Isolation of Viable SARS-CoV-2 Virus from Feces of an Immunocompromised Patient Suggesting a Possible Fecal Mode of Transmission
Abstract
:1. Introduction
2. Materials and Methods
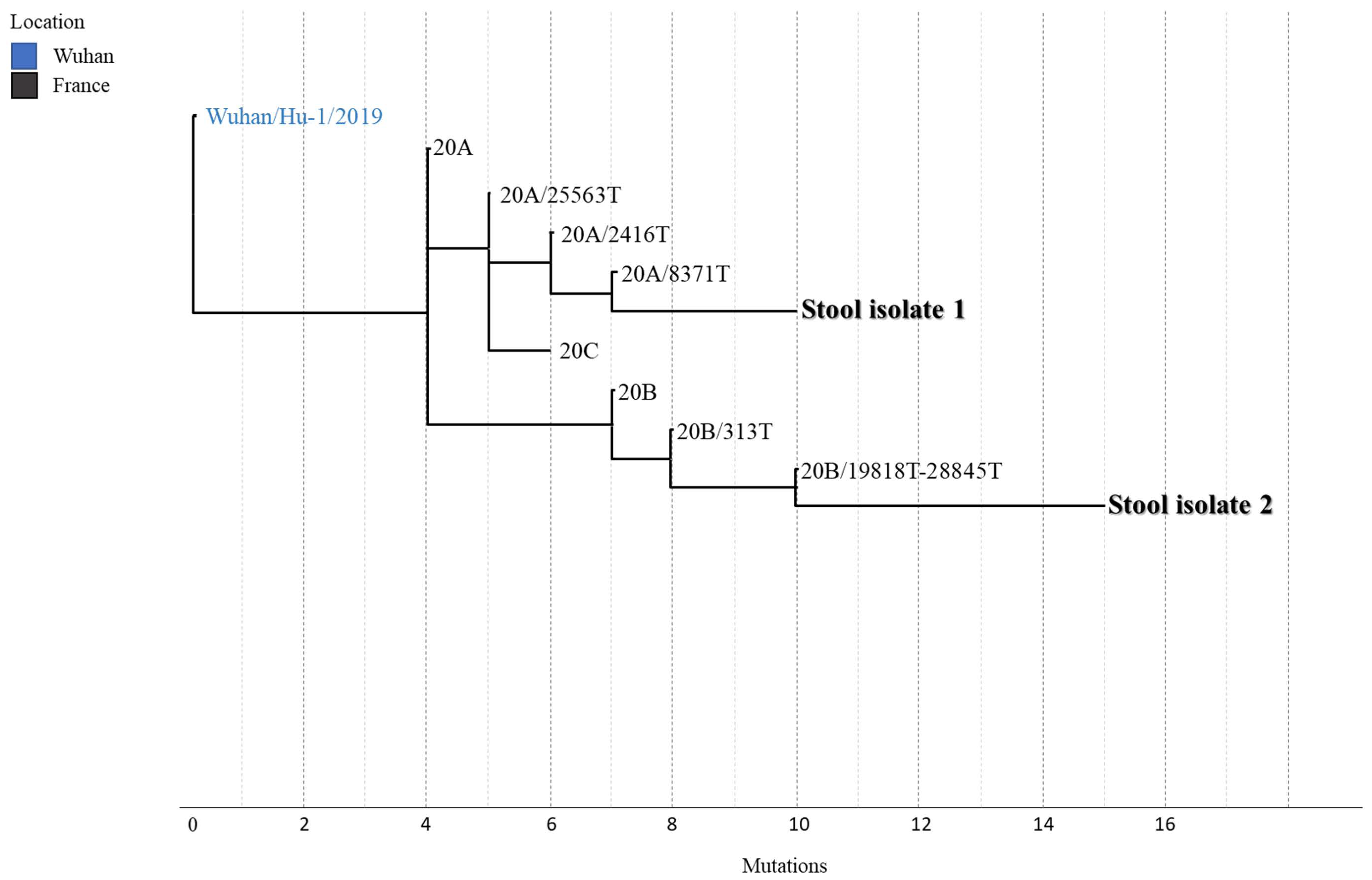
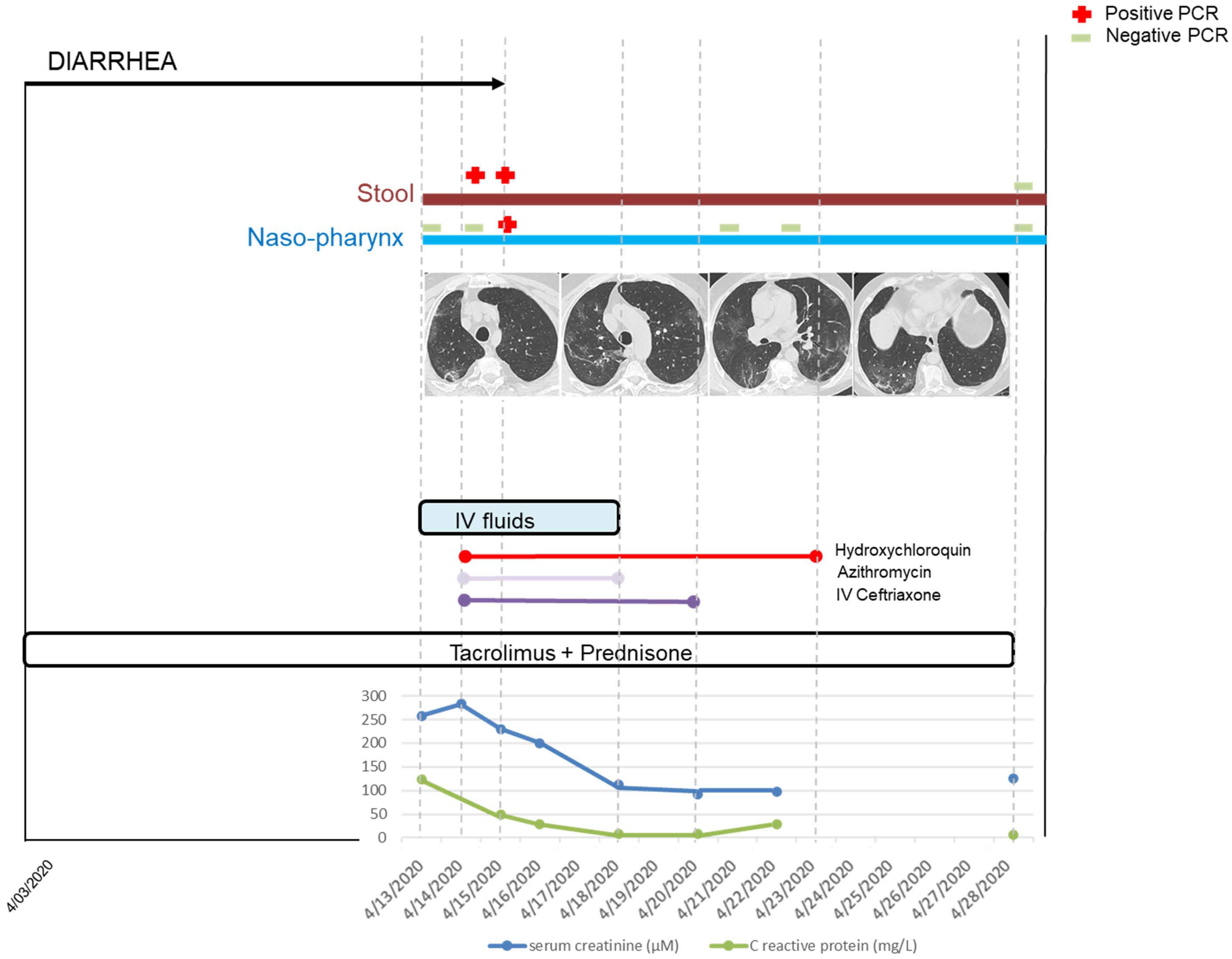
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Coronavirus Disease (COVID-19)—World Health Organization. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 12 June 2021).
- Modes of Transmission of Virus Causing COVID-19: Implications for IPC Precaution Recommendations. Available online: https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations (accessed on 12 June 2021).
- Mao, R.; Qiu, Y.; He, J.S.; Tan, J.Y.; Li, X.H.; Liang, J.; Shen, J.; Zhu, L.R.; Chen, Y.; Iacucci, M.; et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef]
- Wurtz, N.; Penant, G.; Jardot, P.; Duclos, N.; La Scola, B. Culture of SARS-CoV-2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Levasseur, A.; Gautret, P.; Fenollar, F.; Delerce, J.; Bitam, I.; Saile, R.; Maaloum, M.; Padane, A.; Bedotto, M.; et al. Introduction into the Marseille geographical area of a mild SARS-CoV-2 variant originating from sub-Saharan Africa: An investigational study. Travel Med. Infect. Dis. 2021, 40, 101980. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, C.; Zhu, S.; Shu, C.; Wang, D.; Song, J.; Song, Y.; Zhen, W.; Feng, Z.; Wu, G.; et al. Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19). China CDC Wkly. 2020, 2, 123–124. [Google Scholar] [CrossRef]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef]
- Amrane, S.; Tissot-Dupont, H.; Doudier, B.; Eldin, C.; Hocquart, M.; Mailhe, M.; Dudouet, P.; Ormières, E.; Ailhaud, L.; Parola, P.; et al. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, January 31st to March 1st, 2020: A respiratory virus snapshot. Travel Med. Infect. Dis. 2020, 36, 101632. [Google Scholar] [CrossRef]
- La Scola, B.; Le Bideau, M.; Andreani, J.; Grimaldier, C.; Colson, P.; Gautret, P.; Raoult, D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. [Google Scholar] [CrossRef]
- Colson, P.; Lagier, J.-C.; Baudoin, J.-P.; Bou Khalil, J.; La Scola, B.; Raoult, D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1601–1603. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Delerce, J.; Caputo, A.; Brechard, L.; Colson, P.; Lagier, J.C.; Fournier, P.E.; Raoult, D. Genomic diversity and evolution of coronavirus (SARS-CoV-2) in France from 309 COVID-19-infected patients. Genomics 2020. [Google Scholar] [CrossRef]
- Amoah, I.D.; Kumari, S.; Bux, F. Coronaviruses in wastewater processes: Source, fate and potential risks. Environ. Int. 2020, 143, 105962. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Samoilov, A.E.; Kaptelova, V.V.; Bukharina, A.Y.; Shipulina, O.Y.; Korneenko, E.V.; Lukyanov, A.V.; Grishaeva, A.A.; Ploskireva, A.A.; Speranskaya, A.S.; Akimkin, V.G. Change of dominant strain during dual SARS-CoV-2 infection. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Hashim, H.O.; Mohammed, M.K.; Mousa, M.J.; Abdulameer, H.H.; Alhassnawi, A.T.; Hassan, S.A.; Al-Shuhaib, M.B. Infection with different strains of SARS-COV-2 in patients with COVID-19. Arch. Biol. Sci. 2020, 72, 575–585. [Google Scholar] [CrossRef]
- Xu, Y.; Kang, L.; Shen, Z.; Li, X.; Wu, W.; Ma, W.; Fang, C.; Yang, F.; Jiang, X.; Gong, S.; et al. Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J. Genet. Genom. 2020. [Google Scholar] [CrossRef]
- Caillard, S.; Anglicheau, D.; Matignon, M.; Durrbach, A.; Greze, C.; Frimat, L.; Thaunat, O.; Legris, T.; Moal, V.; Westeel, P.F.; et al. An initial report from the French SOT COVID Registry suggests high mortality due to COVID-19 in recipients of kidney transplants. Kidney Int. 2020, 98, 1549–1558. [Google Scholar] [CrossRef]
- Caillard, S.; Chavarot, N.; Francois, H.; Matignon, M.; Greze, C.; Kamar, N.; Gatault, P.; Thaunat, O.; Legris, T.; Frimat, L.; et al. Is COVID-19 infection more severe in kidney transplant recipients? Am. J. Transplant. 2021, 21, 1295–1303. [Google Scholar] [CrossRef]
- Aydillo, T.; Gonzalez-Reiche, A.S.; Aslam, S.; van de Guchte, A.; Khan, Z.; Obla, A.; Dutta, J.; van Bakel, H.; Aberg, J.; García-Sastre, A.; et al. Shedding of Viable SARS-CoV-2 after Immunosuppressive Therapy for Cancer. N. Engl. J. Med. 2020, 383, 2586–2588. [Google Scholar] [CrossRef]
- Moal, V.; Zandotti, C.; Colson, P. Emerging viral diseases in kidney transplant recipients: Emerging viral diseases in renal transplantation. Rev. Med. Virol. 2013, 23, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaafar, R.; Aherfi, S.; Wurtz, N.; Grimaldier, C.; Van Hoang, T.; Colson, P.; Raoult, D.; La Scola, B. Correlation between 3790 Quantitative Polymerase Chain Reaction-Positives Samples and Positive Cell Cultures, Including 1941 Severe Acute Respiratory Syndrome Coronavirus 2 Isolates. Clin. Infect. Dis. 2021, 72, e921. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.W.; Kim, S.M.; Kim, H.S.; Kim, Y.I.; Kim, J.H.; Cho, J.Y.; Kim, S.H.; Kang, H.; Kim, S.G.; Park, S.J.; et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1520–1524. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dergham, J.; Delerce, J.; Bedotto, M.; La Scola, B.; Moal, V. Isolation of Viable SARS-CoV-2 Virus from Feces of an Immunocompromised Patient Suggesting a Possible Fecal Mode of Transmission. J. Clin. Med. 2021, 10, 2696. https://doi.org/10.3390/jcm10122696
Dergham J, Delerce J, Bedotto M, La Scola B, Moal V. Isolation of Viable SARS-CoV-2 Virus from Feces of an Immunocompromised Patient Suggesting a Possible Fecal Mode of Transmission. Journal of Clinical Medicine. 2021; 10(12):2696. https://doi.org/10.3390/jcm10122696
Chicago/Turabian StyleDergham, Julie, Jeremy Delerce, Marielle Bedotto, Bernard La Scola, and Valérie Moal. 2021. "Isolation of Viable SARS-CoV-2 Virus from Feces of an Immunocompromised Patient Suggesting a Possible Fecal Mode of Transmission" Journal of Clinical Medicine 10, no. 12: 2696. https://doi.org/10.3390/jcm10122696





