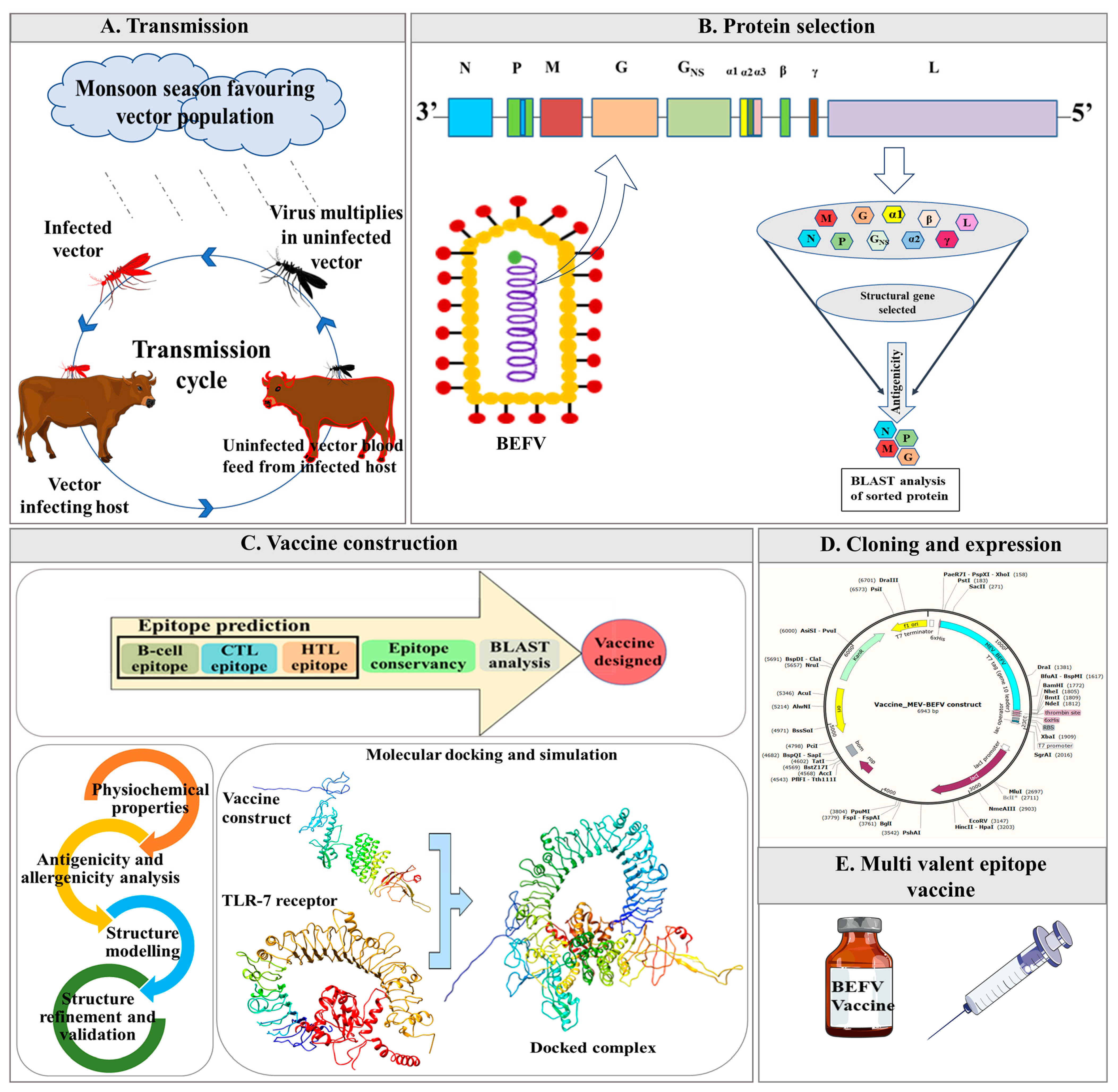
Immunoinformatics Approach to Design Multi-Epitope- Subunit Vaccine against Bovine Ephemeral Fever Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Retrieval and Protein Curation
2.2. Epitope Prediction
2.2.1. Cytotoxic T-Lymphocytes (CTL) Epitope Prediction
2.2.2. Helper T-Lymphocytes (HTL) Epitope Prediction
2.2.3. B-Cell Epitopes Prediction
2.3. Conservancy Analysis with All Global Isolates
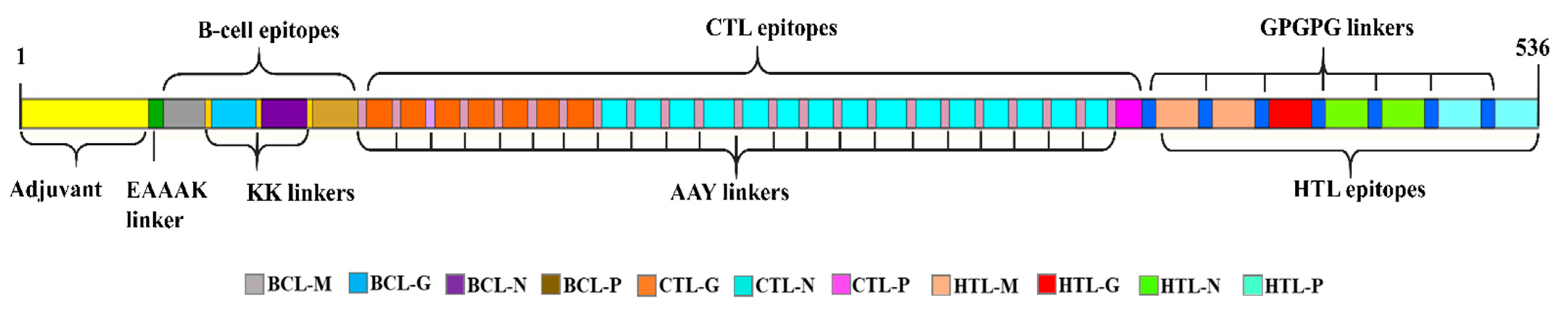
2.4. Multiepitope Vaccine Designing
2.5. Blast Analysis
2.6. Physicochemical and Immunogenic Properties Assessment of the Vaccine Construct
2.7. Secondary and Tertiary Structure Prediction
2.8. Refinement, Model Quality Assessment, and Validation
2.9. Bovine TLR7 Receptor and Vaccine Construct Molecular Docking
2.10. Molecular Dynamic Simulation of the MEV-BEFV and the Vaccine bTLR7
2.11. Codon Adaptation and In Silico Cloning
3. Results
3.1. Sequence Retrieval and Antigenicity Prediction
3.2. Epitope Prediction of B-Cell and T-Cell Epitopes
3.2.1. CTL Epitope Prediction
3.2.2. HTL Epitope Prediction
3.2.3. B-Cell Epitopes Prediction
3.3. Conservancy Analysis
3.4. Multiepitope Vaccine Construction
3.5. Physicochemical and Immunogenic Properties Assessment of the MEV-BEFV
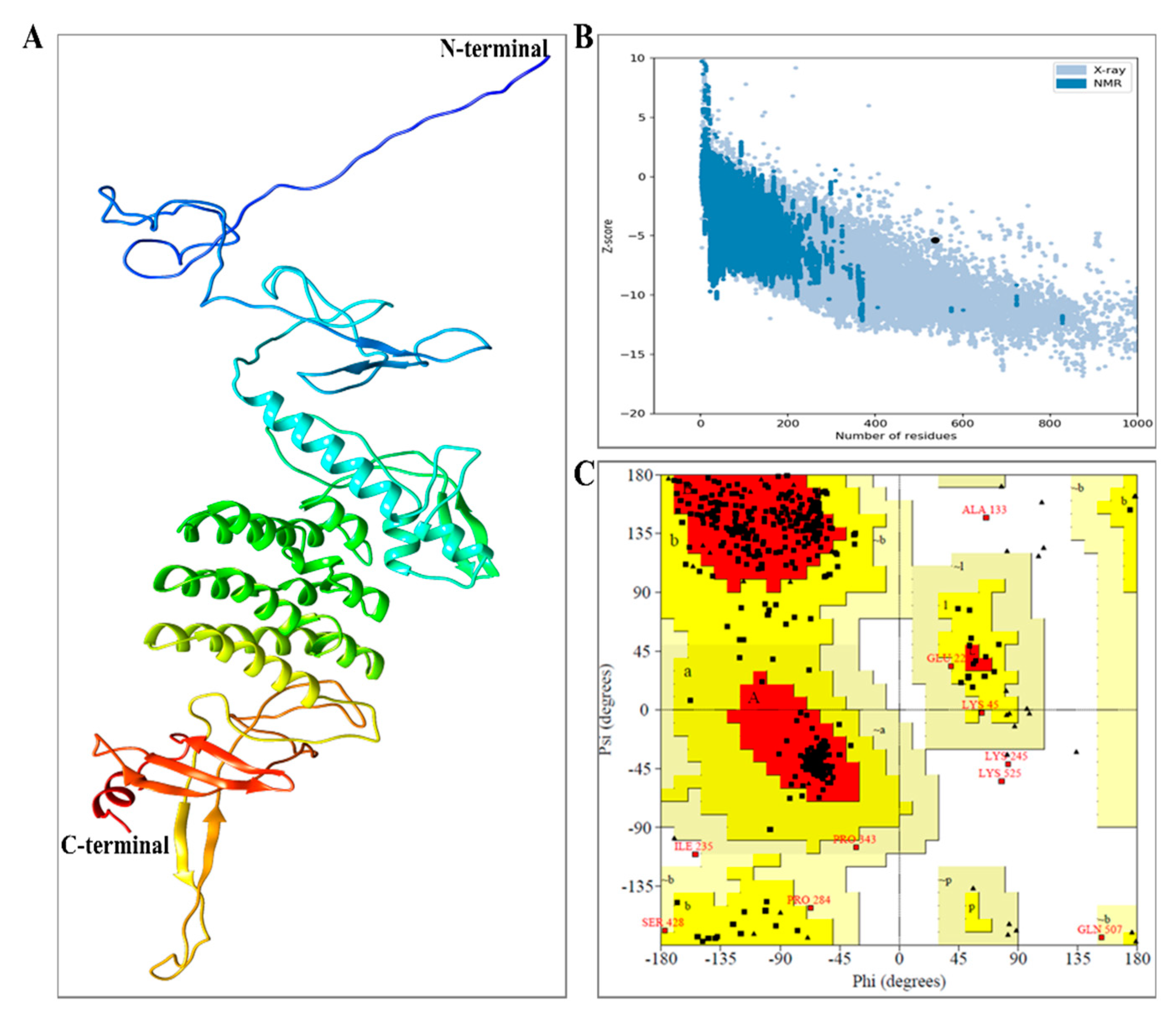
3.6. Structure Prediction of MEV-BEFV
3.7. Structural Evaluations of Vaccine Construct
3.8. Molecular Docking of the MEV-BEFV with bTLR7
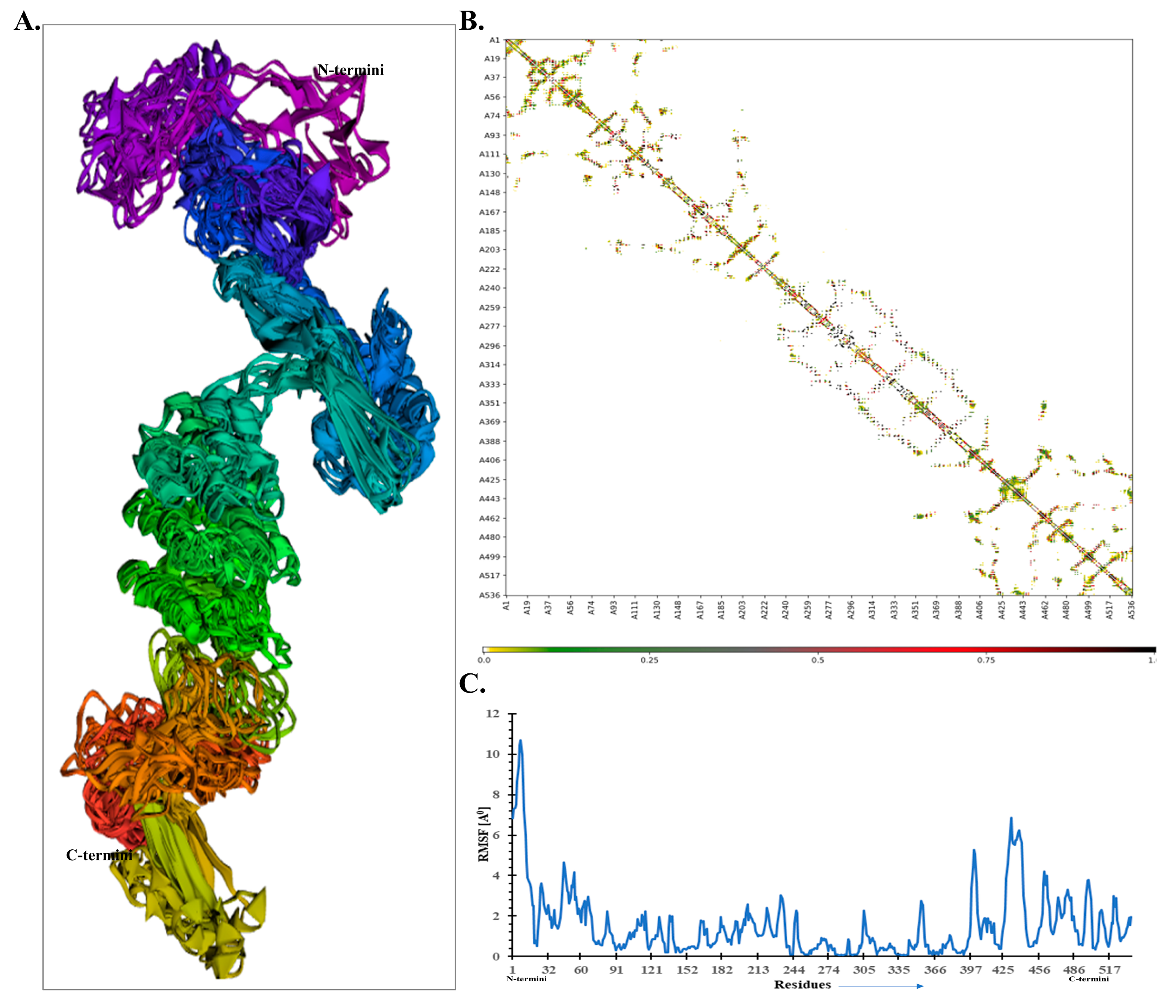
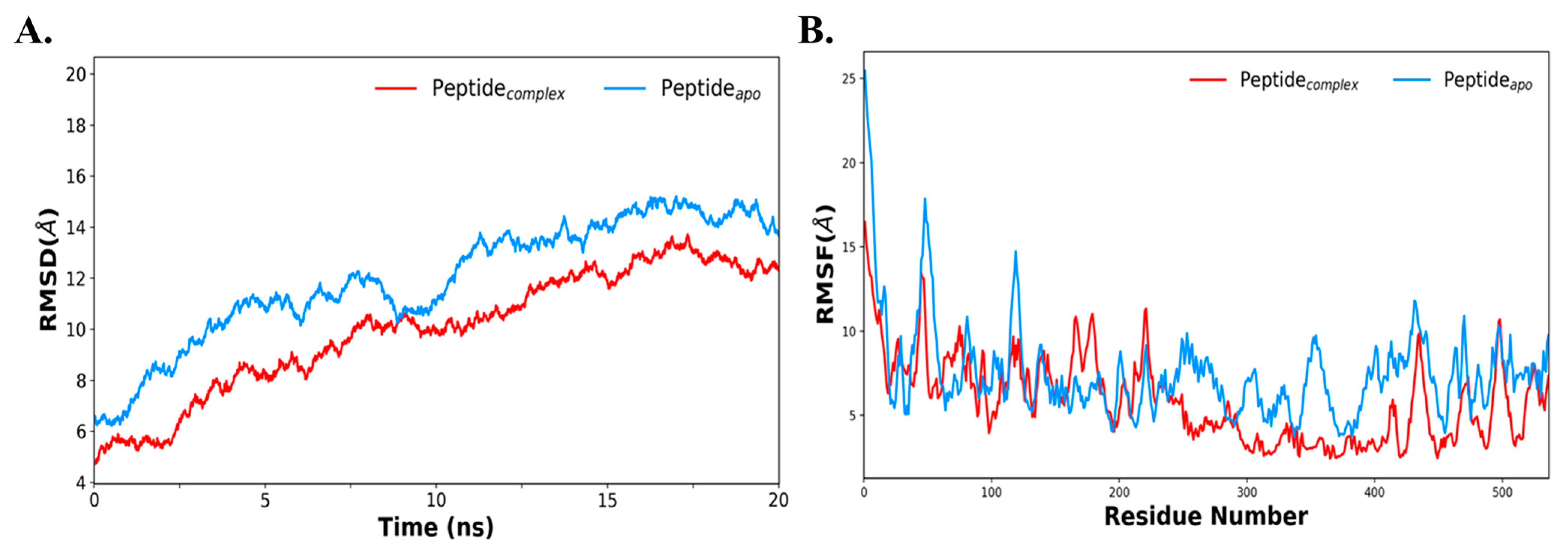
3.9. Molecular Dynamics Simulation of the Docked-Complex
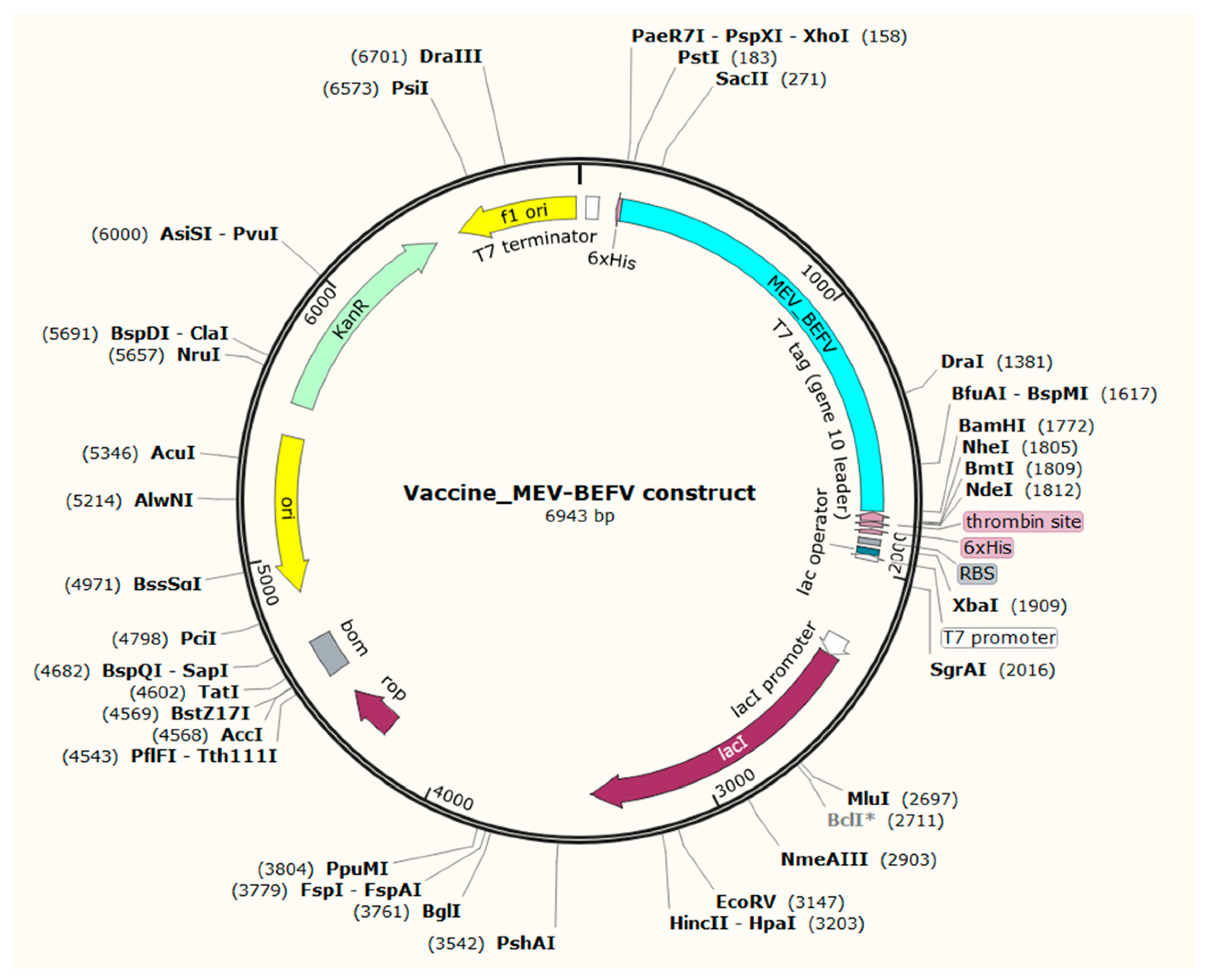
3.10. In Silico Cloning of Vaccine Construct
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nandi, S.; Negi, B.S. Bovine ephemeral fever: A review. Comp. Immunol. Microbiol. Infect. Dis. 1999, 22, 81–91. [Google Scholar] [CrossRef]
- Niwa, T.; Shirafuji, H.; Ikemiyagi, K.; Nitta, Y.; Suzuki, M.; Kato, T.; Yanase, T. Occurrence of bovine ephemeral fever in Okinawa Prefecture, Japan, in 2012 and development of a reverse-transcription polymerase chain reaction assay to detect bovine ephemeral fever virus gene. J. Vet. Med. Sci. 2014. [Google Scholar] [CrossRef][Green Version]
- Abayli, H.; Tonbak, S.; Azkur, A.K.; Bulut, H. Complete genome analysis of highly pathogenic bovine ephemeral fever virus isolated in Turkey in 2012. Arch. Virol. 2017, 162, 3233–3238. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.C.; Chen, S.H.; Chou, C.C.; Ting, L.J.; Itakura, C.; Wang, F.I. Bovine ephemeral fever in Taiwan (2001-2002). J. Vet. Med. Sci. 2005, 67, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kirland, P. The epidemiology of bovine ephemeral fever in Southwestern Australia: Evidence for a mosquito vector. In Proceedings of the 1st International Symposium on Bovine Ephemeral Fever and Related Rhabdoviruses, Beijing, China, 25–27 August 1992. [Google Scholar]
- George, T.D.S.; Standfast, H.A.; Christie, D.G.; Knott, S.G.; Morgan, I.R. The Epizootiology of Bovine Ephemeral Fever in Australia and Papua-New Guinea. Aust. Vet. J. 1977, 53, 17–28. [Google Scholar] [CrossRef] [PubMed]
- St. George, T.D. The epidemiology of bovine ephemeral fever in Australia and its economic effect. In Proceedings of the Fourth Symposium, Brisbane, Australia, 6–9 May 1986. [Google Scholar]
- Malviya, H.K. and Prasad, J. Ephemeral fever—a clinical and epidemiological study in cross bred cows and buffaloes. Indian Vet. J. 1977, 54, 440–444. [Google Scholar]
- Murphy, F.A.; Taylor, W.P.; Mims, C.A.; Whitfield, S.G. Bovine ephemeral fever virus in cell culture and mice. Arch. Gesamte Virusforsch. 1972, 38, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Byrne, K.A.; Cybinski, D.H.; Doolan, D.L.; Wang, Y. Proteins of bovine ephemeral fever virus. J. Gen. Virol. 1991, 72, 67–74. [Google Scholar] [CrossRef]
- Kongsuwan, K.; Cybinski, D.H.; Cooper, J.; Walker, P.J. Location of neutralizing epitopes on the G protein of bovine ephemeral fever rhabdovirus. J. Gen. Virol. 1998, 79, 2573–2581. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uren, M.F.; Walker, P.J.; Zakrzewski, H.; St George, T.D.; Byrne, K.A. Effective vaccination of cattle using the virion G protein of bovine ephemeral fever virus as an antigen. Vaccine 1994, 12, 845–852. [Google Scholar] [CrossRef]
- Uren, M.F.; Zakrzewski, H.; Davis, S.S. Antibody and cell proliferative responses of cattle vaccinated with bovine ephemeral fever virus proteins. In Proceedings of the 1st International Symposium on Bovine Ephemeral Fever and Related Rhabdoviruses, Beijing, China, 25–27 August 1992. [Google Scholar]
- McWilliam, S.M.; Kongsuwan, K.; Cowley, J.A.; Byrne, K.A.; Walker, P.J. Genome organization and transcription strategy in the complex GNS-L intergenic region of bovine ephemeral fever rhabdovirus. J. Gen. Virol. 1997, 78 Pt 6, 1309–1317. [Google Scholar] [CrossRef]
- Yanase, T.; Murota, K.; Hayama, Y. Endemic and Emerging Arboviruses in Domestic Ruminants in East Asia. Front. Vet. Sci. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Aziz-Boaron, O.; Gleser, D.; Yadin, H.; Gelman, B.; Kedmi, M.; Galon, N.; Klement, E. The protective effectiveness of an inactivated bovine ephemeral fever virus vaccine. Veter Microbiol. 2014, 173, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Johal, J.; Gresty, K.; Kongsuwan, K.; Walker, P.J. Antigenic characterization of bovine ephemeral fever rhabdovirus G and GNS glycoproteins expressed from recombinant baculoviruses. Arch. Virol. 2008, 153, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Klement, E. Epidemiology and control of bovine ephemeral fever. Vet. Res. 2015, 46, 1–19. [Google Scholar] [CrossRef]
- María, R.R.; Arturo, C.J.; Alicia, J.A.; Paulina, M.G.; Gerardo, A.O. The Impact of Bioinformatics on Vaccine Design and Development. In Vaccines; IntechOpen: Hamilton, NJ, USA, 2017. [Google Scholar]
- Rana, A.; Akhter, Y. A multi-subunit based, thermodynamically stable model vaccine using combined immunoinformatics and protein structure based approach. Immunobiology 2016, 221, 544–557. [Google Scholar] [CrossRef] [PubMed]
- TAKESHIMA, S.-N.; AIDA, Y. Structure, function and disease susceptibility of the bovine major histocompatibility complex. Anim. Sci. J. 2006, 77, 138–150. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Tahir Ul Qamar, M.; Bari, A.; Adeel, M.M.; Maryam, A.; Ashfaq, U.A.; Du, X.; Muneer, I.; Ahmad, H.I.; Wang, J. Peptide vaccine against chikungunya virus: Immuno-informatics combined with molecular docking approach. J. Transl. Med. 2018, 16, 298. [Google Scholar] [CrossRef]
- Terry, F.; Moise, L.; Martin, R.; Torres, M.; Pilotte, N.; Williams, S.; De Groot, A. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert Rev. Vaccines 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Deb, D.; Basak, S.; Kar, T.; Narsaria, U.; Castiglione, F.; Paul, A.; Pandey, A.; Srivast, A.P. A Candidate Multi-Epitope Vaccine Against Pathogenic Chandipura Vesiculovirus Identified Using Immunoinformatics. Available online: https://assets.researchsquare.com/files/rs-56434/v1/6da142e0-13b0-437b-b7f8-9e621e637b47.pdf (accessed on 19 February 2021).
- Gaafar, B.B.M.; Ali, S.A.; Abd-Elrahman, K.A.; Almofti, Y.A.; Fridkis-Hareli, M. Immunoinformatics Approach for Multiepitope Vaccine Prediction from H, M, F, and N Proteins of Peste des Petits Ruminants Virus. J. Immunol. Res. 2019, 2019. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 1–7. [Google Scholar] [CrossRef]
- Borrego, F. The First Molecular Basis of the “Missing Self” Hypothesis. J. Immunol. 2006, 177, 5759–5760. [Google Scholar] [CrossRef] [PubMed]
- Woolard, S.N.; Kumaraguru, U. Viral vaccines and CTL response. J. Biomed. Biotechnol. 2010, 2010, 141657. [Google Scholar] [CrossRef] [PubMed]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Justesen, S.; Lund, O.; Lundegaard, C.; Buus, S. NetMHCIIpan-2.0-Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 2010, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Fuse, S.; Molloy, M.J.; Usherwood, E.J. Immune responses against persistent viral infections: Possible avenues for immunotherapeutic interventions. Crit. Rev. Immunol. 2008, 28, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Vir, P.; Raghava, G.P.S. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, N.; Pandey, R.K.; Prajapati, V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017, 7, 8285. [Google Scholar] [CrossRef]
- Pandey, R.K.; Sundar, S.; Prajapati, V.K. Differential expression of miRNA regulates T cell differentiation and plasticity during visceral leishmaniasis infection. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.H.; Sidney, J.; Li, W.; Fusseder, N.; Sette, A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mohan, T.; Sharma, C.; Bhat, A.A.; Rao, D.N. Modulation of HIV peptide antigen specific cellular immune response by synthetic α- and β-defensin peptides. Vaccine 2013, 31, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Hajighahramani, N.; Nezafat, N.; Eslami, M.; Negahdaripour, M.; Rahmatabadi, S.S.; Ghasemi, Y. Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017, 48, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook. Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2--a server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Consortium, O.S.D.D.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Magnan, C.N.; Randall, A.; Baldi, P. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics 2009, 25, 2200–2207. [Google Scholar] [CrossRef]
- Kumar, T.A. CFSSP: Chou and Fasman Secondary Structure Prediction server. Wide Spectr. 2013, 1, 15–19. [Google Scholar]
- Källberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. RaptorX server: A resource for template-based protein structure modeling. Methods Mol. Biol. 2014, 1137, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Nowotny, J.; Cao, R.; Cheng, J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016, 44, W406–W409. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.-H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Duke, T.A.D.R.E.; Ghoreishi, D.; Gilson, M.K.; Gohlke, H.; et al. AMBER 18; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Pastor, R.W.; Brooks, B.R.; Szabo, A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988, 65, 1409–1419. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Bohórquez, M.D.; Ordoñez, D.; Suárez, C.F.; Vicente, B.; Vieira, C.; López-Abán, J.; Muro, A.; Ordóñez, I.; Patarroyo, M.A. Major Histocompatibility Complex Class II (DRB3) Genetic Diversity in Spanish Morucha and Colombian Normande Cattle Compared to Taurine and Zebu Populations. Front. Genet. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Mekata, H.; Sekiguchi, S.; Kirino, Y.; Mitoma, S.; Honkawa, K.; Horii, Y.; Norimine, J. Cattle with the BoLA class II DRB3*0902 allele have significantly lower bovine leukemia proviral loads. J. Vet. Med. Sci. 2017, 79, 1552–1555. [Google Scholar] [CrossRef]
- Hernandez, D.; Montes, D.; Alvarez, L.A. Association of BoLA-DRB3.2 alleles with enzootic bovine leukosis: Profiles BLV infection, persistent lymphocytosis and antibody production in Harton del Valle Cattle. Indian J. Sci. Technol. 2018, 11, 1–14. [Google Scholar] [CrossRef]
- Farrell, D.; Jones, G.; Pirson, C.; Malone, K.; Rue-Albrecht, K.; Chubb, A.J.; Vordermeier, M.; Gordon, S.V. Integrated computational prediction and experimental validation identifies promiscuous T cell epitopes in the proteome of Mycobacterium bovis. Microb. Genom. 2016, 2, e000071. [Google Scholar] [CrossRef] [PubMed]
- Maccari, G.; Robinson, J.; Ballingall, K.; Guethlein, L.A.; Grimholt, U.; Kaufman, J.; Ho, C.-S.; de Groot, N.G.; Flicek, P.; Bontrop, R.E.; et al. IPD-MHC 2.0: An improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res. 2017, 45, D860–D864. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- He, C.-Q.; Liu, Y.-X.; Wang, H.-M.; Hou, P.-L.; He, H.-B.; Ding, N.-Z. New genetic mechanism, origin and population dynamic of bovine ephemeral fever virus. Vet. Microbiol. 2016, 182, 50–56. [Google Scholar] [CrossRef]
- Patronov, A.; Doytchinova, I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Shey, R.A.; Ghogomu, S.M.; Esoh, K.K.; Nebangwa, N.D.; Shintouo, C.M.; Nongley, N.F.; Asa, B.F.; Ngale, F.N.; Vanhamme, L.; Souopgui, J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep. 2019, 9, 4409. [Google Scholar] [CrossRef] [PubMed]
- Amanna, I.J.; Slifka, M.K. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology 2011, 411, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Naz, A.; Shahid, F.; Butt, T.T.; Awan, F.M.; Ali, A.; Malik, A. Designing Multi-Epitope Vaccines to Combat Emerging Coronavirus Disease 2019 (COVID-19) by Employing Immuno-Informatics Approach. Front. Immunol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Pavitrakar, D.V.; Atre, N.M.; Tripathy, A.S.; Shil, P. Design of a multi-epitope peptide vaccine candidate against chandipura virus: An immuno-informatics study. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Pandey, R.K.; Ojha, R.; Aathmanathan, V.S.; Krishnan, M.; Prajapati, V.K. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine 2018, 36, 2262–2272. [Google Scholar] [CrossRef]
- Majee, P.; Jain, N.; Kumar, A. Designing of a multi-epitope vaccine candidate against Nipah virus by in silico approach: A putative prophylactic solution for the deadly virus. J. Biomol. Struct. Dyn. 2020, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.A.; Sarkar, B.; Islam, S.S. Exploiting the reverse vaccinology approach to design novel subunit vaccines against Ebola virus. Immunobiology 2020, 225, 151949. [Google Scholar] [CrossRef] [PubMed]
- Kumar Pandey, R.; Ojha, R.; Mishra, A.; Kumar Prajapati, V. Designing B- and T-cell multi-epitope based subunit vaccine using immunoinformatics approach to control Zika virus infection. J. Cell. Biochem. 2018, 119, 7631–7642. [Google Scholar] [CrossRef]
- Bacchetta, R.; Gregori, S.; Roncarolo, M.-G. CD4+ regulatory T cells: Mechanisms of induction and effector function. Autoimmun. Rev. 2005, 4, 491–496. [Google Scholar] [CrossRef]
- Cooper, N.R.; Nemerow, G.R. The Role of Antibody and Complement in the Control of Viral Infections. J. Invest. Dermatol. 1984, 83, S121–S127. [Google Scholar] [CrossRef]
- CaCano RLE, L.H. Introduction to T and B lymphocytes; Anaya, J.M., Shoenfeld, Y., Rojas-Villarraga, A., Eds.; El Rosario University Press: Colombia, UK, 2013; Chapter 5.
- Garcia, K.C.; Teyton, L.; Wilson, I.A. Structural basis of T cell recognition. Annu. Rev. Immunol. 1999, 17, 369–397. [Google Scholar] [CrossRef]
- Arpin, C.; Dechanet, J.; Van Kooten, C.; Merville, P.; Grouard, G.; Briere, F.; Banchereau, J.; Liu, Y.J. Generation of memory B cells and plasma cells in vitro. Science 1995, 268, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.U.; Al Thani, A.A.; Yassine, H.M. Comparative phylogenetic and residue analysis of hepatitis C virus E1 protein from the middle east and North Africa region. Hepat. Mon. 2019, 19. [Google Scholar] [CrossRef]
- Bakhshesh, M.; Ranjbar, M.; Almasi, S. Immunoinformatic analysis of glycoprotein from bovine ephemeral fever virus. Biomed. Biotechnol. Res. J. 2018, 2, 208. [Google Scholar] [CrossRef]
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4+T Cells: Differentiation and Functions. Clin. Dev. Immunol. 2012, 2012, 925135. [Google Scholar] [CrossRef] [PubMed]
- Michel-Todó, L.; Bigey, P.; Reche, P.A.; Pinazo, M.J.; Gascón, J.; Alonso-Padilla, J. Design of an epitope-based vaccine ensemble for animal trypanosomiasis by computational methods. Vaccines 2020, 8, 130. [Google Scholar] [CrossRef]
- Shamriz, S.; Ofoghi, H.; Moazami, N. Effect of linker length and residues on the structure and stability of a fusion protein with malaria vaccine application. Comput. Biol. Med. 2016, 76, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, J.J.; Biragyn, A.; Kwak, L.W.; Yang, D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 2003, 62 (Suppl. 2), ii17–ii21. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie-Dyck, S.; Kovacs-Nolan, J.; Snider, M.; Babiuk, L.A.; van Drunen Littel-van den Hurk, S. Inclusion of the bovine neutrophil beta-defensin 3 with glycoprotein D of bovine herpesvirus 1 in a DNA vaccine modulates immune responses of mice and cattle. Clin. Vaccine Immunol. 2014, 21, 463–477. [Google Scholar] [CrossRef]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 2004, 101, 5598–5603. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Liu, J.; Cheng, Y.; Sun, C.; Wang, C.; Lu, Y.; Ding, J.; Zhou, J.; Xiang, H. Expression of SARS-coronavirus nucleocapsid protein in Escherichia coli and Lactococcus lactis for serodiagnosis and mucosal vaccination. Appl. Microbiol. Biotechnol. 2005, 68, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Nain, Z.; Abdulla, F.; Rahman, M.M.; Karim, M.M.; Khan, M.S.A.; Sayed, S.B.; Mahmud, S.; Rahman, S.M.R.; Sheam, M.M.; Haque, Z.; et al. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J. Biomol. Struct. Dyn. 2020, 38, 4850–4867. [Google Scholar] [CrossRef] [PubMed]


| S. No. | Protein Name | Length (aa) | NCBI Protein ID | VaxiJen Score |
|---|---|---|---|---|
| 1 | Nucleocapsid protein | 432 | QOU09200 | 0.5222 |
| 2 | Phosphoprotein | 279 | QOU09201 | 0.5095 |
| 3 | Matrix protein | 224 | QOU09203 | 0.7226 |
| 4 | Glycoprotein | 624 | QOU09204 | 0.4723 |
| Protein | Selected Epitopes | BoLA Binding Alleles | Position | Prediction Score | %Rank |
|---|---|---|---|---|---|
| RPNTIGKYL | BoLA-2:00501 | 419–427 | 0.492 | 0.04 | |
| BoLA-2:00601 | 0.303 | 0.153 | |||
| ILPEKVTEF | BoLA-2:00602 | 399–407 | 0.260 | 0.097 | |
| LTIEEILDW | BoLA-2:00801 | 238–246 | 0.740 | 0.053 | |
| AMPKSSDPMEW | BoLA-2:00802 | 379–389 | 0.287 | 0.224 | |
| AALTKAFLK | BoLA-2:01201 | 340–348 | 0.942 | 0.009 | |
| GEDVVKIM | BoLA-2:01601 | 253–260 | 0.210 | 0.125 | |
| Nucleoprotein | IPPQYPKEF | BoLA-2:01602 | 19–27 | 0.330 | 0.019 |
| KSSDPMEWF | BoLA-2:04401 | 382–390 | 0.470 | 0.166 | |
| GKLDLPTVREL | BoLA-2:02603 | 43–53 | 0.431 | 0.34 | |
| SHVIRYLYL | BoLA-3:00102 | 66–74 | 0.087 | 0.203 | |
| ILPEKVTEF | BoLA-3:00103 | 399–407 | 0.231 | 0.036 | |
| SCPHIYTFL | BoLA-3:00201 | 291–299 | 0.693 | 0.044 | |
| BoLA-3:00101 | 0.149 | 0.118 | |||
| SNKSPYSSI | BoLA-3:00401 | 282–290 | 0.448 | 0.009 | |
| IQNARPNTI | BoLA-3:01101 | 415–423 | 0.627 | 0.071 | |
| KSSDPMEW | BoLA-4:02402 | 382–389 | 0.605 | 0.2 | |
| Phosphoprotein | KTVEEMIRH | BoLA-1:00901 | 247–255 | 0.812 | 0.08 |
| IPDVRVKEI | BoLA-2:00501 | 192–200 | 0.308 | 0.199 | |
| SELNDTERL | BoLA-2:00601 | 219–227 | 0.217 | 0.1 | |
| EELDIKAEL | BoLA-2:00602 | 296–303 | 0.259 | 0.099 | |
| Glycoprotein | KVLSAVVGW | BoLA-2:00801 | 521–529 | 0.801 | 0.031 |
| NTISKILNK | BoLA-2:01201 | 310–318 | 0.902 | 0.022 | |
| SPHETSQI | BoLA-2:01802 | 499–506 | 0.947 | 0.021 | |
| VKKLDQGAL | BoLA-2:02602 | 116–124 | 0.684 | 0.057 |
| Protein | Epitope | Position | Allele | Predicted Score | IC50 Value | % Rank |
|---|---|---|---|---|---|---|
| FVVSYVKSNKAALTK | 330–344 | BoLA-DRB3*0101 | 0.851 | 4.19 | 0.7 | |
| Nucleoprotein | BoLA-DRB3*0901 | 0.807 | 6.38 | 0.15 | ||
| NLLIKLNAQIKGYRK | 154–168 | BoLA-DRB3*0303 | 0.909 | 2.39 | 0.5 | |
| Phosphoprotein | PLIIKQEAGIYPIEI | 52–66 | BoLA-DRB3*6101 | 0.779 | 8.36 | 7 |
| KKSKSFRSISKTLNV | 258–272 | BoLA-DRB3*0101 | 0.776 | 8.58 | 3 | |
| Matrix protein | MLTLFKKGKSKGGSV | 1–15 | BoLA-DRB3*0101 | 0.658 | 26.79 | 5 |
| NLEVISSKPIERTTD | 63–77 | BoLA-DRB3*0303 | 0.813 | 6.02 | 2 | |
| KVLIITLLVRRLHFE | 3–17 | BoLA-DRB3*0101 | 0.836 | 4.85 | 1 | |
| BoLA-DRB3*2004 | 0.89 | 2.87 | 0.05 | |||
| Glycoprotein | BoLA-DRB3*03021 | 0.865 | 3.66 | 2 | ||
| BoLA-DRB3*0303 | 0.851 | 4.2 | 3 | |||
| BoLA-DRB3*1101 | 0.926 | 2.03 | 0.05 |
| Protein | Predicted Epitope | Position | Score |
|---|---|---|---|
| Nucleoprotein | HGSWVTNSEFCKIAAG | 180–195 | 0.93 |
| Phosphoprotein | CNIPTKDLCMDSGNKE | 112–127 | 0.93 |
| Matrix protein | GGSVDDRNSSYGESDP | 12–27 | 0.94 |
| Glycoprotein | HWECITVKSFRSELND | 208–227 | 0.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyasi, S.; Sharma, V.; Dipti, K.; Jonniya, N.A.; Nayak, D. Immunoinformatics Approach to Design Multi-Epitope- Subunit Vaccine against Bovine Ephemeral Fever Disease. Vaccines 2021, 9, 925. https://doi.org/10.3390/vaccines9080925
Pyasi S, Sharma V, Dipti K, Jonniya NA, Nayak D. Immunoinformatics Approach to Design Multi-Epitope- Subunit Vaccine against Bovine Ephemeral Fever Disease. Vaccines. 2021; 9(8):925. https://doi.org/10.3390/vaccines9080925
Chicago/Turabian StylePyasi, Shruti, Vinita Sharma, Kumari Dipti, Nisha Amarnath Jonniya, and Debasis Nayak. 2021. "Immunoinformatics Approach to Design Multi-Epitope- Subunit Vaccine against Bovine Ephemeral Fever Disease" Vaccines 9, no. 8: 925. https://doi.org/10.3390/vaccines9080925
APA StylePyasi, S., Sharma, V., Dipti, K., Jonniya, N. A., & Nayak, D. (2021). Immunoinformatics Approach to Design Multi-Epitope- Subunit Vaccine against Bovine Ephemeral Fever Disease. Vaccines, 9(8), 925. https://doi.org/10.3390/vaccines9080925






