapsulated in lettuce cells for oral tolerance induction in hemophilia B. Biomaterials 2015, 70, 84–93. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Salazar-González, J.A. Immunological aspects of using plant cells as delivery vehicles for oral vaccines. Expert Rev. Vaccines 2014, 13, 737–749. [Google Scholar] [CrossRef]
- Márquez-Escobar, V.A.; Rosales-Mendoza, S.; Beltrán-López, J.I.; González-Ortega, O. Plant-based vaccines against respiratory diseases: Current status and future prospects. Expert Rev. Vaccines 2017, 16, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Gonzalez, J.A.; Bañuelos-Hernandez, B.; Rosales-Mendoza, S. Current status of viral expression systems in plants and perspectives for oral vaccines development. Plant Mol. Biol. 2015, 87, 203–217. [Google Scholar] [CrossRef]
- Bock, R. Engineering chloroplasts for high-level foreign protein expression. Methods Mol. Biol. 2014, 1132, 93–106. [Google Scholar] [PubMed]
- Bock, R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef]
- Bock, R. Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol. 2001, 312, 425–438. [Google Scholar] [CrossRef]
- Daniell, H. Production of biopharmaceuticals and vaccines in plants via the chloroplast genome. Biotechnol. J. 2006, 1, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Govea-Alonso, D.O.; Cardineau, G.A.; Rosales-Mendoza, S. Principles of plant-based vaccines. In Genetically Engineered Plants as a Source of Vaccines against Wide Spread Diseases—An Integrated View; Mendoza, S.R., Ed.; Springer Science + Business Media: New York, NY, USA, 2014; pp. 1–14. [Google Scholar]
- Hernández, M.; Rosas, G.; Cervantes, J.; Fragoso, G.; Rosales-Mendoza, S.; Sciutto, E. Transgenic plants: A 5-year update on oral antipathogen vaccine development. Expert Rev. Vaccines 2014, 13, 1523–1536. [Google Scholar] [CrossRef]
- Orellana-Escobedo, L.; Korban, S.S.; Rosales-Mendoza, S. Seed-based expression strategies. In Genetically Engineered Plants as a Source of Vaccines against Wide Spread Diseases—An Integrated View; Mendoza, S.R., Ed.; Springer Science + Business Media: New York, NY, USA, 2014; pp. 79–93. [Google Scholar]
- Joung, Y.H.; Park, S.H.; Moon, K.-B.; Jeon, J.-H.; Cho, H.-S.; Kim, H.-S. The Last Ten Years of Advancements in Plant-Derived Recombinant Vaccines against Hepatitis B. Int. J. Mol. Sci. 2016, 17, 1715. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lai, H.; Sun, H.; Chen, Q. Virus-like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci. Rep. 2017, 7, 7679. [Google Scholar] [CrossRef]
- Mobini, S.; Chizari, M.; Mafakher, L.; Rismani, E.; Rismani, E. Computational Design of a Novel VLP-Based Vaccine for Hepatitis B Virus. Front. Immunol. 2020, 11, 2074. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.; Chisari, F.; Block, J.; Dandri, M.; Gehring, A.; Guo, H.; Hu, J.; Kramvis, A.; Lampertico, P.; Janssen, H.L.A.; et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 2019, 4, 545–558. [Google Scholar] [CrossRef]
- Dobrica, M.-O.; Lazar, C.; Paruch, L.; Skomedal, H.; Steen, H.; Haugslien, S.; Tucureanu, C.; Caras, L.; Onu, A.; Ciulean, S.; et al. A novel chimeric Hepatitis B virus S/preS1 antigen produced in mammalian and plant cells elicits stronger humoral and cellular immune response than the standard vaccine-constituent, S protein. Antivir. Res. 2017, 144, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V. Hepatitis B virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Heermann, K.H.; Goldmann, U.; Schwartz, W.; Seyffarth, T.; Baumgarten, H.; Gerlich, W.H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 1984, 52, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Eddleston, A. Modern vaccines. Hepatitis. Lancet 1990, 335, 1142–1145. [Google Scholar] [CrossRef]
- Patient, R.; Hourioux, C.; Vaudin, P.; Pagès, J.C.; Roingeard, P. Chimeric hepatitis B and C viruses envelope proteins can form subviral particles: Implications for the design of new vaccine strategies. New Biotechnol. 2009, 25, 226–234. [Google Scholar] [CrossRef]
- Stirk, H.J.; Thornton, J.M.; Howard, C.R. Atopological Model for Hepatitis B Surface Antigen. Intervirology 1992, 33, 148–158. [Google Scholar] [CrossRef]
- McAleer, W.J.; Buynak, E.B.; Maigetter, R.Z.; Wampler, D.E.; Miller, W.J.; Hilleman, M.R. Human hepatitis B vaccine from recombinant yeast. Nature 1984, 307, 178–180. [Google Scholar] [CrossRef]
- Hayden, C.A.; Fischer, M.E.; Andrews, B.L.; Chilton, H.C.; Turner, D.D.; John, H.; Walker, J.H.; Tizard, I.R.; Howard, J.A. Oral delivery of wafers made from HBsAg-expressing maize germ induces long-term immunological systemic and mucosal responses. Vaccine 2015, 33, 2881–2886. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Anahı’ Tello-Olea, M. Carrot Cells: A Pioneering Platform for Biopharmaceuticals Production. Mol. Biotechnol. 2015, 57, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; LePore, K.; Elkin, G.; Thanavala, Y.; Mason, H.S. High-yield rapid production of hepatitis B surface antigen in plant leaf by a viral expression system. Plant Biotechnol. J. 2008, 5, 202–209. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-based vaccines against viruses. Virol. J. 2014, 11, 205–225. [Google Scholar] [CrossRef]
- Huang, Z.; Elkin, G.; Maloney, B.J.; Beuhner, N.; Arntzen, C.J.; Thanavala, Y.; Mason, H.S. Virus-like particle expression and assembly in plants: Hepatitis B and Norwalk viruses. Vaccine 2005, 23, 1851–1858. [Google Scholar] [CrossRef]
- Huang, Z.; Santi, L.; LePore, K.; Kilbourne, J.; Arntzen, C.J.; Mason, H.S. Rapid, high-level production of hepatitis B core antigen in plant leaf and its immunogenicity in mice. Vaccine 2006, 24, 2506–2513. [Google Scholar] [CrossRef]
- Dobrica, M.-O.; Lazar, C.; Lisa Paruch, L.; Eerde, A.; Clarke, J.L.; Tucureanu, C.; Caras, L.; Ciulean, S.; Onu, A.; Tofan, V.; et al. Oral administration of a chimeric Hepatitis B Virus S/preS1 antigen produced in lettuce triggers infection neutralizing antibodies in mice. Vaccine 2018, 36, 5789–5795. [Google Scholar] [CrossRef]
- Zahmanova, G.; Mazalovska, M.; Takova, K.; Toneva, V.; Minkov, I.; Peyret, H.; Lomonossoff, G. Efficient Production of Chimeric Hepatitis B Virus-Like Particles Bearing an Epitope of Hepatitis E Virus Capsid by Transient Expression in Nicotiana Benthamiana. Life 2021, 11, 64. [Google Scholar] [CrossRef]
- Mohammadzadeh, S.; Roohvand, F.; Memarnejadian, A.; Jafari, A.; Ajdary, S.; Salmanian, A.-H.; Ehsani, P. Co-expression of hepatitis C virus polytope–HBsAg and p19-silencing suppressor protein in tobacco leaves. Pharm. Biol. 2016, 54, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Rolland, S.; Vachon, M.-L. Sofosbuvir for the treatment of hepatitis C virus infection. CMAJ 2015, 187, 203–204. [Google Scholar] [CrossRef][Green Version]
- Fauvelle, C.; Lepiller, Q.; Felmlee, D.J.; Fofana, I.; Habersetzer, F.; Stoll-Keller, F.; Baumert, T.F.; Fafi-Kremer, S. Hepatitis C virus vaccines—Progress and perspectives. Microb. Pathog. 2013, 58, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Chevaliez, S.; Pawlotsky, J.M. Virology of hepatitis C virus infection. Best Pract. Res. Clin. Gastroenterol. 2012, 26, 381–389. [Google Scholar] [CrossRef]
- Simmonds, P. The origin of hepatitis C virus. Curr. Top. Microbiol. Immunol. 2013, 369, 1–15. [Google Scholar]
- Murakami, K.; Abe, M.; Kageyama, T.; Kamoshita, N.; Nomoto, A. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 2001, 146, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef]
- André, P.; Komurian-Pradel, F.; Deforges, S.; Perret, M.; Berland, J.L.; Sodoyer, M.; Pol, S.; Bréchot, C.; Paranhos-Baccalà, G.; Lotteau, V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002, 76, 6919–6928. [Google Scholar] [CrossRef]
- Lindenbach, B.D. Virion Assembly and Release. Curr. Top. Microbiol. Immunol. 2013, 369, 199–218. [Google Scholar]
- Zeisel, M.B.; Felmlee, D.J.; Baumert, T.F. Hepatitis C virus entry. Curr. Top. Microbiol. Immunol. 2013, 369, 87–112. [Google Scholar] [PubMed]
- Blanchard, E.; Belouzard, S.; Goueslain, L.; Wakita, T.; Dubuisson, J.; Wychowski, C.; Rouillé, Y. Hepatitis C virus entry depends on Clathrin-mediated endocytosis. J. Virol. 2006, 80, 6964–6972. [Google Scholar] [CrossRef]
- Rupp, D.; Bartenschlager, R. Targets for antiviral therapy of hepatitis C. Semin. Liver Dis. 2014, 34, 9–21. [Google Scholar] [CrossRef]
- Moradpour, D.; Penin, F. Hepatitis C virus proteins: From structure to function. Curr. Top. Microbiol. Immunol. 2013, 369, 113–142. [Google Scholar] [PubMed]
- Madesis, P.; Osathanunkul, M.; Georgopoulou, U.; Gisby, M.F.; Mudd, E.A.; Nianiou, I.; Tsitoura, P.; Mavromara, P.; Tsaftaris, A.; Day, A. A hepatitis C virus core polypeptide expressed in chloroplasts detects anti-core antibodies in infected human sera. J. Biotechnol. 2010, 145, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, S.; Khabiri, A.; Roohvand, F.; Memarnejadian, A.; Salmanian, A.H.; Ajdary, S.; Ehsani, P. Enhanced-Transient Expression of Hepatitis C Virus Core Protein in Nicotiana tabacum, a Protein with Potential Clinical Applications. Hepat. Mon. 2014, 14, e20524. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, S.; Roohvand, F.; Ehsani, P.; Salmanian, A.H.; Ajdary, S. Canola oilseed- and Escherichia coli- derived hepatitis C virus (HCV) core proteins adjuvanted with oil bodies, induced robust Th1-oriented immune responses in immunized mice. APMIS 2020, 128, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Denis, J.; Majeau, N.; Acosta-Ramirez, E.; Savard, C.; Bedard, M.C.; Simard, S.; Lecours, K.; Bolduc, M.; Pare, C.; Willems, B.; et al. Immunogenicity of papaya mosaic virus-like particles fused to a hepatitis C virus epitope: Evidence for the critical function of multimerization. Virology 2007, 363, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Paruch, L.; Dobrica, M.-O.; Caras, I.; Tucureanu, C.; Onu, A.; Ciulean, S.; Stavaru, C.; Eerde, A.; Wang, Y.; et al. Lettuce-produced hepatitis C virus E1E2 heterodimer triggers immune responses in mice and antibody production after oral vaccination. Plant Biotech. J. 2017, 15, 1611–1621. [Google Scholar] [CrossRef]
- Nemchinov, L.G.; Liang, T.J.; Rifaat, M.M.; Mazyad, H.M.; Hadidi, A.; Keith, J.M. Development of a plant-derived subunit vaccine candidate against hepatitis C virus. Arch. Virol. 2000, 145, 2557–2573. [Google Scholar] [CrossRef] [PubMed]
- Attar, A.E.; Shamloul, A.; Shalaby, A.; Riad, B.; Saad, A.; Mazyad, H.; Keith, J. Expression of chimeric HCV peptide in transgenic tobacco plants infected with recombinant alfalfa mosaic virus for development of a plant-derived vaccine against HCV African J. Biotechnol. 2004, 3, 7. [Google Scholar]
- Natilla, A.; Piazzolla, G.; Nuzzaci, M.; Saldarelli, P.; Tortorella, C.; Antonaci, S.; Piazzolla, P. Cucumber mosaic virus as carrier of a hepatitis C virus-derived epitope. Arch. Virol. 2003, 149, 137–154. [Google Scholar] [CrossRef]
- Piazzolla, G.; Nuzzaci, M.; Tortorella, C.; Panella, E.; Natilla, A.; Boscia, D.; De Stradis, A.; Piazzolla, P.; Antonaci, S. Immunogenic Properties of a Chimeric Plant Virus Expressing a Hepatitis C Virus (HCV)-Derived Epitope: New Prospects for an HCV Vaccine. J. Clin. Immunol. 2005, 25, 142–152. [Google Scholar] [CrossRef]
- Nuzzaci, M.; Piazzolla, G.; Vitti, A.; Lapelosa, M.; Tortorella, C.; Stella, I.; Natilla, A.; Antonaci, S.; Piazzolla, P. Cucumber mosaic virus as a presentation system for a double hepatitis C virus-derived epitope. Arch. Virol. 2007, 152, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Nuzzaci, M.; Vitti, A.; Condelli, V.; Lanorte, M.T.; Tortorella, C.; Boscia, D.; Piazzolla, P.; Piazzolla, G. In vitro stability of Cucumber mosaic virus nanoparticles carrying a Hepatitis C virus-derived epitope under simulated gastrointestinal conditions and in vivo efficacy of an edible vaccine. J. Virol. Meth. 2010, 165, 211–215. [Google Scholar] [CrossRef]
- Yusibov, V.; Kushnir, N.; Streatfield, S.J. Advances and challenges in the development and production of effective plant-based influenza vaccines. Expert Rev. Vaccines 2015, 14, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Krug, R.M. Orthomyxoviridae: The Viruses and Their Replication. In Fields Virology, 4th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 1487–1531. [Google Scholar]
- Noda, T.; Sagara, H.; Yen, A.; Takada, A.; Kida, H.; Cheng, R.H.; Kawaoka, Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 2006, 439, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Pielak, R.M.; Chou, J.J. Influenza M2 proton channels. Biochim. Biophys. Acta 2011, 1808, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Puertas, P.; Albo, C.; Pérez-Pastrana, E.; Vivo, A.; Portela, A. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 2000, 74, 11538–11547. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.P.; Balogun, R.A.; Yamada, H.; Zhou, Z.H.; Barman, S. Influenza virus morphogenesis and budding. Virus Res. 2009, 143, 147–161. [Google Scholar] [CrossRef]
- Han, T.; Marasco, W.A. Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 2011, 1217, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Gamblin, S.J.; Skehel, J.J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 2010, 285, 28403–28409. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.R.; Barr, G.C., Jr.; Mackenzie, R.S.; Rosenau, A.M.; Weaver, K.R.; Ortiz, M. Building an effective ED influenza vaccine program. Am. J. Emerg. Med. 2009, 27, 1154–1157. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.F.; Wang, H.Q.; Wang, J.Z.; Fang, H.H.; Wu, J.; Zhu, F.C.; Li, R.C.; Xia, S.L.; Zhao, Y.L.; Li, F.J.; et al. Safety and immunogenicity of 2009 pandemic influenza A H1N1 vaccines in China: A multicentre, double-blind, randomised, placebo-controlled trial. Lancet 2010, 375, 56–66. [Google Scholar] [CrossRef]
- Plennevaux, E.; Sheldon, E.; Blatter, M.; Reeves-Hoché, M.-K.; Denis, M. Immune response after a single vaccination against 2009 influenza A H1N1 in USA: A preliminary report of two randomised controlled phase 2 trials. Lancet 2010, 375, 41–48. [Google Scholar] [CrossRef]
- Keitel, W.A.; Campbell, J.D.; Treanor, J.J.; Walter, E.B.; Patel, S.M.; He, F.; Noah, D.L.; Hill, H. Safety and immunogenicity of an inactivated influenza A/H5N1 vaccine given with or without aluminum hydroxide to healthy adults: Results of a phase I-II randomized clinical trial. J. Infect. Dis. 2008, 198, 1309–1316. [Google Scholar] [CrossRef]
- Nolan, T.M.; Richmond, P.C.; Skeljo, M.V.; Pearce, G.; Hartel, G.; Formica, N.T.; Höschler, K.; Bennet, J.; Ryan, D.; Papanaoum, K.; et al. Phase I and II randomised trials of the safety and immunogenicity of a prototype adjuvanted inactivated split-virus influenza A (H5N1) vaccine in healthy adults. Vaccine 2008, 26, 4160–4167. [Google Scholar] [CrossRef] [PubMed]
- Schultz-Cherry, S.; Jones, J.C. Influenza vaccines: The good, the bad, and the eggs. Adv. Virus Res. 2010, 77, 63–84. [Google Scholar] [PubMed]
- Michiels, B.; Govaerts, F.; Remmen, R.; Vermeire, E.; Coenen, S. A systematic review of the evidence on the effectiveness and risks of inactivated influenza vaccines in different target groups. Vaccine 2011, 29, 9159–9170. [Google Scholar] [CrossRef]
- Osterholm, M.T.; Kelley, N.S.; Sommer, A.; Belongia, E.A. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 36–44. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Ravin, N.V. Plant-produced Recombinant Influenza A Vaccines Based on the M2e Peptide. Curr. Pharm. Des. 2018, 24, 1317–1324. [Google Scholar] [CrossRef]
- Pellerin, C. DARPA Effort Speeds Biothreat Response. In American Forces Press Service; U.S. Department of Defense: Washington, DC, USA, 2010. [Google Scholar]
- DARPA Makes 10 Million Strides in the Race to Contain a Hypothetical Pandemic. 2012. Available online: http://www.darpa.mil/NewsEvents/Releases/2012/07/25.aspx (accessed on 25 July 2012).
- Landry, N.; Ward, B.J.; Trepanier, S.; Montomoli, E.; Dargis, M.; Lapini, G.; Vezina, L.P. Preclinical and clinical development of plant-made virus-like particle vaccine against avian H5N1 influenza. PLoS ONE 2010, 5, e15559. [Google Scholar] [CrossRef] [PubMed]
- Pillet, S.; Aubin, É.; Trépanier, S.; Poulin, J.F.; Yassine-Diab, B.; Ter Meulen, J.; Ward, B.J.; Landry, N. Humoral and cell-mediated immune responses to H5N1 plant-made virus-like particle vaccine are differentially impacted by alum and GLA-SE adjuvants in a Phase 2 clinical trial. NPJ Vaccines 2018, 3, 3. [Google Scholar] [CrossRef]
- Pillet, S.; Racine, T.; Nfon, C.; Di Lenardo, T.Z.; Babiuk, S.; Ward, B.J.; Kobinger, G.P.; Landry, N. Plant-derived H7 VLP vaccine elicits protective immune response against H7N9 influenza virus in mice and ferrets. Vaccine 2015, 33, 6282–6289. [Google Scholar] [CrossRef] [PubMed]
- Pillet, S.; Aubin, É.; Trépanier, S.; Bussière, D.; Dargis, M.; Poulin, J.F.; Yassine-Diab, B.; Ward, B.J.; Landry, N. A plant-derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin. Immunol. 2016, 168, 72–87. [Google Scholar] [CrossRef]
- Pillet, S.; Couillard, J.; Trépanier, S.; Poulin, J.F.; Yassine-Diab, B.; Guy, B.; Ward, B.J.; Landry, N. Immunogenicity and safety of a quadrivalent plant-derived virus like particle influenza vaccine candidate-Two randomized Phase II clinical trials in 18 to 49 and ≥50 years old adults. PLoS ONE 2019, 14, e0216533. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, immuno-genicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): Two multicentre, randomised phase 3 trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Tregoning, J.S. First human efficacy study of a plant derived influenza vaccine. Lancet 2020, 396, 1464–1465. [Google Scholar] [CrossRef]
- Medicago. Available online: https://www.nature.com/articles/d43747-020-00537-y (accessed on 1 June 2018).
- Medicago’s Plant-Based COVID-19 Vaccine Shows Positive Phase 2 Results. Available online: https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=18790 (accessed on 26 May 2021).
- Mapp Biopharmaceutical; Gantz, S. Emergent BioSolutions among three under consideration for Ebola drug manufacturing. Baltim. Bus. J. Available online: https://www.bizjournals.com/baltimore/news/2014/10/20/emergent-biosolutions-among-three-under.html. (accessed on 20 October 2014).
- Medicago successfully produces plant-based Rotavirus VLP vaccine candidate. Available online: https://www.prnewswire.com/news-releases/medicago-successfully-produces-plant-based-rotavirus-vlp-vaccine-candidate-212290651.html (accessed on 20 June 2013).
- Tacket, C.O.; Mason, H.S.; Losonsky, G.; Estes, M.K.; Levine, M.M.; Arntzen, C.J. Human immune responses to a novel Norwalk virus vaccine delivered in transgenic potatoes. J. Infect. Dis. 2000, 182, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Yusibov, V.; Hooper, D.C.; Spitsin, S.V.; Fleysh, N.; Kean, R.B.; Mikheeva, T.; Deka, D.; Karasev, A.; Cox, S.; Randall, J.; et al. Expression in plants and immunogenicity of plant virus-based experimental rabies vaccine. Vaccine 2002, 20, 3155–3164. [Google Scholar] [CrossRef]
- Kapusta, J.; Modelska, A.; Figlerowicz, M.; Pniewski, T.; Letellier, M.; Lisowa, O.; Yusibov, V.; Koprowski, H.; Plucienniczak, A.; Legocki, A.B. A plant-derived edible vaccine against hepatitis B virus. FASEB J. 1999, 13, 1796–1799. [Google Scholar] [CrossRef]
- Thanavala, Y.; Mahoney, M.; Pal, S.; Scott, A.; Richter, L.; Natarajan, N.; Goodwin, P.; Arntzen, C.J.; Mason, H.S. Immunogenicity in humans of an edible vaccine for hepatitis B. Proc. Natl. Acad. Sci. USA 2005, 102, 3378–3382. [Google Scholar] [CrossRef]
- Nochi, T.; Yuki, Y.; Katakai, Y.; Shibata, H.; Tokuhara, D.; Mejima, M.; Kurokawa, S.; Takahashi, Y.; Nakanishi, U.; Ono, F.; et al. A rice-based oral cholera vaccine induces macaque-specific systemic neutralizing antibodies but does not influence pre-existing intestinal immunity. J. Immunol. 2009, 183, 6538–6544. [Google Scholar] [CrossRef]
- Yuki, Y.; Mejima, M.; Kurokawa, S.; Hiroiwa, T.; Takahashi, Y.; Tokuhara, D.; Nochi, T.; Katakai, Y.; Kuroda, M.; Takeyama, N.; et al. Induction of toxin-specific neutralizing immunity by molecularly uniform rice-based oral cholera toxin B subunit vaccine without plant-associated sugar modification. Plant Biotechnol. J. 2013, 11, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Kurup, V.M.; Thomas, J. Edible Vaccines: Promises and Challenges. Mol. Biotechnol. 2020, 62, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Mallajosyula, J.K.; Hiatt, E.; Hume, S.; Johnson, A.; Jeevan, T.; Chikwamba, R.; Pogue, G.P.; Bratcher, B.; Haydon, H.; Webby, R.J.; et al. Single-dose monomeric HA subunit vaccine generates full protection from influenza challenge. Hum. Vaccin Immunother. 2014, 10, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Parkhouse, K.; Gumina, M.E.; Kim, K.; Perez, S.D.; Wilson, P.C.; Treanor, J.J.; Sant, A.J.; Cobey, S.; Hensley, S.E. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. USA 2017, 114, 12578–12583. [Google Scholar] [CrossRef]
- Lindsay, B.J.; Bonar, M.M.; Costas-Cancelas, I.N.; Hunt, K.; Makarkov, A.I.; Chierzi, S.; Krawczyk, C.M.; Landry, N.; Ward, B.J.; Rouiller, I. Morphological characterization of a plant-made virus-like particle vaccine bearing influenza virus hemagglutinins by electron microscopy. Vaccine 2018, 36, 2147–2154. [Google Scholar] [CrossRef]
- Pushko, P.; Tretyakova, I. Influenza Virus Like Particles (VLPs): Opportunities for H7N9 Vaccine Development. Viruses 2020, 12, 518. [Google Scholar] [CrossRef]
- Won, S.-Y.; Hunt, K.; Guak, H.; Hasaj, B.; Charland, N.; Landry, N.; Ward, B.J.; Krawczyk, C.M. Characterization of the innate stimulatory capacity of plant-derived virus-like particles bearing influenza hemagglutinin. Vaccine 2018, 36, 8028–8038. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Kotlyarov, R.Y.; Kuprianov, V.V.; Stepanova, L.A.; Tsybalova, L.M.; Lomonosoff, G.P.; Ravin, N.V. Rapid high-yield expression of a candidate influenza vaccine based on the ectodomain of M2 protein linked to flagellin in plants using viral vectors. BMC Biotechnol. 2015, 15, 42. [Google Scholar] [CrossRef]
- Ameghi, A.; Pilehvar-Soltanahmadi, Y.; Baradaran, B.; Barzegar, A.; Taghizadeh, M.; Zarghami, N.; Aghaiypour, K. Protective Immunity Against Homologous and Heterologous Influenza Virus Lethal Challenge by Immunization with New Recombinant Chimeric HA2-M2e Fusion Protein in BALB/C Mice. Viral Immunol. 2016, 29, 228–234. [Google Scholar] [CrossRef]
- Deng, L.; Kim, J.R.; Chang, T.Z.; Zhang, H.; Mohan, T.; Champion, J.A.; Wang, B.-Z. Protein nanoparticle vaccine based on flagellin carrier fused to influenza conserved epitopes confers full protection against influenza A virus challenge. Virology 2017, 509, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, L.A.; Mardanova, E.S.; Shuklina, M.A.; Blokhina, E.A.; Kotlyarov, R.Y.; Potapchuk, M.V.; Kovaleva, A.A.; Vidyaeva, I.G.; Korotkov, A.V.; Eletskaya, E.I.; et al. Flagellin-fused protein targeting M2e and HA2 induces potent humoral and T-cell responses and protects mice against various influenza viruses a subtypes. J. Biomed. Sci. 2018, 25, 33. [Google Scholar] [CrossRef] [PubMed]
- Neirynck, S.; Deroo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, L.A.; Kotlyarov, R.Y.; Shuklina, M.A.; Blochina, E.A.; Sergeeva, M.V.; Potapchuk, M.V.; Kovaleva, A.A.; Ravin, N.V.; Tsybalova, L.M. Influence of the Linking Order of Fragments of HA2 and M2e of the influenza A Virus to Flagellin on the Properties of Recombinant Proteins. Acta Nat. 2018, 10, 85–94. [Google Scholar] [CrossRef]
- Ward, B.J.; Landry, N.; Trépanier, S.; Mercier, G.; Dargis, M.; Couture, M.; D’Aoust, M.-A.; Vézina, L.-P. Human antibody response to N-glycans present on plant-made influenza virus-like particle (VLP) vaccines. Vaccine 2014, 32, 6098–6106. [Google Scholar] [CrossRef]
- Makarkov, A.I.; Chierzi, S.; Pillet, S.; Murai, K.K.; Landry, N.; Ward, B.J. Plant-made virus-like particles bearing influenza hemagglutinin (HA) recapitulate early interactions of native influenza virions with human monocytes/macrophages. Vaccine 2017, 35, 4629–4636. [Google Scholar] [CrossRef]
- Carignan, D.; Thérien, A.; Rioux, G.; Paquet, G.; Gagné, M.-E.L.; Bolduc, M.; Savard, P.; Leclerc, D. Engineering of the PapMV vaccine platform with a shortened M2e peptide leads to an effective one dose influenza vaccine. Vaccine 2015, 33, 7245–7253. [Google Scholar] [CrossRef]
- Thérien, A.; Bédard, M.; Carignan, D.; Rioux, G.; Gauthier-Landry, L.; Laliberté-Gagné, M.-E.; Bolduc, M.; Savard, P.; Leclerc, D. A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. J. Nanobiotechnology 2017, 15, 54. [Google Scholar] [CrossRef]
- Bolduc, M.; Baz, M.; Laliberté-Gagné, M.-E.; Carignan, D.; Garneau, C.; Russel, A.; Boivin, G.; Savard, P.; Leclerc, D. The quest for a nanoparticle-based vaccine inducing broad protection to influenza viruses. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2563–2574. [Google Scholar] [CrossRef]
- Denis, J.; Acosta-Ramirez, E.; Zhao, Y.; Hamelin, M.-E.; Koukavica, I.; Baz, M.; Abed, Y.; Savard, C.; Pare, P.; Macias, C.L.; et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine 2008, 26, 3395–3403. [Google Scholar] [CrossRef]
- Babin, C.; Majeau, N.; Leclerc, D. Engineering of papaya mosaic virus (PapMV) nanoparticles with a CTL epitope derived from influenza NP. J. Nanobiotechnol. 2013, 11, 10. [Google Scholar] [CrossRef]
- Laliberté-Gagné, M.-E.; Bolduc, M.; Thérien, A.; Garneau, C.; Casault, P.; Savard, P.; Estaquier, J.; Leclerc, D. Increased Immunogenicity of Full-Length Protein Antigens through Sortase-Mediated Coupling on the PapMV Vaccine Platform. Vaccines 2019, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, D.; Beauseigle, D.; Denis, J.; Morin, H.; Paré, C.; Lamarre, A.; Lapointe, R. Proteasome-independent major histocompatibility complex class I cross-presentation mediated by papaya mosaic virus-like particles leads to expansion of specific human T cells. J. Virol. 2007, 81, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Zahmanova, G.G.; Mazalovska, M.; Takova, K.H.; Toneva, V.T.; Minkov, I.N.; Mardanova, E.S.; Ravin, N.V.; Lomonossoff, G.P. Rapid High-Yield Transient Expression of Swine Hepatitis E ORF2 Capsid Proteins in Nicotiana benthamiana Plants and Production of Chimeric Hepatitis E Virus-Like Particles Bearing the M2e Influenza Epitope. Plants 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, E.A.; Mardanova, E.S.; Stepanova, L.A.; M Tsybalova, L.M.; Ravin, N.V. Plant-Produced Recombinant Influenza A Virus Candidate Vaccine Based on Flagellin Linked to Conservative Fragments of M2 Protein and Hemagglutintin. Plants 2020, 9, 162. [Google Scholar] [CrossRef]
- Pham, N.B.; Ho, T.T.; Nguyen, G.T.; Le, T.T.; Le, N.T.; Chang, H.-C.; Pham, M.D.; Conrad, U.; Chu, H.H. Nanodiamond enhances immune responses in mice against recombinant HA/H7N9 protein. J. Nanobiotechnol. 2017, 15, 69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, T.; O’Kennedy, M.M.; Wandrag, D.B.R.; Adeyemi, M.; Abolnik, C. Efficacy of a plant-produced virus-like particle vaccine in chickens challenged with Influenza A H6N2 virus. Plant Biotechnol. J. 2020, 18, 502–512. [Google Scholar] [CrossRef]
- Schiller, J.T.; Castellsagué, X.; Villa, L.L.; Hildesheim, A. An update of prophylactic human papillomavirus L1 virus-like particle vaccine clinical trial results. Vaccine 2008, 26, K53–K61. [Google Scholar] [CrossRef]
- Petrosky, E.; Bocchini, J.A., Jr.; Hariri, S.; Chesson, H.; Curtis, C.R.; Saraiya, M.; Unger, E.R.; Markowitz, L.E. Use of 9-valent human papillomavirus (HPV) vaccine: Updated HPV vaccination recommendations of the advisory committee on immunization practices. MMWR 2015, 64, 300. [Google Scholar]
- McKee, S.J.; Bergot, A.S.; Leggatt, G.R. Recent progress in vaccination against human papillomavirus-mediated cervical cancer. Rev. Med. Virol. 2015, 25, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Biemelt, S.; Sonnewald, U.; Galmbacher, P.; Willmitzer, L.; Müller, M. Production of human papillomavirus type 16 virus-like particles in transgenic plants. J. Virol. 2003, 77, 9211–9220. [Google Scholar] [CrossRef]
- Williams, M.G.; Howatson, A.F.; Almeida, J.D. Morphological characterization of the virus of the human common wart (verruca vulgaris). Nature 1961, 189, 895–897. [Google Scholar] [CrossRef]
- Howley, P.M.; Lowy, D.R. Fields Virology, 2nd ed.; Bernard, F.N., Diane, G.E., Peter, H.M., David, K.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 2197–2229. [Google Scholar]
- Fehrmann, F.; Laimins, L.A. Human papillomaviruses: Targeting differentiating epithelial cells for malignant transformation. Oncogene 2003, 22, 5201–5207. [Google Scholar] [CrossRef]
- Braspenning, J.; Gissmanna, L. Chimeric papillomavirus like particles. Virology 1997, 234, 93–111. [Google Scholar]
- Villa, L.L.; Costa, R.L.; Petta, C.A.; Andrade, R.P.; Paavonen, J.; Iversen, O.E.; Olsson, S.E.; Hoye, J.; Steinwall, M.; Riis, J.G.; et al. High sustained efficacy of a prophylactic quadrivalent human papillomavirus types 6/11/16/18 L1 virus-like particle vaccine through 5 years of follow-up. Br. J. Cancer 2006, 95, 1459–1466. [Google Scholar] [CrossRef]
- Breitburd, F.; Kirnbauer, R.; Hubbert, N.L.; Nonnenmacher, B.; Desmarquet, T.D.; Orth, G.; Schiller, J.T.; Lowy, D.R. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 1995, 69, 3959–3963. [Google Scholar] [CrossRef]
- Suzich, J.A.; Ghim, S.J.; Palmer, F.J.; White, W.I.; Tamura, J.K.; Bell, J.A.; Newsome, J.A.; Jenson, A.B.; Schlegel, R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc. Natl. Acad. Sci. USA 1995, 92, 11553–11557. [Google Scholar] [CrossRef] [PubMed]
- Waheed, M.T.; Gottschamel, J.; Hassan, S.W.; Lössl, A.G. Plant-derived vaccines: An approach for affordable vaccines against cervical cancer. Hum. Vaccines Immunother. 2012, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Rybicki, E.P. Plant made vaccines for humans and animals. Plant Biotechnol. J. 2010, 8, 620–637. [Google Scholar] [CrossRef]
- Fischer, R.; Stoger, E.; Schillberg, S.; Christou, P.; Twyman, R.M. Plant-based production of biopharmaceuticals. Curr. Opin. Plant Biol. 2004, 7, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Chabeda, A.; van Zyl, A.R.; Rybicki, E.P.; Hitzeroth, I.I. Substitution of Human Papillomavirus Type 16 L2 Neutralizing Epitopes into L1 Surface Loops: The Effect on Virus-Like Particle Assembly and Immunogenicity. Front. Plant Sci. 2019, 10, 779. [Google Scholar] [CrossRef]
- Naupu, P.N.; van Zyl, A.R.; Rybicki, E.P.; Hitzeroth, I.I. Immunogenicity of Plant-Produced Human Papillomavirus (HPV) Virus-Like Particles (VLPs). Vaccine 2020, 8, 740. [Google Scholar] [CrossRef] [PubMed]
- Latif, S.; Gottschamel, J.; Syed, T.; Younus, I.; Gull, K.; Sameeullah, M.; Batool, N.; Lössl, A.G.; Mariz, F.; Müller, M.; et al. Inducible expression of human papillomavirus-16 L1 capsomeres in the plastomes of Nicotiana tabacum: Transplastomic plants develop normal flowers and pollen. Biotechnol. Appl. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Salyaev, R.K.; Rekoslavskaya, N.I.; Stolbikov, A.S. The New Plant Expression System for the Development of Vaccines against Papillomaviruses. Dokl. Biochem. Biophys. 2019, 484, 52–54. [Google Scholar] [CrossRef]
- Massa, S.; Paolini, F.; Marino, C.; Franconi, R.; Venuti, A. Bioproduction of a Therapeutic Vaccine Against Human Papillomavirus in Tomato Hairy Root Cultures. Front. Plant Sci. 2019, 10, 452. [Google Scholar] [CrossRef]
- Yazdani, R.; Shams-Bakhsh, M.; Hassani-Mehraban, A.; Arab, S.S.; Thelen, N.; Thiry, M.; Crommen, J.; Fillet, M.; Jacobs, N.; Brans, A.; et al. Production and characterization of virus like particles of grapevine fanleaf virus presenting L2 epitope of human papillomavirus minor capsid protein. BMC Biotechnol. 2019, 19, 81. [Google Scholar] [CrossRef]
- Diamos, A.G.; Larios, D.; Brown, L.; Kilbourne, J.; Kim, H.S.; Saxena, D.; Palmer, K.E.; Mason, H.S. Vaccine synergy with virus-like particle and immune complex platforms for delivery of human papillomavirus L2 antigen. Vaccine 2019, 37, 137–144. [Google Scholar] [CrossRef]
- Tremouillaux-Guiller, J.; Moustafa, K.; Hefferon, K.; Gaobotse, G.; Makhzoum, A. Plant-made HIV vaccines and potential candidates. Curr. Opin. Biotechnol. 2020, 61, 209–216. [Google Scholar] [CrossRef]
- Stephenson, K.E.; Neubauer, G.H.; Reimer, U.; Pawlowski, N.; Knaute, T.; Zerweck, J.; Korber, B.T.; Barouch, D.H. Quantification of the epitope diversity of HIV-1-specific binding antibodies by peptide microarrays for global HIV-1 vaccine development. J. Immunol. Methods 2015, 416, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Haddox, H.K.; Dingens, A.S.; Hilton, S.K.; Overbaugh, J.; Bloom, J.D. Mapping mutational effects along the evolutionary landscape of HIV envelope. eLife 2018, 7, e34420. [Google Scholar] [CrossRef] [PubMed]
- Rathore, U.; Purwar, M.; Vignesh, V.S.; Das, R.; Kumar, A.A.; Bhattacharyya, S.; Arendt, H.; DeStefano, J.; Wilson, A.; Parks, C.; et al. Bacterially expressed HIV-1 gp120 outer-domain fragment immunogens with improved stability and affinity for CD4- binding site neutralizing antibodies. J. Biol. Chem. 2018, 293, 15002–15020. [Google Scholar] [CrossRef]
- Marusic, C.; Vitale, A.; Pedrazzini, E.; Donini, M.; Frigerio, L.; Bock, R.; Dix, P.J.; McCabe, M.S.; Bellucci, M.; Benvenuto, E. Plant-based strategies aimed at expressing HIV antigens and neutralizing antibodies at high levels. Nef as a case study. Transgenic Res. 2009, 18, 499. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seber Kasinger, L.E.; Dent, M.W.; Mahajan, G.; Hamorsky, K.T.; Matoba, N. A novel anti-HIV-1 bispecific bNAb-lectin fusion protein engineered in a plant-based transient expression system. Plant Biotechnol. J. 2019, 17, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.K.C.; Drossard, J.; Lewis, D.; Altmann, F.; Boyle, J.; Christou, P.; Cole, T.; Dale, P.; van Dolleweerd, C.J.; Isitt, V.; et al. Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 2015, 13, 1106–1120. [Google Scholar] [CrossRef]
- Govea-Alonso, D.O.; Gómez-Cardona, E.E.; Rubio-Infante, N.; García-Hernández, A.L.; Varona-Santos, J.T.; Salgado-Bustamante, M.; Korban, S.S.; Moreno-Fierros, L.; Rosales-Mendoza, S. Production of an antigenic C4 (V3) 6 multiepitopic HIV protein in bacterial and plant systems. Plant Cell Tissue Organ. Cult. 2013, 113, 73–79. [Google Scholar]
- Rosales-Mendoza, S.; Rubio-Infante, N.; Monreal-Escalante, E.; Govea-Alonso, D.O.; García-Hernández, A.L.; Salazar-González, J.A.; González-Ortega, O.; Paz-Maldonado, L.T.; Moreno-Fierros, L. Chloroplast expression of an HIV envelop-derived multiepitope protein: Towards a multivalent plant-based vaccine. Plant Cell Tissue Organ. Cult. 2014, 116, 111–123. [Google Scholar]
- Loh, H.-S.; Green, B.J.; Yusibov, V. Using transgenic plants and modified plant viruses for the development of treatments for human diseases. Curr. Opin. Virol. 2017, 26, 81–89. [Google Scholar]
- Orellana-Escobedo, L.; Rosales-Mendoza, S.; Romero- Maldonado, A.; Parsons, J.; Decker, E.L.; Monreal-Escalante, E.; Moreno-Fierros, L.; Reski, R. An Env-derived multi-epitope HIV chimeric protein produced in the moss Physcomitrella patens is immunogenic in mice. Plant Cell Rep. 2015, 34, 425–433. [Google Scholar] [CrossRef]
- Ruhl, C.; Knodler, M.; Opdensteinen, P.; Buyel, J.F. A linear epitope coupled to DsRed provides an affinity ligand for the capture of monoclonal antibodies. J. Chromatogr. A 2018, 1571, 55–64. [Google Scholar] [CrossRef]
- Rubio-Infante, N.; Govea-Alonso, D.O.; Romero-Maldonado, A.; Garcia-Hernandez, A.L.; Ilhuicatzi-Alvarado, D.; Salazar-Gonzalez, J.A.; Korban, S.S.; Rosales-Mendoza, S.; Moreno-Fierros, L. A plant-derived derived multi-HIV antigen induces broad immune responses in orally immunized mice. Mol. Biotechnol. 2015, 57, 662–674. [Google Scholar] [CrossRef]
- Vamvaka, E.; Farre, G.; Molinos-Albert, L.M.; Evans, A.; Canela- Xandri, A.; Twyman, R.M.; Carrillo, J.; Ordonez, R.A.; Shattock, R.J.; O’Keefe, B.R.; et al. Unexpected synergistic HIV neutralization by a triple microbicide produced in rice endosperm. Proc. Natl. Acad. Sci. USA 2018, 115, E7854–E7862. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, J.L.; Wanga, V.; Palmer, K.E. Improving the large scale purification of the HIV microbicide, griffithsin. BMC Biotechnol. 2015, 15, 12. [Google Scholar] [CrossRef]
- Alam, A.; Jiang, L.; Kittleson, G.A.; Steadman, K.D.; Nandi, S.; Fuqua, J.L.; Palmer, K.E.; Tusé, D.; McDonald, K.A. Technoeconomic modeling of plant-based Griffithsin manufacturing. Front. Bioeng. Biotechnol. 2018, 6, 102. [Google Scholar] [CrossRef]
- Nandi, S.; Kwong, A.T.; Holtz, B.R.; Erwin, R.L.; Marcel, S.; McDonald, K.A. Techno-economic analysis of a transient plant-based platform for monoclonal antibody production. MAbs 2016, 8, 1456–1466. [Google Scholar] [CrossRef]
- Opdensteinen, P.; Clodt, J.I.; Müschen, C.R.; Filiz, V.M.; Buyel, J.F. A combined ultrafiltration/diafiltration step facilitates the purification of cyanovirin-N from transgenic tobacco extracts. Front. Bioeng. Biotechnol. 2018, 6, 206. [Google Scholar] [CrossRef] [PubMed]
- Swanson, M.D.; Winter, H.C.; Goldstein, I.J.; Markovitz, D.M. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 2010, 285, 8646–8655. [Google Scholar] [CrossRef] [PubMed]
- Hopper, J.T.; Ambrose, S.; Grant, O.C.; Krumm, S.A.; Allison, T.M.; Degiacomi, M.T.; Tully, M.D.; Pritchard, L.K.; Ozorowski, G.; Ward, A.B. The tetrameric plant lectin BanLec neutralizes HIV through bidentate binding to specific viral glycans. Structure 2017, 25, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Margolin, E.; Chapman, R.; Meyers, A.; van Diepen, M.; Ximba, P.; Hermanus, T.; Crowther, C.; Weber, B.; Morris, L.; Williamson, A.-L. Production and immunogenicity of soluble plant-produced HIV-1 subtype C envelope gp140 immunogens. Front. Plant Sci. 2019, 10, 1378. [Google Scholar] [CrossRef]
- Barahimipour, R.; Neupert, J.; Bock, R. Efficient expression of nuclear transgenes in the green alga Chlamydomonas: Synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 2016, 90, 403–418. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, M.A.; Couture, M.M.; Lavoie, P.O.; Vezina, L.P. Virus Like Particle Production in Plants. Patent number WO2012083445 (A1), 28 June 2012. [Google Scholar]
- Wang, B.Z.; Liu, W.; Kang, S.M.; Alam, M.; Huang, C.; Ye, L.; Sun, Y.; Li, Y.; Kothe, D.L.; Pushko, P.; et al. Incorporation of high levels of chimeric human immunodeficiency virus envelope glycoproteins into virus-like particles. J. Virol. 2007, 81, 10869–10878. [Google Scholar] [CrossRef]
- Kessans, S.A.; Linhart, M.D.; Matoba, N.; Mor, T. Biological and biochemical characterization of HIV-1 Gag/dgp41 virus-like particles expressed in Nicotiana benthamiana. Plant Biotechnol. J. 2013, 11, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Scotti, N.; Alagna, F.; Ferraiolo, E.; Formisano, G.; Sannino, L.; Buonaguro, L.; De Stradis, A.; Vitale, A.; Monti, L.; Grillo, S.; et al. High-level expression of the HIV-1 Pr55gag polyprotein in transgenic tobacco chloroplasts. Planta 2009, 229, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Porta, C.; Spall, V.E.; Loveland, J.; Johnson, J.E.; Barker, P.J.; Lomonossoff, G.P. Development of cowpea mosaic virus as a high-yielding system for the presentation of foreign peptides. Virology 1994, 202, 949–955. [Google Scholar] [CrossRef] [PubMed]
- McLain, L.; Porta, C.; Lomonossoff, G.P.; Durrani, Z.; Dimmock, N.J. Human immunodeficiency virus type 1-neutralizing anti-bodies raised to a glycoprotein 41 peptide expressed on the surface of a plant virus. AIDS Res. Hum. Retrovir. 1995, 11, 327–334. [Google Scholar] [CrossRef] [PubMed]
- McLain, L.; Durrani, Z.; Wisniewski, L.A.; Porta, C.; Lomonossoff, G.P.; Dimmock, N.J. Stimulation of neutralizing antibodies to human immunodeficiency virus type 1 in three strains of mice immunized with a 22 amino acid peptide of gp41 expressed on the surface of a plant virus. Vaccine 1996, 14, 799–810. [Google Scholar] [CrossRef]
- Buratti, E.; McLain, L.; Tisminetzky, S.; Cleveland, S.M.; Dimmock, N.J.; Baralle, F.E. The neutralizing antibody response against a conserved region of human immunodeficiency virus type 1 gp41 (amino acid residues 731–752) is uniquely directed against a conformational epitope. J. Gen. Virol. 1998, 79, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.G.; Rodrigues, L.; Rovinski, B.; White, K.A. Production of HIV-1 p24 protein in transgenic tobacco plants. Mol. Biotechnol. 2002, 20, 131–136. [Google Scholar] [CrossRef]
- Karasev, A.V.; Foulke, S.; Wellens, C.; Rich, A.; Shon, K.J.; Zwierzynski, I.; Hone, D.; Koprowski, H.; Reitz, M. Plant based HIV-1 vaccine candidate: Tat protein produced in spinach. Vaccine 2005, 23, 1875–1880. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S. Will plant-made biopharmaceuticals play a role in the fight against COVID-19? Expert Opin. Biol. Ther. 2020, 20, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 26, 102433. [Google Scholar] [CrossRef]
- Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020, 81, 537–540. [Google Scholar] [CrossRef]
- Shen, C.; Wang, Z.; Zhao, F.; Yang, Y.; Li, J.; Yuan, J.; Wang, F.; Li, D.; Yang, M.; Xing, L.; et al. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA 2020, 323, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H., III; Leist, S.R.; Yount, B.L., Jr.; McAnarney, E.T.; Graham, R.L.; Waters, K.M.; Baric, R.S. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018, 92, e00710-18. [Google Scholar] [CrossRef]
- Kotomina, T.; Isakova-Sivak, I.; Matyushenko, V.; Kim, K.H.; Lee, Y.; Jung, Y.J.; Kang, S.M.; Rudenko, L. Recombinant live attenuated influenza vaccine viruses carrying CD8 T-cell epitopes of respiratory syncytial virus protect mice against both pathogens without inflammatory disease. Antivir. Res. 2019, 168, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kobinger, G.P.; Figueredo, J.M.; Rowe, T.; Zhi, Y.; Gao, G.; Sanmiguel, J.C.; Bell, P.; Wivel, N.A.; Zitzow, L.A.; Flieder, D.B.; et al. Adenovirus-based vaccine prevents pneumonia in ferrets challenged with the SARS coronavirus and stimulates robust immune responses in macaques. Vaccine 2007, 25, 5220–5231. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Wu, T.; Guan, J.; Handel, A.; Tscharke, D.C.; Sidney, J.; Sette, A.; Wakim, L.M.; Sng, X.Y.X.; Thomas, P.G.; Croft, N.P.; et al. Quantification of epitope abundance reveals the effect of direct and cross-presentation on influenza CTL responses. Nat. Commun. 2019, 10, 2846. [Google Scholar] [CrossRef]
- Armbruster, N.; Jasny, E.; Petsch, B. Advances in RNA vaccines for preventive indications: A case study of a vaccine against rabies. Vaccines 2019, 7, 132. [Google Scholar] [CrossRef]
- CureVac Focuses on the Development of mRNA-Based Coronavirus Vaccine to Protect People Worldwide. Available online: https://www.curevac.com/news/curevac-focuses-on-the-development-of-mrna-basedcoronavirus-vaccine-to-protect-people-worldwide (accessed on 25 March 2020).
- Mahmood, N.; Nasir, S.B.; Hefferon, K. Plant-Based Drugs and Vaccines for COVID-19. Vaccines 2021, 9, 15. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Menachery, V.D. Return of the coronavirus: 2019-nCoV. Viruses 2020, 12, 135. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J. Virol. 2020, 9, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Jiang, S.; He, Y.; Liu, S. SARS vaccine development. Emerg. Infect. Dis. 2005, 11, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Regla-Nava, J.A.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Fernandez-Delgado, R.; Fett, C.; Castaño-Rodríguez, C.; Perlman, S.; Enjuanes, L.; DeDiego, M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015, 89, 3870–3887. [Google Scholar] [CrossRef]
- Takano, T.; Yamada, S.; Doki, T.; Hohdatsu, T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 2019, 81, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Capell, T.; Twyman, R.M.; Armario-Najera, V.; Ma, J.K.-C.; Schillberg, S.; Christou, P. Potential Applications of Plant Biotechnology against SARS-CoV-2. Trends Plant Sci. 2020, 25, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.P.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. Nature 2020. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Guan, X.-H.; Li, Y.-H.; Huang, J.-Y.; Jiang, T.; Hou, L.-H.; Li, J.-X.; Yang, B.-F.; Wang, L.; Wang, W.-J.; et al. Immunogenicity and Safety of a Recombinant Adenovirus Type-5-Vectored COVID-19 Vaccine in Healthy Adults Aged 18 Years or Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 2 Trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Novavax.com. Clinical Stage Pipeline–Novavax−Creating Tomorrow’s Vaccines Today. 2021. Available online: https://novavax.com/ourpipeline#nvx-cov2373 (accessed on 1 July 2021).
- Gretler, C. Tobacco-Based Coronavirus Vaccine Poised for Human Tests Bloomberg. 2020. Available online: https://www.bloomberg.com/news/articles/2020-05-15/cigarette-maker-s-coronavirus-vaccine-poised-for-human-tests (accessed on 24 December 2020).
- Palca, J. Tobacco Plants Contribute Key Ingredient For COVID-19 Vaccine. 2020. Available online: https://www.npr.org/sections/health-shots/2020/10/15/923210562/tobacco-plants-contribute-key-ingredient-for-covid-19-vaccine#:~{}:text=Historically%2C%20tobacco%20plants%20are%20responsible,be%20used%20in%20a%20vaccine (accessed on 20 December 2020).
- Krokhin, O.; Li, Y.; Andonov, A.; Feldmann, H.; Flick, R.; Jones, S.; Stroeher, U.; Bastien, N.; Dasuri, K.V.N.; Cheng, K.; et al. Mass spectrometric characterization of proteins from the SARS virus: A preliminary report. Mol. Cell. Proteom. 2003, 2, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Mullan, K. Tobacco Giant BAT Says It Could be Making 1 to 3 Million COVID-19 Vaccines a Week by June. 2020. Available online: https://www.derryjournal.com/news/people/tobacco-giant-bat-says-it-could-be-making-1-3-million-covid-19-vaccines-week-june-2526933 (accessed on 24 December 2020).
- Krenek, P.; Šamajová, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Šamaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 trial of a Candidate Recombinant Virus-Like Particle Vaccine for Covid-19 Disease Produced in Plants. MedRxiv 2020. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Márquez-Escobar, V.A.; González-Ortega, O.; Nieto-Gómez, R.; Arévalo-Villalobos, J.I. What Does Plant-Based Vaccine Technology Offer to the Fight against COVID-19? Vaccines 2020, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Phillip Morris International. 2020. Available online: https://www.pmi.com/media-center/news/pmi-announces-medicago-to-supply-up-to-76-million-doses-of-its-plant-derived-covid-19-vaccine-candidate (accessed on 24 October 2020).
- Makay, C. Algae Tasked with Making COVID-19 Kits. 2020. Available online: https://phys.org/news/2020-04-algae-taskedcovid-kits.html (accessed on 24 December 2020).
- Available online: https://www.biospace.com/article/ibio-s-fastpharming-platform-produces-decoy-therapeutic-to-bind-to-sars-cov-2/ (accessed on 16 September 2020).
- Nogrady, B. How SARS-CoV-2 TestsWork and What’s Next in COVID-19 Diagnostics. The Scientist 2020. Available online: https://www.the-scientist.com/news-opinion/how-sars-cov-2-tests-work-and-whats-next-in-covid-19-diagnostics-67210 (accessed on 24 December 2020).
- Chung, Y.H.; Cai, H.; Steinmetz, N.F. Viral nanoparticles for drug delivery, imaging, immunotherapy, and theranostic applications. Adv. Drug Deliv. Rev. 2020, 156, 214–235. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ramirez, M.A.; Soto, F.; Wang, C.; Rueda, R.; Shukla, S.; Silva-Lopez, C.; Kupor, D.; McBride, D.A.; Pokorski, J.K.; Nourhani, A.; et al. Built-In Active Microneedle Patch with Enhanced Autonomous Drug Delivery. Adv. Mater. 2020, 32, e1905740. [Google Scholar] [CrossRef]
- Zhang, W.; Bailey-Elkin, B.A.; Knaap, R.C.M.; Khare, B.; Dalebout, T.J.; Johnson, G.G.; Van Kasteren, P.B.; McLeish, N.J.; Gu, J.; He, W.; et al. Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLoS Pathog. 2017, 13, e1006372. [Google Scholar] [CrossRef] [PubMed]
- Clemente, V.; D’Arcy, P.; Bazzaro, M. Deubiquitinating Enzymes in Coronaviruses and Possible Therapeutic Opportunities for COVID-19. Int. J. Mol. Sci. 2020, 21, 3492. [Google Scholar] [CrossRef] [PubMed]
- Hefferon, K.L. DNA Virus Vectors for Vaccine Production in Plants: Spotlight on Geminiviruses. Vaccines 2014, 2, 642–653. [Google Scholar] [CrossRef]
- Rattanapisit, K.; Shanmugaraj, B.; Manopwisedjaroen, S.; Purwono, P.B.; Siriwattananon, K.; Khorattanakulchai, N.; Hanittinan, O.; Boonyayothin, W.; Thitithanyanont, A.; Ferguson-Smith, A.C.; et al. Rapid production of SARS-CoV-2 receptor binding domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Diamos, A.G.; Pardhe, M.D.; Sun, H.; Hunter, J.G.L.; Mor, T.; Meador, L.; Kilbourne, J.; Chen, Q.; Mason, H.S. Codelivery of improved immune complex and virus-like particle vaccines containing Zika virus envelope domain III synergistically enhances immunogenicity. Vaccine 2020, 38, 3455–3463. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Vannice, K.; Durbin, A.; Hombach, J.; Thomas, S.J.; Thevarjan, I.; Simmons, C.P. Zika vaccines and therapeutics: Landscape analysis and challenges ahead. BMC Med. 2018, 16, 84. [Google Scholar] [CrossRef]
- Durbin, A.; Wilder-Smith, A. An update on Zika vaccine developments. Expert Rev. Vaccines 2017, 16, 781–787. [Google Scholar] [CrossRef] [PubMed]
- (NIAID) NI of A and, ID. VRC 705: A Zika Virus DNA Vaccine in Healthy Adults and Adolescents (DNA) n.d. Available online: https://clinicaltrials.gov/ct2/show/NCT03110770 (accessed on 3 May 2019).
- Attar, N. ZIKA virus circulates in new regions. Nat. Rev. Microbiol. 2016, 14, 62. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.-M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: A case control study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef]
- Lazear, H.M.; Diamond, M.S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 2016, 90, 4864–4875. [Google Scholar] [CrossRef]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 Å resolution cryo-EM structure of Zika virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Foo, S.-S.; Bruzzone, R.; Vu Dinh, L.; King, N.J.C.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef] [PubMed]
- Mason, H.S. Recombinant immune complexes as versatile and potent vaccines. Hum. Vaccines Immunother. 2016, 12, 988–989. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-M.; Mu, L.; Shi, Y. Immunoregulatory functions of immune complexes in vaccine and therapy. EMBO Mol. Med. 2016, 8, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.I.; Camps, M.G.M.; De Haas, E.F.E.; Trouw, L.A.; Verbeek, J.S.; Ossendorp, F. C1qdependent dendritic cell cross-presentation of in vivo-formed antigen–antibody complexes. J. Immunol. 2017, 198, 4235–4243. [Google Scholar] [CrossRef]
- Fletcher, E.A.K.; van Maren, W.; Cordfunke, R.; Dinkelaar, J.; Codee, J.D.C.; van der Marel, G.; Melief, C.J.M.; Ossendorp, F.; Drijfhout, J.W.; Mangsbo, S.M. Formation of immune complexes with a tetanus-derived B Cell epitope boosts human T cell responses to covalently linked peptides in an ex vivo blood loop system. J. Immunol. 2018, 201, 87–97. [Google Scholar] [CrossRef]
- Cabral-Miranda, G.; Lim, S.M.; Mohsen, M.O.; Pobelov, I.V.; Roesti, E.S.; Heath, M.D.; Skinner, M.A.; Kramer, M.F.; Martina, B.E.E.; Bachmann, M.F. Zika Virus-Derived E-DIII Protein Displayed on Immunologically Optimized VLPs Induces Neutralizing Antibodies without Causing Enhancement of Dengue Virus Infection. Vaccines 2019, 7, 72. [Google Scholar] [CrossRef]
- Diamos, A.G.; Hunter, J.G.L.; Pardhe, M.D.; Rosenthal, S.H.; Sun, H.; Foster, B.C.; DiPalma, M.P.; Chen, Q.; Mason, H.S. High Level Production of Monoclonal Antibodies Using an Optimized Plant Expression System. Front. Bioeng. Biotechnol. 2020, 7, 472. [Google Scholar] [CrossRef]
- Chichester, J.A.; Green, B.J.; Jones, R.M.; Shoji, Y.; Miura, K.; Long, C.A.; Lee, C.K.; Ockenhouse, C.F.; Morin, M.J.; Streatfield, S.J.; et al. Safety and immunogenicity of a plant produced Pfs25 virus-like particle as a transmission blocking vaccine against malaria: A phase 1 dose-escalation study in healthy adults. Vaccine 2018, 36, 5865–5871. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.P.; Malhotra, P.V.; Lalitha, S.; Guha-Mukherjee Chauhan, V.S. Expression of Plasmodium falciparum C-terminal region of merozoite surface protein (PfMSP119), a potential malaria vaccine candidate, in tobacco. Plant Sci. 2002, 162, 335–343. [Google Scholar] [CrossRef]
- Targett, G.A.; Greenwood, B.M. Malaria vaccines and their potential role in the elimination of malaria. Malar. J. 2008, 7, S10. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.P.; Reed, Z.H.; Friede, M.; Kieny, M.P. A review of human vaccine research and development: Malaria. Vaccine 2007, 25, 1567–1580. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Huang, D.; Zhang, Q.; Qu, L.; Zhang, D.; Zhang, X.; Xue, X.; Qian, F. Fusion of two malaria vaccine candidate antigens enhances product yield, immunogenicity, and antibody mediated inhibition of parasite growth in vitro. J. Immunol. 2004, 172, 6167–6174. [Google Scholar] [CrossRef] [PubMed]
- Hamid, M.M.; Remarque, E.J.; van Duivenvoorde, L.M.; van der Werff, N.; Walraven, V.; Faber, B.W.; Kocken, C.H.M.; Thomas, A.W. Vaccination with Plasmodium knowlesi AMA1 formulated in the novel adjuvant co-vaccine HT™ protects against blood-stage challenge in Rhesus macaques. PLoS ONE 2011, 6, e20547. [Google Scholar]
- Wang, L.; Webster, D.E.; Campbell, A.E.; Dry, A.B.; Wesselingh, S.L.; Coppel, R.L. Immunogenicity of Plasmodium yoelii merozoite surface protein 4/5 produced in transgenic plants. Int. J. Parasitol. 2008, 38, 103–110. [Google Scholar] [CrossRef]
- Chitnis, C.E.; Mukherjee, P.; Mehta, S.; Yazdani, S.S.; Dhawan, S.; Shakri, A.R.; Bhardwaj, R.; Gupta, P.K.; Hans, D.; Mazumdar, S.; et al. Phase I clinical trial of a recombinant blood stage vaccine candidate for Plasmodium falciparum malaria based on MSP1 and EBA175. PLoS ONE 2015, 10, e0117820. [Google Scholar]
- Boes, A.; Spiegel, H.; Voepel, N.; Edgue, G.; Beiss, V.; Kapelski, S.; Fendel, R.; Scheuermayer, M.; Pradel, G.; Bolscher, J.M.; et al. Analysis of a multi-component multi-stage malaria vaccine candidate—Tackling the cocktail challenge. PLoS ONE 2015, 10, e0131456. [Google Scholar] [CrossRef][Green Version]
- Remarque, E.; Faber, B.; Kocken, C.; Thomas, A. Apical membrane antigen 1: A malaria vaccine candidate in review. Trends Parasitol. 2008, 24, 74–84. [Google Scholar] [CrossRef]
- Paul, G.; Deshmukh, A.; Chourasia, B.K.; Kalamuddin, M.; Panda, A.; Singh, S.K.; Gupta, P.K.; Mohmmed, A.; Chauhan, V.S.; Theisen, M.; et al. Protein–protein interaction studies reveal the Plasmodium falciparum merozoite surface protein-1 region involved in a complex formation that binds to human erythrocytes. Biochem. J. 2018, 475, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Daly, T.; Long, C. A recombinant 15-kilodalton carboxyl-terminal fragment of Plasmodium yoelii yoelii 17XL merozoite surface protein 1 induces a protective immune response in mice. Infect. Immun. 1993, 61, 2462–2467. [Google Scholar] [CrossRef] [PubMed]
- Crewther, P.; Matthew, M.; Flegg, R.; Anders, R. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 1996, 64, 3310–3317. [Google Scholar] [CrossRef] [PubMed]
- Hirunpetcharat, C.; Tian, J.; Kaslow, D.; van Rooijen, N.; Kumar, S.; Berzofsky, J. Complete protective immunity induced in mice by immunization with the 19-kilodalton carboxyl-terminal fragment of the merozoite surface protein-1 (MSP1[19]) of Plasmodium yoelii expressed in Saccharomyces cerevisiae: Correlation of protection with antigen-specific antibody titer, but not with effector CD4+ T cells. J. Immunol. 1997, 159, 3400–3411. [Google Scholar] [PubMed]
- Draper, S.J.; Sack, B.K.; King, C.R.; Nielsen, C.M.; Rayner, J.C.; Higgins, M.K.; Long, C.A.; Seder, R.A. Malaria vaccines: Recent advances and new horizons. Cell Host Microbe 2018, 24, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Milán-Noris, E.M.; Monreal-Escalante, E.; Rosales-Mendoza, S.; Soria-Guerra, R.E.; Radwan, O.; Juvik, J.A.; Korban, S.S. An AMA1/MSP119 Adjuvanted Malaria Transplastomic Plant-Based Vaccine Induces Immune Responses in Test Animals. Mol. Biotechnol. 2020, 62, 534–545. [Google Scholar] [CrossRef]
- Kim, H.; Kwon, K.W.; Park, J.; Kang, H.; Lee, Y.; Sohn, E.-J.; Hwang, I.; Eum, S.-Y.; Shin, S.J. Plant-Produced N-glycosylated Ag85A Exhibits Enhanced Vaccine Efficacy Against Mycobacterium tuberculosis HN878 Through Balanced Multifunctional Th1 T Cell Immunity. Vaccines 2020, 8, 189. [Google Scholar] [CrossRef]
- Saba, K.; Sameeullah, M.; Asghar, A.; Gottschamel, J.; Latif, S.; Lössl, A.G.; Mirza, B.; Mirza, O.; Waheed, M.T. Expression of ESAT-6 antigen from Mycobacterium tuberculosis in broccoli: An edible plant. Biotech. Appl. Biochem. 2020, 67, 148–157. [Google Scholar] [CrossRef]
- Sabaa, K.; Gottschamelb, J.; Younusa, I.; Syeda, T.; Gulla, K.; Lössl, A.G.; Mirzaa, B.; Waheed, M.T. Chloroplast-based inducible expression of ESAT-6 antigen for development of a plant-based vaccine against tuberculosis. J. Biotechnol. 2019, 305, 1–10. [Google Scholar] [CrossRef]
- Xisto, M.F.; Dias, R.S.; Feitosa-Araujo, E.; Prates, J.W.O.; da Silva, C.C.; de Paula, S.O. Efficient Plant Production of Recombinant NS1 Protein for Diagnosis of Dengue. Front. Plant Sci. 2020, 11, 581100. [Google Scholar] [CrossRef]
- Ponndorf1, D.; Meshcheriakova, Y.; Thuenemann, E.C.; Alonso, A.D.; Overman, R.; Holton, N.; Dowall, S.; Kennedy, E.; Stocks, M.; Lomonossoff, G.P.; et al. Plant-made dengue virus-like particles produced by coexpression of structural and non-structural proteins induce a humoral immune response in mice. Plant Biotech. J. 2021, 19, 745–756. [Google Scholar] [CrossRef]
- Grilo, A.L.; Mantalaris, A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 2019, 37, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Davis, K.R. The potential of plants as a system for the development and production of human biologics. F1000Research 2016, 5, 912. [Google Scholar] [CrossRef]
- Tusé, D.; Tu, T.; McDonald, K.A. Manufacturing economics of plant made biologics: Case studies in therapeutic and industrial enzymes. Biomed. Res. Int. 2014, 2014, 256135. [Google Scholar] [CrossRef] [PubMed]
- Walwyn, D.R.; Huddy, S.M.; Rybicki, E.P. Techno-economic analysis of horseradish peroxidase production using a transient expression system in Nicotiana benthamiana. Appl. Biochem. Biotechnol. 2015, 175, 841–854. [Google Scholar] [CrossRef]
- Mir-Artigues, P.; Twyman, R.M.; Alvarez, D.; Cerda Bennasser, P.; Balcells, M.; Christou, P.; Capell, T. A simplified techno-economic model for the molecular pharming of antibodies. Biotechnol. Bioeng. 2019, 116, 2526–2539. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.K.C.; Christou, P.; Chikwamba, R.; Haydon, H.; Paul, M.; Ferrer, M.P.; Ramalingam, S.; Rech, E.; Rybicki, E.; Wigdorovitz, A.; et al. Realising the value of plant molecular pharming to benefit the poor in developing countries and emerging economies. Plant Biotechnol. J. 2013, 11, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Montero-Morales, L.; Steinkellner, H. Advanced plant-based glycan engineering. Front. Bioeng. Biotechnol. 2018, 9, 81. [Google Scholar] [CrossRef]
- Mastrangeli, R.; Palinsky, W.; and Bierau, H. Glycoengineered antibodies: Towards the next-generation of immunotherapeutics. Glycobiology 2019, 29, 199–210. [Google Scholar] [CrossRef]
- Zeitlin, L.; Pettitt, J.; Scully, C.; Bohorova, N.; Kim, D.; Pauly, M.; Hiatt, A.; Ngo, L.; Steinkellner, H.; Whaley, K.J.; et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc. Natl. Acad. Sci. USA 2011, 108, 20690–20694. [Google Scholar] [CrossRef]
- Marusic, C.; Pioli, C.; Stelter, S.; Novelli, F.; Lonoce, C.; Morrocchi, E.; Benvenuto, E.; Maria Salzano, A.; Scaloni, A.; Donini, M. N-glycan engineering of a plant-produced anti-CD20-hIL-2 immunocytokine significantly enhances its effector functions. Biotechnol. Bioeng. 2018, 115, 565–576. [Google Scholar] [CrossRef]
- Castilho, A.; Strasser, R.; Stadlmann, J.; Grass, J.; Jez, J.; Gattinger, P.; Kunert, R.; Quendler, H.; Pabst, M.; Leonard, R.; et al. In planta protein sialylation through overexpression of the respective mammalian pathway. J. Biol. Chem. 2010, 285, 15923–15930. [Google Scholar] [CrossRef]
- Castilho, A.; Neumann, L.; Daskalova, S.; Mason, H.S.; Steinkellner, H.; Altmann, F.; Strasser, R. Engineering of sialylated mucin-type O-glycosylation in plants. J. Biol. Chem. 2012, 287, 36518–36526. [Google Scholar] [CrossRef]
- Olinger, G.G.; Pettitt, J.; Kim, D.; Working, C.; Bohorov, O.; Bratcher, B.; Hiatt, E.; Hume, S.D.; Johnson, A.K.; Morton, J.; et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. USA 2012, 109, 18030–18035. [Google Scholar] [CrossRef]
- Lyon, G.M.; Mehta, A.K.; Varkey, J.B.; Brantly, K.; Plyler, L.; McElroy, A.K.; Kraft, C.S.; Towner, J.S.; Spiropoulou, C.; Ströher, U.; et al. Clinical care of two patients with Ebola virus disease in the United States. N. Engl. J. Med. 2014, 371, 2402–2409. [Google Scholar] [CrossRef]
- Qiu, X.; Wong, G.; Audet, J.; Bello, A.; Fernando, L.; Alimonti, J.B.; Fausther-Bovendo, H.; Wei, H.; Aviles, J.; Hiatt, E.; et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514, 47–53. [Google Scholar] [CrossRef]
- Sun, H.; Chen, Q.; Lai, H. Development of antibody therapeutics against flaviviruses. Int. J. Mol. Sci. 2017, 19, 54. [Google Scholar]
- Dent, M.; Hurtado, J.; Paul, A.M.; Sun, H.; Lai, H.; Yang, M.; Esqueda, A.; Bai, F.; Steinkellner, H.; Chen, Q. Plant produced anti-dengue virus monoclonal antibodies exhibit reduced antibody dependent enhancement of infection activity. J. Gen. Virol. 2016, 97, 3280–3290. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, J.; Acharya, D.; Lai, H.; Sun, H.; Kallolimath, S.; Steinkellner, H.; Bai, F.; Chen, Q. In vitro and in vivo efficacy of anti-chikungunya virus monoclonal antibodies produced in wild-type and glycoengineered Nicotiana benthamiana plants. Plant Biotechnol. J. 2019, 18, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, Q.; Hjelm, B.; Arntzen, C.; and Mason, H. A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnol. Bioeng. 2009, 103, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Phoolcharoen, W.; Lai, H.; Piensook, K.; Cardineau, G.; Zeitlin, L.; Whaley, K.J.; Arntzen, C.J.; Mason, H.S.; Chen, Q. High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnol. Bioeng. 2010, 106, 9–17. [Google Scholar] [CrossRef]
- Diamos, A.G.; Mason, H.S. High-level expression and enrichment of norovirus virus-like particles in plants using modified geminiviral vectors. Protein Exp. Purif. 2018, 151, 86–92. [Google Scholar] [CrossRef]
- Hunter, J.G.; Wilde, S.; Tafoya, A.M.; Horsman, J.; Yousif, M.; Diamos, A.G.; Kenneth L Roland, K.L.; Hugh S Mason, H.S. Evaluation of a toxoid fusion protein vaccine produced in plants to protect poultry against necrotic enteritis. PeerJ 2019, 7, e6600. [Google Scholar] [CrossRef]
- Diamos, A.G.; Mason, H.S. Chimeric 3′ flanking regions strongly enhance gene expression in plants. Plant Biotechnol. J. 2018, 16, 1971–1982. [Google Scholar] [CrossRef]
- van Dolleweerd, C.J.; Teh, A.Y.; Banyard, A.C.; Both, L.; Lotter-Stark, H.C.; Tsekoa, T.; Phahladira, B.; Shumba, W.; Chakauya, E.; Sabeta, C.T.; et al. Engineering, expression in transgenic plants and characterisation of e559, a rabies virus-neutralising monoclonal antibody. J. Infect. Dis. 2014, 210, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.; Dellorto, D. 9 Questions about This New Ebola Drug; CNN: Atlanta, GA, USA, 2014. [Google Scholar]
- Rademacher, T.; Sack, M.; Arcalis, E.; Stadlmann, J.; Balzer, S.; Altmann, F.; Quendler, H.; Stiegler, G.; Kunert, R.; Fischer, R.; et al. Recombinant antibody 2G12 produced in maize endosperm efficiently neutralizes HIV-1 and contains predominantly single-GlcNAc N-glycans. Plant Biotechnol. J. 2008, 6, 189–201. [Google Scholar] [CrossRef]
- Ramessar, K.; Rademacher, T.; Sack, M.; Stadlmann, J.; Platis, D.; Stiegler, G.; Labrou, N.; Altmann, F.; Ma, J.; Stöger, E.; et al. Cost-effective production of a vaginal protein microbicide to prevent HIV transmission. Proc. Natl. Acad. Sci. USA 2008, 105, 3727–3732. [Google Scholar] [CrossRef] [PubMed]
- Vamvaka, E.; Twyman, R.M.; Murad, A.M.; Melnik, S.; Teh, A.Y.-H.; Arcalis, E.; Altmann, F.; Stoger, E.; Rech, E.; Ma, J.K.C.; et al. Rice endosperm produces an underglycosylated and potent form of the HIV-neutralizing monoclonal antibody 2G12. Plant Biotechnol. J. 2016, 14, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.A.; Pooe, O.; Kwezi, L.; Lotter-Stark, T.; Stoychev, S.H.; Alexandra, K.; Gerber, I.; Bhiman, J.N.; Vorster, J.; Pauly, M.; et al. Plant-based production of highly potent anti-HIV antibodies with engineered posttranslational modifications. Sci. Rep. 2020, 10, 6201. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Marsian, J.; Lomonossoff, G.P. Molecular pharming—VLPs made in plants. Curr. Opin. Biotechnol. 2016, 37, 201–206. [Google Scholar] [CrossRef]
- D’Aoust, M.A.; Couture, M.M.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.-P. The production of hemagglutinin-based virus-like particles in plants: A rapid, efficient and saferesponse to pandemic influenza. Plant Biotechnol. J. 2010, 8, 607–619. [Google Scholar] [CrossRef]
- McCarthy, M. US signs contract with ZMapp maker to accelerate development of the Ebola drug. BMJ 2014, 349, g5488. [Google Scholar] [CrossRef]
- Monreal-Escalante, E.; Ramos-Vega, A.A.; Salazar-González, J.A.; Bañuelos-Hernández, B.; Angulo, C.; Rosales-Mendoza, S. Expression of the VP40 antigen from the Zaire ebolavirus in tobacco plants. Planta 2017, 246, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Nieto-Gómez, R.; Angulo, C. A perspective on the development of plant-made vaccines in the fight against ebola virus. Front. Immunol. 2017, 8, 252. [Google Scholar] [CrossRef] [PubMed]
- Phoolcharoen, W.; Dye, J.M.; Kilbourne, J.; Piensook, K.; Pratt, W.D.; Arntzen, C.J.; Chen, Q.; Mason, H.S.; Herbst-Kralovetz, M.M. A nonreplicating subunit vaccine protects mice against lethal Ebola virus challenge. Proc. Natl. Acad. Sci. USA 2011, 108, 20695–20700. [Google Scholar] [CrossRef] [PubMed]
- Hoelscher, M.; Tiller, N.; Teh, A.Y.; Wu, G.-Z.; Ma, J.K.-C.; Bock, R. High-level expression of the HIV entry inhibitor griffithsin from the plastid genome and retention of biological activity in dried tobacco leaves. Plant Mol. Biol. 2018, 97, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Cervera, L.; Gòdia, F.; Tarrés-Freixas, F.; Aguilar-Gurrieri, C.; Carrillo, J.; Blanco, J.; Gutiérrez-Granados, S. Production of HIV-1-based virus-like particles for vaccination: Achievements and limits. Appl. Microbiol. Biotechnol. 2019, 103, 7367–7384. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Rubio-Infante, N.; Govea-Alonso, D.O.; Moreno-Fierros, L. Current status and perspectives of plant-based candidate vaccines against the human immunodeficiency virus (HIV). Plant Cell Rep. 2012, 31, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Lotter-Stark, H.C.; Rybicki, E.P.; Chikwamba, R.K. Plant made anti-HIV microbicides–a field of opportunity. Biotechnol. Adv. 2012, 30, 1614–1626. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-made vaccines and reagents for the one health initiative. Hum. Vaccines Immunother. 2017, 13, 2912–2917. [Google Scholar] [CrossRef]
- Yang, M.; Sun, H.; Lai, H.; Hurtado, J.; Chen, Q. Plant-produced Zika virus envelope protein elicits neutralizing immune responses that correlate with protective immunity against Zika virus in mice. Plant Biotechnol. J. 2018, 16, 572–580. [Google Scholar] [CrossRef]
- Lai, H.; Paul, A.M.; Sun, H.; He, J.; Ming Yang, M.; Bai, F.; Chen, Q. A plant-produced vaccine protects mice against lethal West Nile virus infection without enhancing Zika or dengue virus infectivity. Vaccine 2018, 36, 1846–1852. [Google Scholar] [CrossRef]
- Hefferon, K.L. The role of plant expression platforms in biopharmaceutical development: Possibilities for the future. Expert Rev. Vaccines 2019, 18, 1301–1308. [Google Scholar] [CrossRef]
- McInerney, T.L.; Brennan, F.R.; Jones, T.D.; Dimmock, N.J. Analysis of the ability of five adjuvants to enhance immune responses to a chimeric plant virus displaying an HIV-1 peptide. Vaccine 1999, 17, 1359–1368. [Google Scholar] [CrossRef]
- Durrani, Z.; McInerney, T.L.; McLain, L.; Jones, T.; Bellaby, T.; Brennan, F.R.; Dimmock, N.J. Intranasal immunization with a plant virus expressing a peptide from HIV-1 gp41 stimulates better mucosal and systemic HIV-1-specific IgA and IgG than oral immunization. J. Immunol. Meth. 1998, 220, 93–103. [Google Scholar] [CrossRef]
- Marusic, C.; Rizza, P.; Lattanzi, L.; Mancini, C.; Spada, M.; Belardelli, F.; Benvenuto, E.; Capone, I. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J. Virol. 2001, 75, 8434–8439. [Google Scholar] [CrossRef] [PubMed]
- Lico, C.; Mancini, C.; Italiani, P.; Betti, C.; Boraschi, D.; Benvenuto, E.; Baschieri, S. Plant-produced potato virus X chimeric particles displaying an influenza virus-derived peptide activate specific CD8+ T cells in mice. Vaccine 2009, 27, 5069–5076. [Google Scholar] [CrossRef]
- Uhde-Holzem, K.; Schlosser, V.; Viazov, S.; Fischer, R.; Commandeur, U. Immunogenic properties of chimeric potato virus X particles displaying the hepatitis C virus hypervariable region I peptide R9. J. Virol. Meth. 2010, 166, 12–20. [Google Scholar] [CrossRef]
- Cabral-Miranda, G.; Heath, M.D.; Mohsen, M.O.; Gomes, A.C.; Engeroff, P.; Flaxman, A.; Leoratti, F.M.S.; El-Turabi, A.; Reyes-Sandoval, A.; Skinner, M.A.; et al. Virus-like particle (VLP) plus microcrystalline tyrosine (MCT) adjuvants enhance vaccine efficacy improving T and B cell immunogenicity and protection against Plasmodium berghei/vivax. Vaccines 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Yusibov, V.; Mett, V.; Mett, V.; Davidson, C.; Musiychuk, K.; Gilliam, S.; Farese, A.; Macvittie, T.; Mann, D. Peptide-based candidate vaccine against respiratory syncytial virus. Vaccine 2005, 23, 2261–2265. [Google Scholar] [CrossRef] [PubMed]
- Joelson, T.; Akerblom, L.; Oxelfelt, P.; Strandberg, B.; Tomenius, K.; Morris, T.J. Presentation of a foreign peptide on the surface of tomato bushy stunt virus. J. Gen. Virol. 1997, 78, 1213–1217. [Google Scholar] [CrossRef]
- Hanafi, L.A.; Bolduc, M.; Gagne, M.E.; Dufour, F.; Langelier, Y.; Boulassel, M.R.; Routy, J.-P.; Leclerc, D.; Lapointe, R. Two distinct chimeric potexviruses share antigenic cross-presentation properties of MHC class I epitopes. Vaccine 2010, 28, 5617–5626. [Google Scholar] [CrossRef]
- Butkovich, N.; Li, E.; Ramirez, A.; Burkhardt, A.M.; Wang, S.-W. Advancements in protein nanoparticle vaccine platforms to combat infectious disease. WIREs Nanomed Nanobiotechnology 2021, 13, e1681. [Google Scholar] [CrossRef]
- Masavuli, M.G.; Wijesundara, D.K.; Torresi, J.; Gowans, E.J.; Grubor-Bauk, B. Preclinical Development and Production of Virus-Like Particles as Vaccine Candidates for Hepatitis C. Front. Microbiol. 2017, 8, 2413. [Google Scholar] [CrossRef]
- Kirk, D.D.; McIntosh, K.; Walmsley, A.M.; Peterson, R.K.D. Risk analysis for plant-made vaccines. Transgenic Res. 2005, 14, 449–462. [Google Scholar] [CrossRef]
- Xu, J.; Dolan, M.C.; Medrano, G.; Cramer, C.L.; Weathers, P.J. Green factory: Plants as bioproduction platforms for recombinant proteins. Biotechnol. Adv. 2012, 30, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Tabll, A.; Abbas, A.T.; El-Kafrawy, S.; Wahid, A. Monoclonal antibodies: Principles and applications of immmunodiagnosis and immunotherapy for hepatitis C virus. World J. Hepatol. 2015, 7, 2369–2383. [Google Scholar] [CrossRef]
- Castilho, A.; Bohorova, N.; Grass, J.; Bohorov, O.; Zeitlin, L.; Whaley, K.; Altmann, F.; Steinkellner, H. Rapid high yield production of different glycoforms of Ebola virus monoclonal antibody. PLoS ONE 2011, 6, e26040. [Google Scholar] [CrossRef] [PubMed]
- Virdi, V.; Depicker, A. Role of plant expression systems in antibody production for passive immunization. Int. J. Dev. Biol. 2013, 57, 587–593. [Google Scholar] [CrossRef] [PubMed]
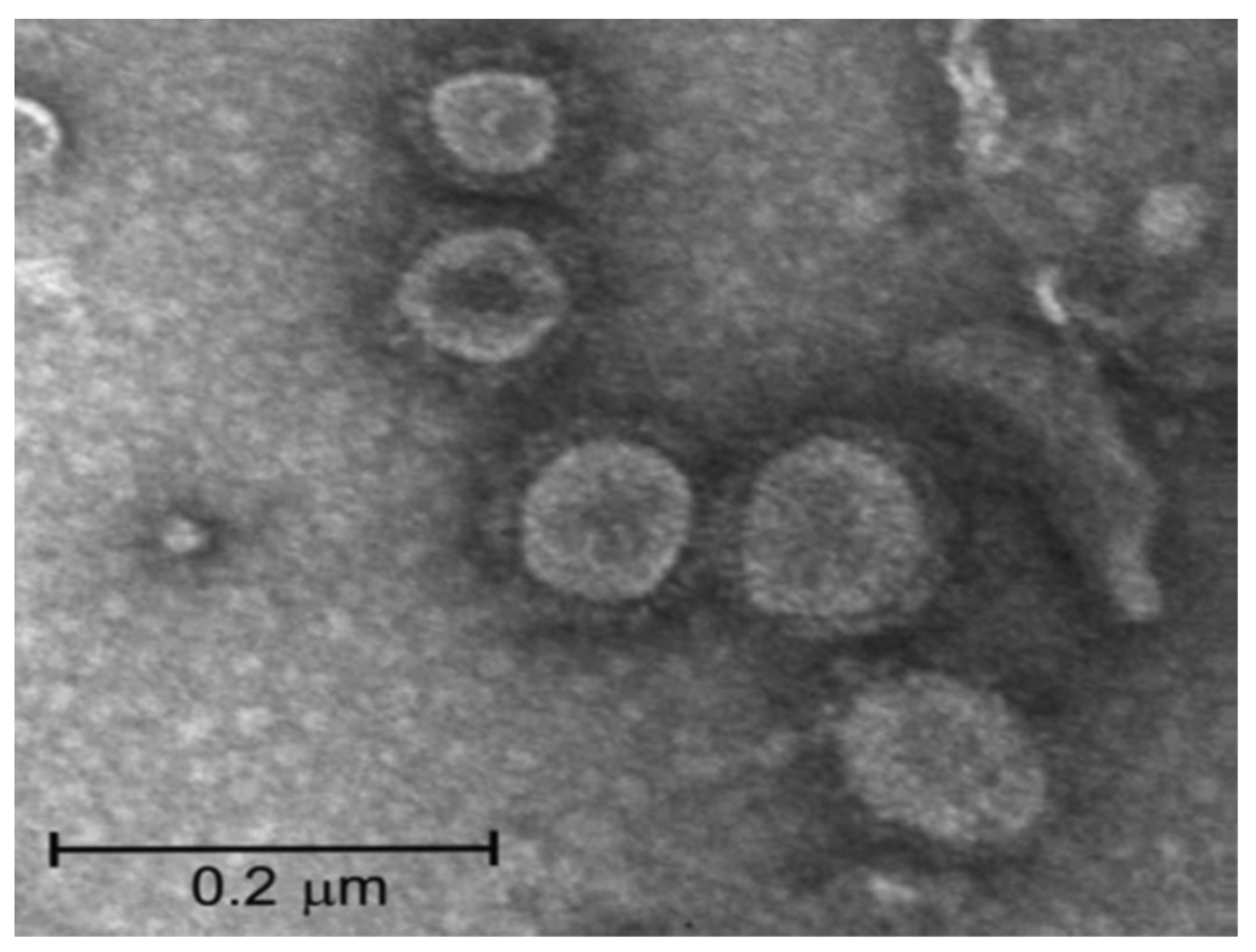
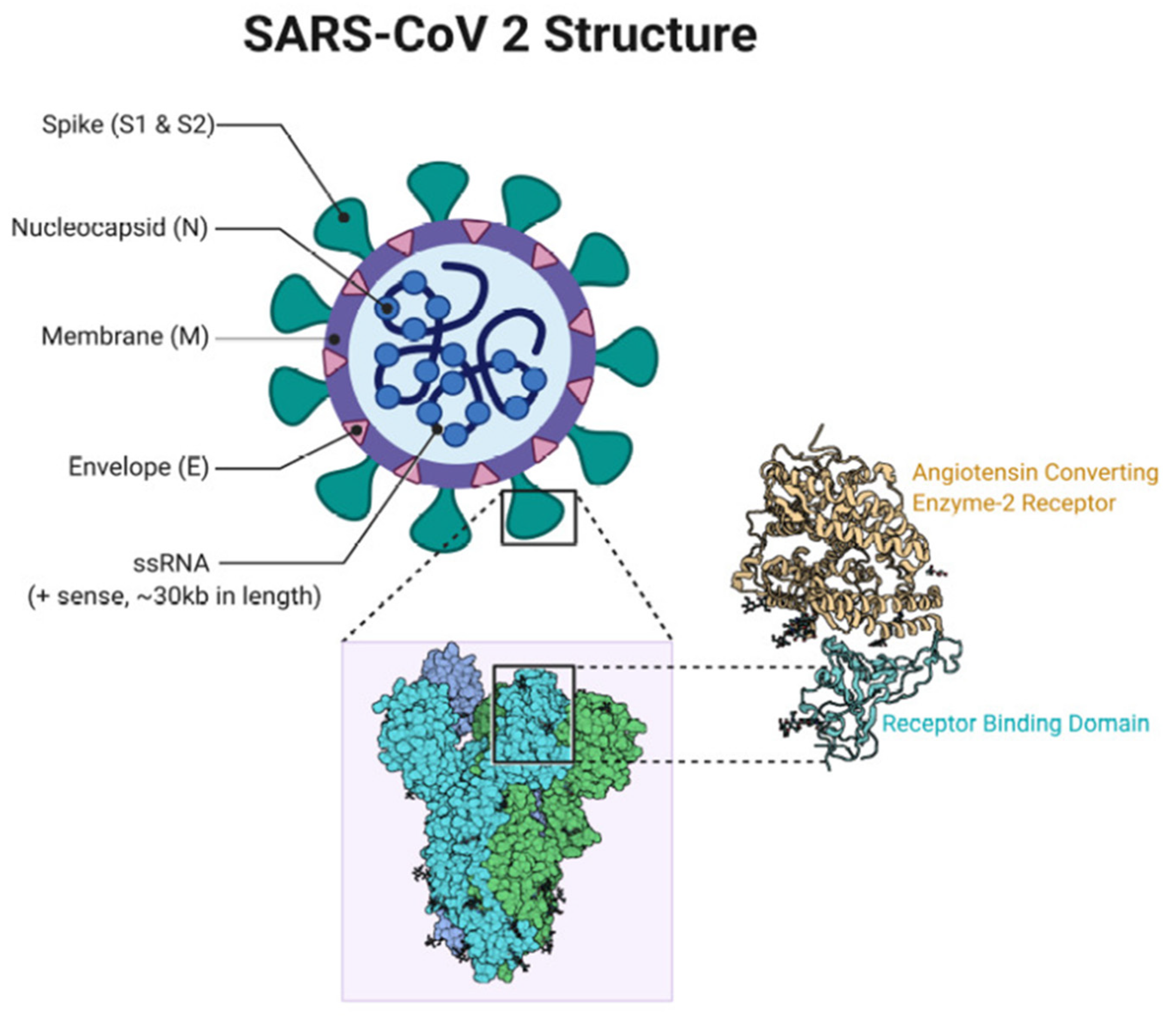

| Viral Vaccine | Antigen/Expression System | Stage of Clinical Trial | Reference |
|---|---|---|---|
| Quadrivalent Influenza vaccine | Mix of recombinant H1, H3, and two B hemagglutinin proteins expressed as VLPs transiently in N. benthamiana | Phase 3 ongoing | [81,82] |
| SARS-CoV-2 vaccine | Recombinant spike (S) glycoprotein expressed as VLPs transiently in N. benthamiana | Phase 1 successfully completed; Phase 2/3 ongoing | [83] |
| Ebola virus vaccine | ZMapp produced by rapid transient expression of 3 plant-based neutralizing monoclonal antibody cocktails against Ebola; made in N. benthamiana | Phase 2/3 completed; FDA approved | [84] |
| Rotavirus vaccine | Four structural antigens of rotavirus (VP2, VP4, VP6, and VP7) expressed as VLPs in plants | Phase 1 | [85] |
| Norwalk virus vaccine | CP expressed in potato | Early phase 1 | [86] |
| Rabies virus vaccine | GP/NP antigens expressed in spinach | Early phase 1 | [87] |
| Hepatitis B virus vaccine | HBsAg expressed in lettuce | Early phase 1 | [88] |
| Hepatitis B virus vaccine | HBsAg expressed in potato | Phase 1 | [89] |
| Bacterial vaccine: Vibrio cholera vaccine | CTB antigen expressed in rice | Phase 1 | [90,91] |
| Virus. | Plant-Derived Biopharmaceutical | Technology for Expression In Plants | Reference |
|---|---|---|---|
| Influenza | VLPs | Plant virus vector | Yusibov et al., 2015 [56] Marsian and Lomonosoff, 2016 [281] D’Aoust et al., 2010 [282] Lindsay et al., 2018 [95] Marquez-Escobar et al., 2017 [4] |
| Ebola | mAbs | Transgenic plants, plant virus vector | McCarthy, 2014 [283] Zeitlin et al., 2011 [258] Monreal-Escalante et al., 2017 [284] Rosales-Mendoza et al., 2017 [285] Phoolcharoen et al., 2011 [286] |
| HIV | Griffithsin | Transgenic plants | Hoelscher et al., 2018 [287] Vamvaka et al., 2018 [152] |
| HIV | VLPs | Transiently and transgenic plants | Cervera et al., 2019 [288] |
| HIV | Antigens derived from Env, Gag proteins | Transiently and transgenic plant | Rosales-Mendoza et al., 2012 [289] |
| HIV | mAbs | Transgenic plant | Lotter-Stark et al., 2012 [290] |
| West Nile, Zika, Chickungunya | mAbs | Transgenic plants | Rybicki, 2017 [291] |
| WNV, Zika | Envelope protein | Transgenic plant | Yang et al., 2018 [292] Lai et al., 2018 [293] |
| Scaffold Platform | Viral Disease | In Vitro/In Vivo | Elicited Immune Response | Specific Immune Reactions | References |
|---|---|---|---|---|---|
| CPMV | HIV-1 | In vivo | Humoral | Neutralizing antibodies | McInerney et al. (1999) [295] Durrani et al. (1998) [296] |
| 4 | HIV-1 | In vivo | Cellular | Antigen-specific T-cell proliferation | McInerney et al. (1999) [295] |
| PVX | HIV-1 | In vivo | Humoral | Neutralizing antibodies | Marusic et al. (2001) [297] |
| Influenza A {H1N1) | In vivo | Cellular | Antigen-specific CD8+ T-cell activation | Lico et al. (2009) [298] | |
| HCV | In vitro, In vivo | Humoral | Antigen-specific Abs | Uhde-Holzem et al. (2010) [299] | |
| TMV | Influenza (H1N1) | In vivo | Humoral | Antigen-specific Abs, protection against challenge | Mallajosyula et al. (2014) [93] |
| CMV | HCV | In vitro, In vivo | Humoral | Antigen-specific Abs | Piazzolla et al. (2005) [53] Nuzzaci et al. (2007) [54] |
| HCV | In vitro, In vivo | Cellular | Cytokine release (IFN-γ, IL-12, and IL-15) | Piazzolla et al. (2005) [53] Nuzzaci et al. (2007) [54] | |
| Zika virus | In vitro, In vivo | Humoral | Neutralizing antibodies | Cabral-Miranda et al. (2017) [300] | |
| AlMV | RSV | In vivo | Humoral | Antigen-specific Abs | Yusibov et al. (2005) [301] |
| RSV | In vivo | Cellular | CD4+ and CD8+ T-cell responses | Yusibov et al. (2005) [301] | |
| TBSV | HIV-1 | In vitro, In vivo | Humoral | Antigen-specific Abs | Joelson et al. (1997) [302] |
| PapMV | Influenza | In vitro, In vivo | Humoral | Antigen-specific Abs, B-cell expansion, protection against challenge | Denis et al. (2008) [109] Hanafi et al. (2010) [303] Bolduc et al. (2018) [108] Carignan et al. (2015) [106] Therien et al. (2017) [107] |
| Influenza | In vitro, In vivo | Cellular | Antigen-specific CD8+ T-cell expansion and response | Babinet al. (2013), [110] Leclercet al. (2007) [112] Hanafi et al. (2010) [303] Laliberte-Gagne et al. (2019) [111] | |
| HCV | In vivo | Humoral | Antigen-specific Abs | Denis et al. (2007) [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkataraman, S.; Hefferon, K.; Makhzoum, A.; Abouhaidar, M. Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer? Vaccines 2021, 9, 761. https://doi.org/10.3390/vaccines9070761
Venkataraman S, Hefferon K, Makhzoum A, Abouhaidar M. Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer? Vaccines. 2021; 9(7):761. https://doi.org/10.3390/vaccines9070761
Chicago/Turabian StyleVenkataraman, Srividhya, Kathleen Hefferon, Abdullah Makhzoum, and Mounir Abouhaidar. 2021. "Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer?" Vaccines 9, no. 7: 761. https://doi.org/10.3390/vaccines9070761
APA StyleVenkataraman, S., Hefferon, K., Makhzoum, A., & Abouhaidar, M. (2021). Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer? Vaccines, 9(7), 761. https://doi.org/10.3390/vaccines9070761






