Efficient Isolation of Outer Membrane Vesicles (OMVs) Secreted by Gram-Negative Bacteria via a Novel Gradient Filtration Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Apparatus
2.2. Measuring Growth Curve for EcN
2.3. EcN Culture and Isolation of EcN-Derived OMVs
2.3.1. EcN-Derived OMV Isolation via Gradient Filtration Method
2.3.2. EcN-Derived OMV Isolation via Ultracentrifugation
2.4. TEM of EcN-Derived OMV Samples
2.5. NTA of EcN-Derived OMV Samples
2.6. Sodium Dodecyl Sulfate Poly-Acrylamide Gel Electrophoresis (SDS-PAGE)
2.7. BCA Protein Quantification of EcN-Derived OMVs
2.8. EcN-Derived OMV Internalization by RAW264.7 Macrophages
2.9. Effect of the EcN-Derived OMVs on Cytokine Secretion in RAW264.7 Macrophages
2.10. Statistical Analysis
3. Results
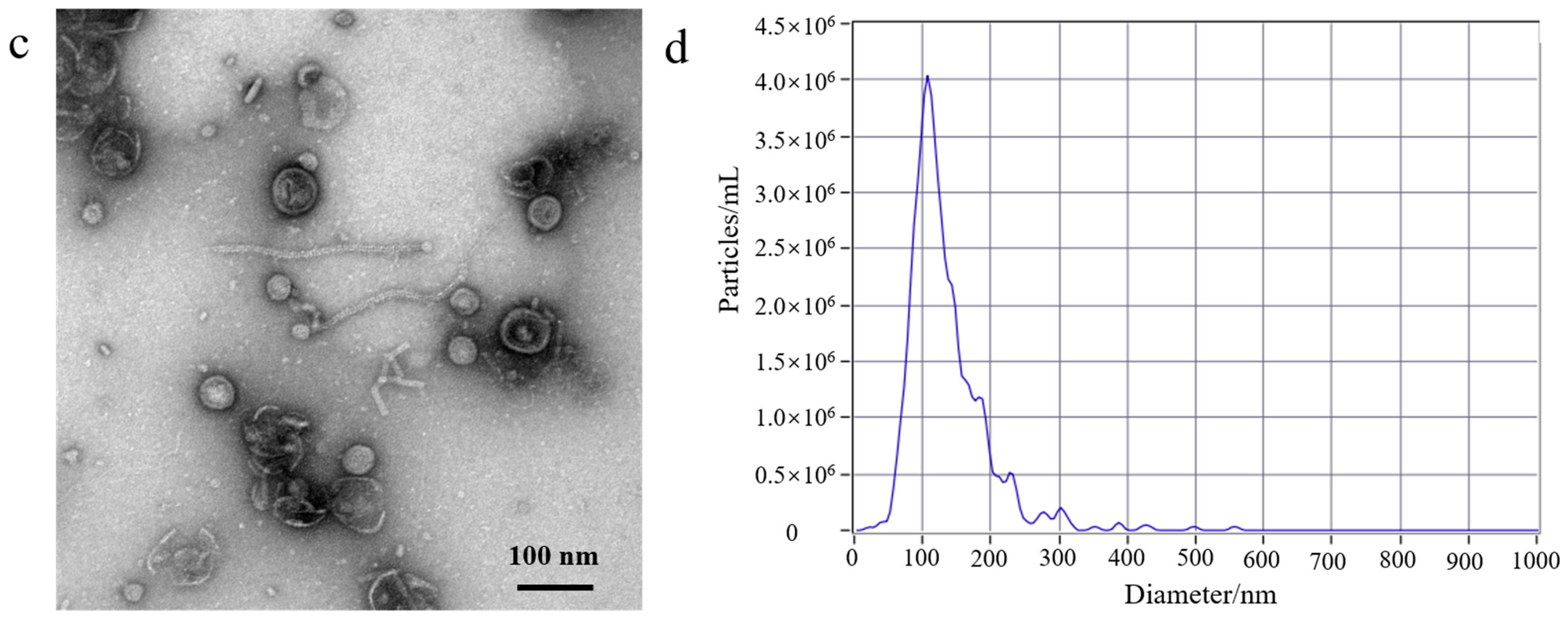
3.1. EcN-Derived OMV Isolation via Traditional Ultracentrifugation and Gradient Filtration Method
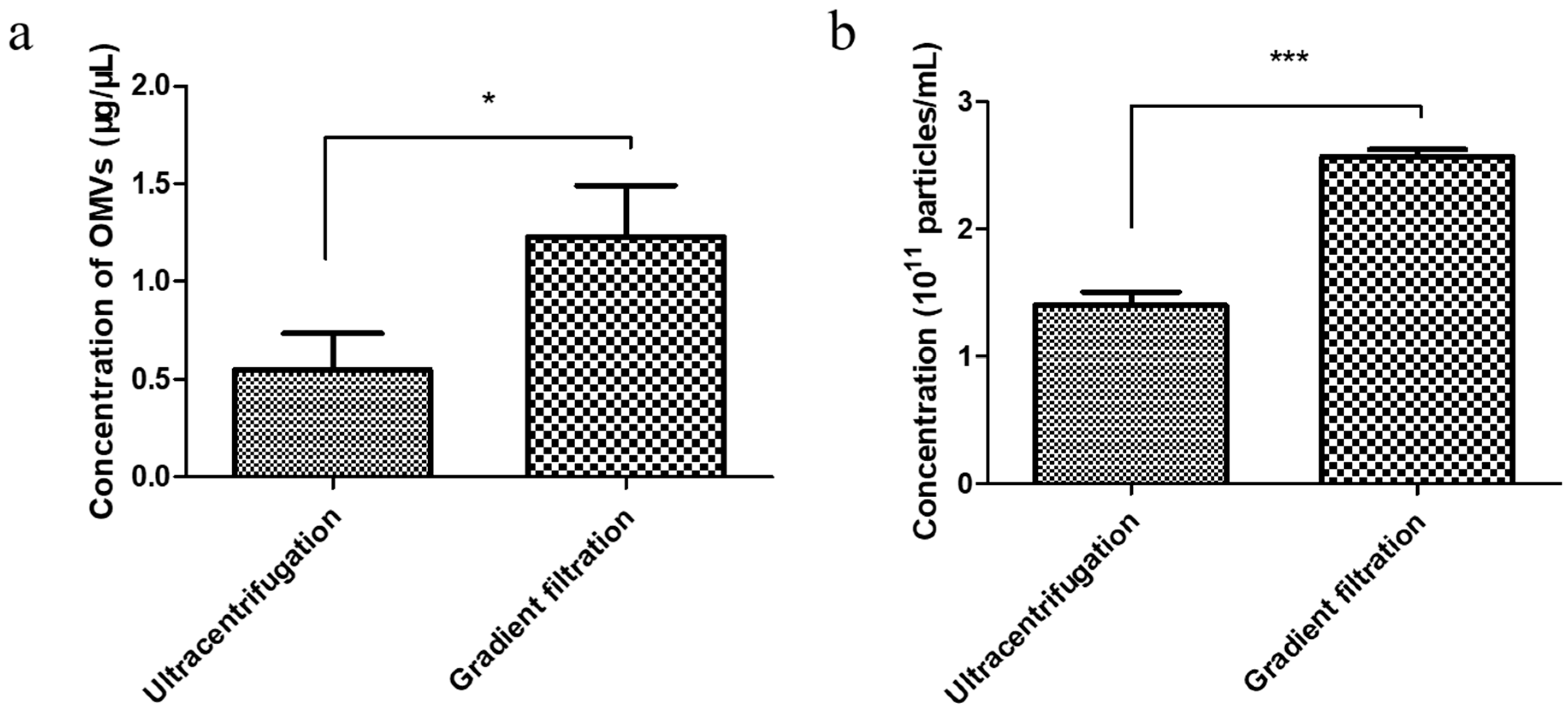
3.2. Comparison of the Yield of EcN-Derived OMVs Isolated via Ultracentrifugation and the Gradient Filtration Method
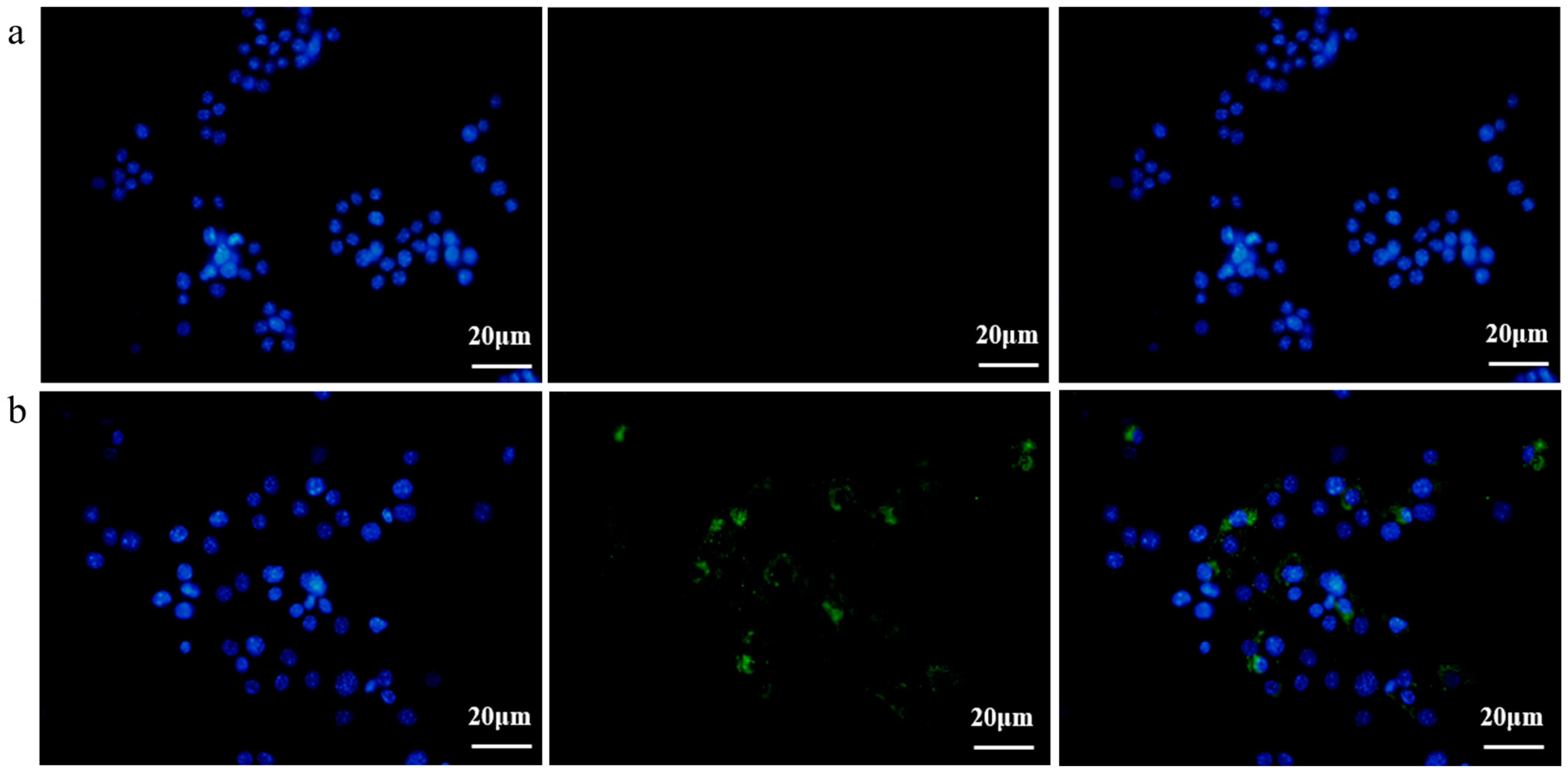
3.3. EcN-Derived OMVs Obtained via the Gradient Filtration Method Were Taken up by RAW264.7 Macrophages
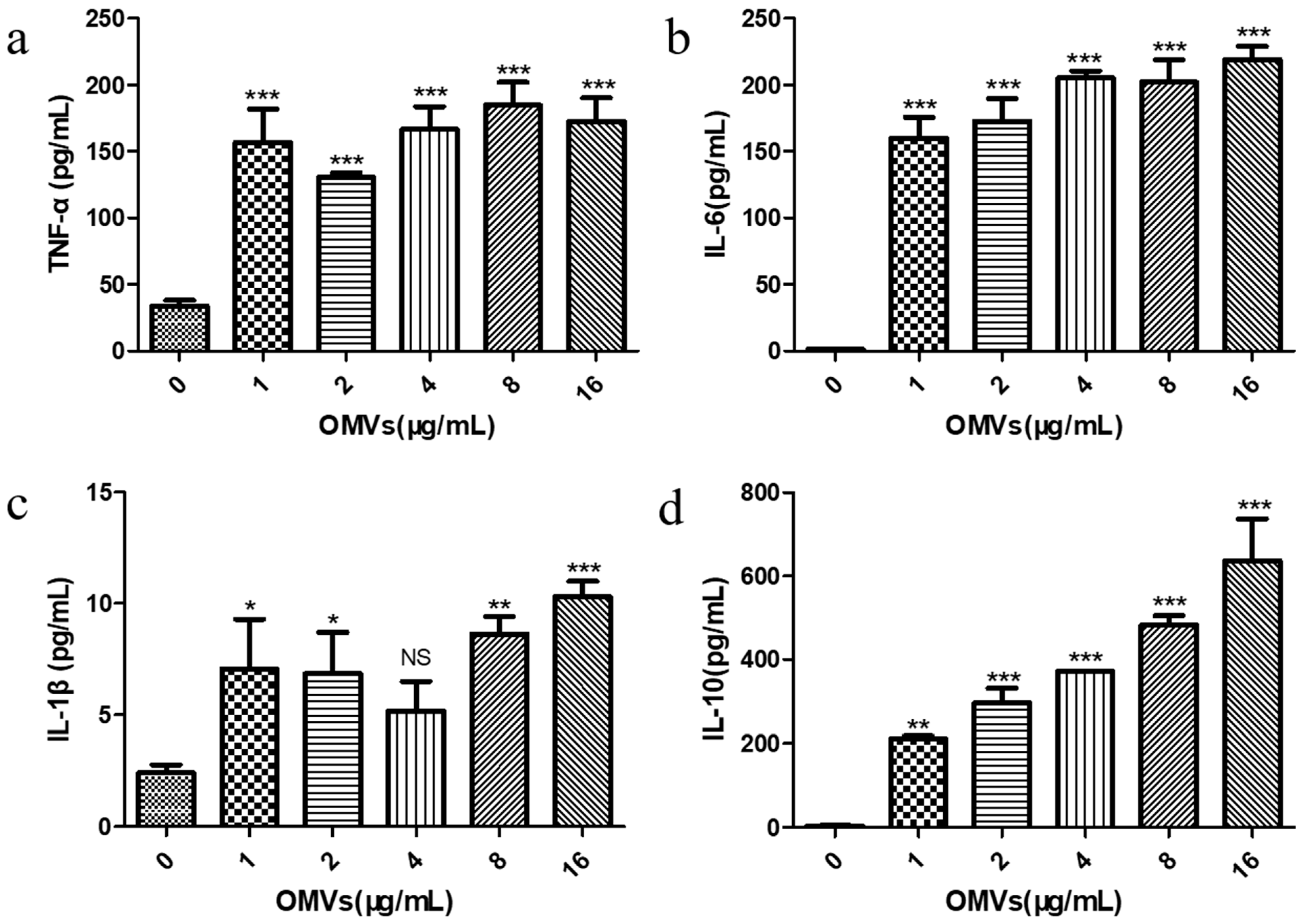
3.4. EcN-Derived OMVs Obtained via the Gradient Filtration Method Induced Secretion of Inflammatory Cytokines by RAW264.7 Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| bEVs | bacterial extracellular vesicles |
| OMVs | outer membrane vesicles |
| EcN | Escherichia coli Nissle 1917 |
| TEM | transmission electron microscope |
| NTA | nanoparticle tracking analysis |
| SDS-PAGE | sodium dodecyl sulfate poly-acrylamide gel electrophoresis |
| BCA | bicinchoninic acid |
| DiO | 3,3′-dioctadecyloxacarbocyanine perchlorate |
| TNF-α | tumor necrosis factor-alpha |
| IL-6 | interleukin-6 |
| IL-1β | interleukin-1β |
| IL-10 | interleukin-10 |
| LPS | lipopolysaccharide |
| NO | nitric oxide |
| ELISA | enzyme-linked immunosorbent assay |
| SDs | standard deviations |
References
- Xie, J.; Li, Q.; Haesebrouck, F.; Van Hoecke, L.; Vandenbroucke, R.E. The tremendous biomedical potential of bacterial extracellular vesicles. Trends Biotechnol. 2022, 40, 1173–1194. [Google Scholar] [CrossRef]
- Xie, J.; Haesebrouck, F.; Van Hoecke, L.; Vandenbroucke, R.E. Bacterial extracellular vesicles: An emerging avenue to tackle diseases. Trends Microbiol. 2023, 31, 1206–1224. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological Functions and Biogenesis of Secreted Bacterial Outer Membrane Vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2018, 17, 13–24. [Google Scholar] [CrossRef]
- Oh, J.; Ñahui Palomino, R.A.; Vanpouille, C.; Costantini, P.E.; Margolis, L. Microbiota–host communications: Bacterial extracellular vesicles as a common language. PLoS Pathog. 2021, 17, e1009508. [Google Scholar] [CrossRef]
- Cecil, J.D.; Sirisaengtaksin, N.; O'Brien-Simpson, N.M.; Krachler, A.M. Outer Membrane Vesicle-Host Cell Interactions. Microbiol. Spectrum. 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Volgers, C.; Savelkoul, P.H.M.; Stassen, F.R.M. Gram-negative bacterial membrane vesicle release in response to the host-environment: Different threats, same trick? Crit. Rev. Microbiol. 2017, 44, 258–273. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Yang, C.; Wu, Y.; Zhou, X.; Liu, H.; Wang, Y. Bacterial outer membrane vesicles as a platform for biomedical applications: An update. J. Control. Release 2020, 323, 253–268. [Google Scholar] [CrossRef]
- Grandi, A.; Tomasi, M.; Zanella, I.; Ganfini, L.; Caproni, E.; Fantappiè, L.; Irene, C.; Frattini, L.; Isaac, S.J.; König, E.; et al. Synergistic Protective Activity of Tumor-Specific Epitopes Engineered in Bacterial Outer Membrane Vesicles. Front. Oncol. 2017, 7, 253. [Google Scholar] [CrossRef]
- Tan, K.; Li, R.; Huang, X.; Liu, Q. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front. Microbiol. 2018, 9, 783. [Google Scholar] [CrossRef]
- Wang, S.; Huang, W.; Li, K.; Yao, Y.; Yang, X.; Bai, H.; Sun, W.; Liu, C.; Ma, Y. Engineered outer membrane vesicle is potent to elicit HPV16E7-specific cellular immunity in a mouse model of TC-1 graft tumor. Int. J. Nanomed. 2017, 12, 6813–6825. [Google Scholar] [CrossRef]
- Liu, Q.; Tan, K.; Yuan, J.; Song, K.; Li, R.; Huang, X.; Liu, Q. Flagellin-deficient outer membrane vesicles as adjuvant induce cross-protection of Salmonella Typhimurium outer membrane proteins against infection by heterologous Salmonella serotypes. Int. J. Med. Microbiol. 2018, 308, 796–802. [Google Scholar] [CrossRef]
- Zhang, K.; Chu, P.; Song, S.; Yang, D.; Bian, Z.; Li, Y.; Gou, H.; Jiang, Z.; Cai, R.; Li, C. Proteome Analysis of Outer Membrane Vesicles From a Highly Virulent Strain of Haemophilus parasuis. Front. Vet. Sci. 2021, 8, 756764. [Google Scholar] [CrossRef]
- Rad, A.T.; Chen, C.W.; Aresh, W.; Xia, Y.; Lai, P.S.; Nieh, M.P. Combinational Effects of Active Targeting, Shape, and Enhanced Permeability and Retention for Cancer Theranostic Nanocarriers. ACS Appl. Mater. Interfaces 2019, 11, 10505–10519. [Google Scholar]
- Sun, J.; Huang, Y.; Li, X.; Xu, X.; Cui, X.; Hao, F.; Ji, Q.; Chen, C.; Bao, G.; Liu, Y. Characterization and immunological effect of outer membrane vesicles from Pasteurella multocida on macrophages. Appl. Microbiol. Biotechnol. 2024, 108, 238. [Google Scholar] [CrossRef]
- Zariri, A.; Beskers, J.; van de Waterbeemd, B.; Hamstra, H.J.; Bindels, T.H.E.; van Riet, E.; van Putten, J.P.M.; van der Ley, P.; Palmer, G.H. Meningococcal Outer Membrane Vesicle Composition-Dependent Activation of the Innate Immune Response. Infect. Immun. 2016, 84, 3024–3033. [Google Scholar] [CrossRef]
- Huang, W.; Shu, C.; Hua, L.; Zhao, Y.; Xie, H.; Qi, J.; Gao, F.; Gao, R.; Chen, Y.; Zhang, Q.; et al. Modified bacterial outer membrane vesicles induce autoantibodies for tumor therapy. Acta Biomater. 2020, 108, 300–312. [Google Scholar] [CrossRef]
- Wo, J.; Lv, Z.-Y.; Sun, J.-N.; Tang, H.; Qi, N.; Ye, B.-C. Engineering probiotic-derived outer membrane vesicles as functional vaccine carriers to enhance immunity against SARS-CoV-2. iScience 2023, 26, 105772. [Google Scholar] [CrossRef]
- Cheng, K.; Zhao, R.; Li, Y.; Qi, Y.; Wang, Y.; Zhang, Y.; Qin, H.; Qin, Y.; Chen, L.; Li, C.; et al. Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via Plug-and-Display technology. Nat. Commun. 2021, 12, 2041. [Google Scholar] [CrossRef]
- Kuerban, K.; Gao, X.; Zhang, H.; Liu, J.; Dong, M.; Wu, L.; Ye, R.; Feng, M.; Ye, L. Doxorubicin-loaded bacterial outer-membrane vesicles exert enhanced anti-tumor efficacy in non-small-cell lung cancer. Acta Pharm. Sin. B 2020, 10, 1534–1548. [Google Scholar] [CrossRef]
- Gujrati, V.; Kim, S.; Kim, S.H.; Min, J.J.; Choy, H.E.; Kim, S.C.; Jon, S. Bioengineered Bacterial Outer Membrane Vesicles as Cell-Specific Drug-Delivery Vehicles for Cancer Therapy. ACS Nano 2014, 8, 1525–1537. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.; Wang, S.; Weng, W.; Jing, Y.; Su, J. Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: Advances and perspectives. Bioact. Mater. 2022, 14, 169–181. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Dell’Annunziata, F.; Ilisso, C.P.; Dell’Aversana, C.; Greco, G.; Coppola, A.; Martora, F.; Dal Piaz, F.; Donadio, G.; Falanga, A.; Galdiero, M.; et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the miRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020, 8, 1985. [Google Scholar] [CrossRef]
- Bhar, S.; Edelmann, M.J.; Jones, M.K. Characterization and proteomic analysis of outer membrane vesicles from a commensal microbe, Enterobacter cloacae. J. Proteom. 2021, 231, 103994. [Google Scholar] [CrossRef]
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Carol, A.F.; Timothy, M.W. Cytokines in acute and chronic inflammation. Front. Biosci. 1997, 2, d12–d26. [Google Scholar]
- Hu, R.; Lin, H.; Li, J.; Zhao, Y.; Wang, M.; Sun, X.; Min, Y.; Gao, Y.; Yang, M. Probiotic Escherichia coli Nissle 1917-derived outer membrane vesicles enhance immunomodulation and antimicrobial activity in RAW264.7 macrophages. BMC Microbiol. 2020, 20, 268. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2018, 43, 273–303. [Google Scholar] [CrossRef]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Jacobi, C.A.; Malfertheiner, P. Escherichia coli Nissle 1917 (Mutaflor): New insights into an old probiotic bacterium. Dig. Dis. 2011, 29, 600–607. [Google Scholar] [CrossRef]
- Scaldaferri, F.; Gerardi, V.; Mangiola, F.; Lopetuso, L.R.; Pizzoferrato, M.; Petito, V.; Papa, A.; Stojanovic, J.; Poscia, A.; Cammarota, G. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients: An update. World J. Gastroenterol. 2016, 22, 5505. [Google Scholar] [CrossRef]
- Zyrek, A.A.; Cichon, C.; Helms, S.; Enders, C.; Sonnenborn, U.; Schmidt, M.A. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCζ redistribution resulting in tight junction and epithelial barrier repair. Cell. Microbiol. 2007, 9, 804–816. [Google Scholar] [CrossRef]
- Grabig, A.; Paclik, D.; Guzy, C.; Dankof, A.; Baumgart, D.; Erckenbrecht, J.; Raupach, B.; Sonnenborn, U.; Eckert, J.; Schumann, R. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via toll-like receptor 2-and toll-like receptor 4-dependent pathways. Infect. Immun. 2006, 74, 4075–4082. [Google Scholar] [CrossRef]
- Schultz, M.; Strauch, U.G.; Linde, H.-J.R.; Watzl, S.; Obermeier, F.; Göttl, C.; Dunger, N.; Grunwald, N.; Schölmerich, J.R.; Rath, H.C. Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin. Vaccine Immunol. 2004, 11, 372–378. [Google Scholar] [CrossRef]
- Remer, K.A.; Bartrow, M.; Roeger, B.; Moll, H.; Sonnenborn, U.; Oelschlaeger, T.A. Split immune response after oral vaccination of mice with recombinant Escherichia coli Nissle 1917 expressing fimbrial adhesin K88. Int. J. Med. Microbiol. 2009, 299, 467–478. [Google Scholar] [CrossRef]
- Kaparakis-Liaskos, M.; Ferrero, R.L. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. [Google Scholar] [CrossRef]
- Arai, K. Cytokines: Coordinations of immune and inflammatory response. Ann.Rev.Biochem 1990, 59, 783–836. [Google Scholar] [CrossRef]
- Budhu, A.; Wang, X.W. The role of cytokines in hepatocellular carcinoma. J. Leukoc. Biol. 2006, 80, 1197–1213. [Google Scholar] [CrossRef]
- Mosser, D.M.; Zhang, X. Interleukin-10: New perspectives on an old cytokine. Immunol. Rev. 2008, 226, 205–218. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Wu, M.; Wang, L.; Tang, M.; Xin, H.; Deng, K. Efficient Isolation of Outer Membrane Vesicles (OMVs) Secreted by Gram-Negative Bacteria via a Novel Gradient Filtration Method. Membranes 2024, 14, 135. https://doi.org/10.3390/membranes14060135
Li N, Wu M, Wang L, Tang M, Xin H, Deng K. Efficient Isolation of Outer Membrane Vesicles (OMVs) Secreted by Gram-Negative Bacteria via a Novel Gradient Filtration Method. Membranes. 2024; 14(6):135. https://doi.org/10.3390/membranes14060135
Chicago/Turabian StyleLi, Ning, Minghui Wu, Lu Wang, Mengyu Tang, Hongbo Xin, and Keyu Deng. 2024. "Efficient Isolation of Outer Membrane Vesicles (OMVs) Secreted by Gram-Negative Bacteria via a Novel Gradient Filtration Method" Membranes 14, no. 6: 135. https://doi.org/10.3390/membranes14060135
APA StyleLi, N., Wu, M., Wang, L., Tang, M., Xin, H., & Deng, K. (2024). Efficient Isolation of Outer Membrane Vesicles (OMVs) Secreted by Gram-Negative Bacteria via a Novel Gradient Filtration Method. Membranes, 14(6), 135. https://doi.org/10.3390/membranes14060135





