The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review
Abstract
:1. Introduction
2. Artificial Intelligence
3. Machine Learning
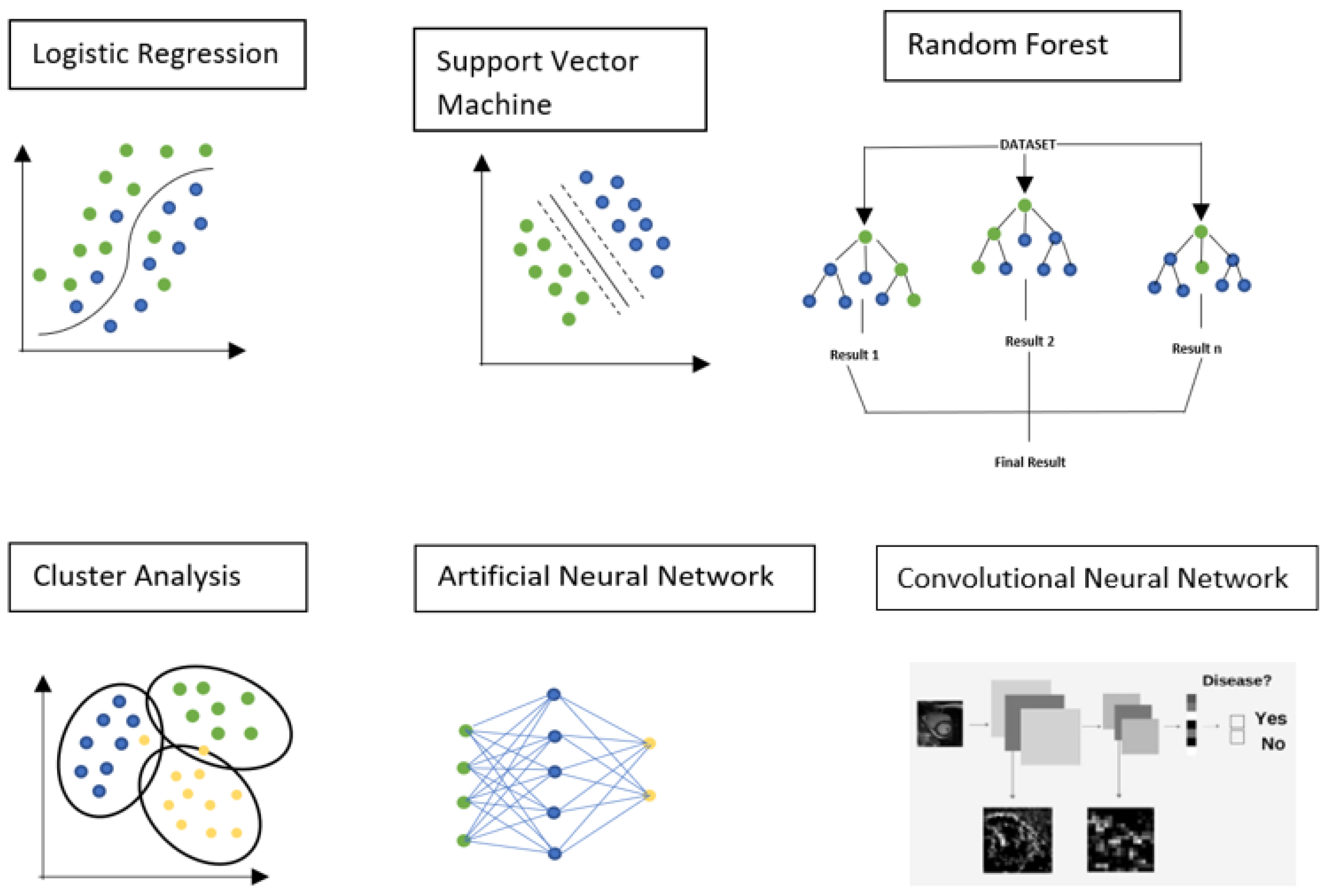
3.1. Logistic Regression
3.2. Support Vector Machine
3.3. Random Forest
3.4. Cluster Analysis
3.5. Artificial Neural Network
3.6. Convolutional Neural Network
4. Deep Learning
- Input layer: the input layer is used to take input data from sources and then pass it to the hidden layers of the neural network. It does not perform any calculations.
- Hidden level: this level consists of many hidden levels. All the calculation is performed at this level. After all the calculations are complete, it proceeds to the output level.
- Output level: this level is used to provide the output to the outside world.
5. Current Applications of Artificial Intelligence
6. Image Acquisition
6.1. Slice Position
6.2. Image Quality
6.3. Image Speed Acquisition
7. Image Segmentation
8. Myocardial Tissue Characterization
9. Diagnosis
9.1. Myocardial Infarction
9.2. Cardiomyopathies
9.3. Heart Failure
9.4. Abnormal Wall Motion
10. Prognosis
11. Limitations
12. Future Perspectives
13. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CMR | Cardiovascular Magnetic Resonance |
| ML | Machine Learning |
| DL | Deep Learning |
| SVM | Support Vector Machine |
| ANN | Artificial Neural Network |
| CNN | Convolutional Neural Network |
| CS | Compressed Sensing |
| LGE | Late Gadolinium Enhancement |
| EF | Ejection Fraction |
| HCM | Hypertrophic Cardiomyopathy |
| DCM | Dilated Cardiomyopathy |
| MI | Myocardial Infarction |
| HF | Heart Failure |
References
- Timmis, A.; Townsend, N.; Gale, C.P.; Torbica, A.; Lettino, M.; Petersen, S.E.; Mossialos, E.A.; Maggioni, A.P.; Kazakiewicz, K.; May, H.T.; et al. European Society of Cardiology Cardiovascular Disease Statistics 2019. Eur. Heart. J. 2020, 41, 12–85. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Andreini, D.; Bertella, E.; Baggiano, A.; Mushtaq, S.; Loguercio, M.; Segurini, C.; Conte, E.; Beltrama, V.; Annoni, A.; et al. Impact of an intra-cycle motion correction algorithm on overall evaluability and diagnostic accuracy of computed tomography coronary angiography. Eur. Radiol. 2016, 26, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Guaricci, A.I.; Maffei, E.; Brunetti, N.D.; Montrone, D.; Di Biase, L.; Tedeschi, C.; Gentile, G.; Macarini, L.; Midiri, M.; Cademartiri, F.; et al. Heart rate control with oral ivabradine in computed tomography coronary angiography: A randomized comparison of 7.5 mg vs 5 mg regimen. Int. J. Cardiol. 2013, 168, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Weir-McCall, J.R.; Baggiano, A.; Del Torto, A.; Fusini, L.; Guglielmo, M.; Muscogiuri, G.; Guaricci, A.I.; Andreini, D.; Patel, M.; et al. Determinants of Rejection Rate for Coronary CT Angiography Fractional Flow Reserve Analysis. Radiology 2019, 292, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Andreini, D.; Guaricci, A.I.; Guglielmo, M.; Baggiano, A.; Muscogiuri, G.; Fusini, L.; Soldi, M.; Fazzari, F.; Berzovini, C.; et al. Quantitative vs. qualitative evaluation of static stress computed tomography perfusion to detect haemodynamically significant coronary artery disease. Eur. Heart. J. Cardiovasc. Imaging 2018, 19, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, M.; Fusini, L.; Muscogiuri, G.; Baessato, F.; Loffreno, A.; Cavaliere, A.; Rizzon, G.; Baggiano, A.; Rabbat, M.G.; Muratori, M.; et al. T1 mapping and cardiac magnetic resonance feature tracking in mitral valve prolapse. Eur. Radiol. 2021, 31, 1100–1119. [Google Scholar] [CrossRef] [PubMed]
- Baggiano, A.; Fusini, L.; Del Torto, A.; Vivona, P.; Guglielmo, M.; Muscogiuri, G.; Soldi, M.; Martini, C.; Fraschini, E.; Rabbat, M.G.; et al. Sequential Strategy Including FFR(CT) Plus Stress-CTP Impacts on Management of Patients with Stable Chest Pain: The Stress-CTP RIPCORD Study. J. Clin. Med. 2020, 9, 2147. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Muscogiuri, G.; Andreini, D.; Guaricci, A.I.; Guglielmo, M.; Baggiano, A.; Fazzari, F.; Mushtaq, S.; Conte, E.; Annoni, A.; et al. Impact of a New Adaptive Statistical Iterative Reconstruction (ASIR)-V Algorithm on Image Quality in Coronary Computed Tomography Angiography. Acad. Radiol. 2018, 25, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Guaricci, A.I.; Palmer, S.C.; Andreini, D.; Verdecchia, M.; Fusini, L.; Lorenzoni, V.; Guglielmo, M.; Muscogiuri, G.; Baggiano, A.; et al. Diagnostic performance of non-invasive imaging for stable coronary artery disease: A meta-analysis. Int. J. Cardiol. 2020, 300, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial Intelligence in Cardiovascular Imaging: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef] [PubMed]
- Al’Aref, S.J.; Anchouche, K.; Singh, G.; Slomka, P.J.; Kolli, K.K.; Kumar, A.; Pandey, M.; Maliakal, G.; Van Rosendael, A.R.; Beecy, A.N.; et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur. Heart J. 2019, 40, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Visco, V.; Ferruzzi, G.J.; Nicastro, F.; Virtuoso, N.; Carrizzo, A.; Galasso, G.; Vecchione, C.; Ciccarelli, M. Artificial Intelligence as a Business Partner in Cardiovascular Precision Medicine: An Emerging Approach for Disease Detection and Treatment Optimization. Curr. Med. Chem. 2021, 28, 6569–6590. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; de Vos, B.D.; Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Chiesa, M.; Trotta, M.; Gatti, M.; Palmisano, V.; Dell’Aversana, S.; Baessato, F.; Cavaliere, A.; Cicala, G.; Loffreno, A.; et al. Performance of a deep learning algorithm for the evaluation of CAD-RADS classification with CCTA. Atherosclerosis 2020, 294, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mintz, Y.; Brodie, R. Introduction to artificial intelligence in medicine. Minim Invasive Ther. Allied Technol. 2019, 28, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Kilic, A. Artificial Intelligence and Machine Learning in Cardiovascular Health Care. Ann. Thorac. Surg. 2020, 109, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [PubMed]
- Syeda-Mahmood, T. Role of Big Data and Machine Learning in Diagnostic Decision Support in Radiology. J. Am. Coll. Radiol. 2018, 15, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef] [PubMed]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Liu, Z.; Adeli, E.; Pohl, K.M. Longitudinal self-supervised learning. Med. Image Anal. 2021, 71, 102051. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Liu, X.; Liu, F.; Wang, C. Comparison of machine learning and logistic regression models in predicting acute kidney injury: A systematic review and meta-analysis. Int. J. Med. Inform. 2021, 151, 104484. [Google Scholar] [CrossRef] [PubMed]
- Habehh, H.; Gohel, S. Machine Learning in Healthcare. Curr. Genom. 2021, 22, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Waring, J.; Lindvall, C.; Umeton, R. Automated machine learning: Review of the state-of-the-art and opportunities for healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef] [PubMed]
- Battleday, R.M.; Peterson, J.C.; Griffiths, T.L. From convolutional neural networks to models of higher-level cognition (and back again). Ann. N. Y. Acad. Sci. 2021, 1505, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.M.; Barkhausen, J.; Bucciarelli-Ducci, C.; Flamm, S.D.; Kim, R.J.; Nagel, E. Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update. J. Cardiovasc. Magn. Reson. 2020, 22, 17. [Google Scholar] [CrossRef] [PubMed]
- Schlemper, J.; Caballero, J.; Hajnal, J.V.; Price, A.N.; Rueckert, D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans. Med. Imaging 2018, 37, 491–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kido, T.; Kido, T.; Nakamura, M.; Watanabe, K.; Schmidt, M.; Forman, C.; Mochizuki, T. Compressed sensing real-time cine cardiovascular magnetic resonance: Accurate assessment of left ventricular function in a single-breath-hold. J. Cardiovasc. Magn. Reson. 2016, 18, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basha, T.A.; Akçakaya, M.; Liew, C.; Tsao, C.W.; Delling, F.N.; Addae, G.; Ngo, L.; Manning, W.J.; Nezafat, R. Clinical performance of high-resolution late gadolinium enhancement imaging with compressed sensing. J. Magn. Reson. Imaging 2017, 46, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Vermersch, M.; Longère, B.; Coisne, A.; Schmidt, M.; Forman, C.; Monnet, A.; Pagniez, J.; Silvestri, V.; Simeone, A.; Cheasty, E.; et al. Compressed sensing real-time cine imaging for assessment of ventricular function, volumes and mass in clinical practice. Eur. Radiol. 2020, 30, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Van Assen, M.; Muscogiuri, G.; Caruso, D.; Lee, S.J.; Laghi, A.; De Cecco, C.N. Artificial intelligence in cardiac radiology. Radiol. Med. 2020, 125, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Martini, C.; Gatti, M.; Dell’Aversana, S.; Ricci, F.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Bracciani, A.; Scafuri, S.; et al. Feasibility of late gadolinium enhancement (LGE) in ischemic cardiomyopathy using 2D-multisegment LGE combined with artificial intelligence reconstruction deep learning noise reduction algorithm. Int. J. Cardiol. 2021, 343, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Forman, C.; Piccini, D.; Grimm, R.; Hutter, J.; Hornegger, J.; Zenge, M.O. High-resolution 3D whole-heart coronary MRA: A study on the combination of data acquisition in multiple breath-holds and 1D residual respiratory motion compensation. Magma 2014, 27, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Forman, C.; Piccini, D.; Grimm, R.; Hutter, J.; Hornegger, J.; Zenge, M.O. Reduction of respiratory motion artifacts for free-breathing whole-heart coronary MRA by weighted iterative reconstruction. Magn. Reson. Med. 2015, 73, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kido, T.; Kido, T.; Watanabe, K.; Schmidt, M.; Forman, C.; Mochizuki, T. Non-contrast compressed sensing whole-heart coronary magnetic resonance angiography at 3T: A comparison with conventional imaging. Eur. J. Radiol. 2018, 104, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Frick, M.; Paetsch, I.; den Harder, C.; Kouwenhoven, M.; Heese, H.; Dries, S.; Schnackenburg, B.; de Kok, W.; Gebker, R.; Fleck, E.; et al. Fully automatic geometry planning for cardiac MR imaging and reproducibility of functional cardiac parameters. J. Magn. Reson. Imaging 2011, 34, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, K.; Nitta, S.; Kuhara, S.; Ishimura, R.; Kariyasu, T.; Imai, M.; Nitatori, T.; Takeguchi, T.; Shiodera, T. Automatic slice-alignment method in cardiac magnetic resonance imaging for evaluation of the right ventricle in patients with pulmonary hypertension. AIP Adv. 2015, 5, 097182. [Google Scholar] [CrossRef] [Green Version]
- Nitta, S.; Takeguchi, T.; Matsumoto, N.; Kuhara, S.; Yokoyama, K.; Ishimura, R.; Nitatori, T. Automatic slice alignment method for cardiac magnetic resonance imaging. Magma 2013, 26, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Oktay, O.; Rueckert, D.; Bai, W.; Guerrero, R.; Rajchl, M.; de Marvao, A.; O’Regan, D.P.; Cook, S.A.; Heinrich, M.P.; Glocker, B. Stratified Decision Forests for Accurate Anatomical Landmark Localization in Cardiac Images. IEEE Trans. Med. Imaging 2017, 36, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Jolly, M.P.; Georgescu, B.; Haye, C.; Speier, P.; Schmidt, M.; Bi, X.; Kroeker, R.; Comaniciu, D.; Kellman, P.; et al. Automatic view planning for cardiac MRI acquisition. Med. Image Comput. Comput. Assist. Interv. 2011, 14, 479–486. [Google Scholar] [PubMed] [Green Version]
- Blansit, K.; Retson, T.; Masutani, E.; Bahrami, N.; Hsiao, A. Deep Learning-based Prescription of Cardiac MRI Planes. Radiol. Artif. Intell. 2019, 1, e180069. [Google Scholar] [CrossRef] [PubMed]
- Lebel, R.M. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. arXiv 2020, arXiv:2008.06559. [Google Scholar]
- Van der Velde, N.; Hassing, H.C.; Bakker, B.J.; Wielopolski, P.A.; Lebel, R.M.; Janich, M.A.; Kardys, I.; Budde, R.P.J.; Hirsch, A. Improvement of late gadolinium enhancement image quality using a deep learning-based reconstruction algorithm and its influence on myocardial scar quantification. Eur Radiol. 2021, 31, 3846–3855. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, A.; Arridge, S.; Lucka, F.; Muthurangu, V.; Steeden, J.A. Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning-proof of concept in congenital heart disease. Magn. Reson. Med. 2019, 81, 1143–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandino, C.M.; Lai, P.; Vasanawala, S.S.; Cheng, J.Y. Accelerating cardiac cine MRI using a deep learning-based ESPIRiT reconstruction. Magn. Reson. Med. 2021, 85, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Kustner, T.; Fuin, N.; Hammernik, K.; Bustin, A.; Qi, H.; Hajhosseiny, R.; Masci, P.G.; Neji, R.; Rueckert, D.; Botnar, R.M.; et al. CINENet: Deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions. Sci. Rep. 2020, 10, 13710. [Google Scholar] [CrossRef] [PubMed]
- Ursuleanu, T.F.; Luca, A.R.; Gheorghe, L.; Grigorovici, R.; Iancu, S.; Hlusneac, M.; Preda, C.; Grigorovici, A. Deep Learning Application for Analyzing of Constituents and Their Correlations in the Interpretations of Medical Images. Diagnostics 2021, 11, 1373. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Gatti, M.; Dell’Aversana, S.; Pica, S.; Andreini, D.; Guaricci, A.I.; Guglielmo, M.; Baggiano, A.; Mushtaq, S.; Conte, E.; et al. Reliability of single breath hold three-dimensional cine kat-ARC for the assessment of biventricular dimensions and function. Eur. J. Radiol. 2020, 124, 108820. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.D.; Cheng, H.M. Primer and Historical Review on Rapid Cardiac CINE MRI. J. Magn. Reson. Imaging 2022, 55, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Menger, J.; Bluemke, D.A.; Bremerich, J.; Flamm, S.D.; Fogel, M.A.; Friedrich, M.G.; Kim, R.J.; Von Knobelsdorff-Brenkenhoff, F.; Kramer, C.M.; Pennell, D.J.; et al. Standardized image interpretation and post processing in cardiovascular magnetic resonance: Society for Cardiovascular Magnetic Resonance (SCMR) board of trustees task force on standardized post processing. J. Cardiovasc. Magn. Reson. 2013, 15, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guaricci, A.I.; Masci, P.G.; Lorenzoni, V.; Schwitter, J.; Pontone, G. CarDiac MagnEtic Resonance for Primary Prevention Implantable CardioVerter DebrillAtor ThErapy international registry: Design and rationale of the DERIVATE study. Int. J. Cardiol. 2018, 261, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Guaricci, A.I.; Masci, P.G.; Muscogiuri, G.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Lorenzoni, V.; Martini, C.; Andreini, D.; Pavon, A.G.; et al. CarDiac magnEtic Resonance for prophylactic Implantable-cardioVerter defibrillAtor ThErapy in Non-Ischaemic dilated CardioMyopathy: An international Registry. Europace 2021, 23, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Romaguera, L.V.R.F.; Fernandes Costa Filho, C.F.; Fernandes Costa, M.G. Myocardial segmentation in cardiac magnetic resonance images using fully convolutional neural networks. Biomed. Signal Process Control 2018, 44, 48–57. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.-A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 2018, 20, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penso, M.; Moccia, S.; Scafuri, S.; Muscogiuri, G.; Pontone, G.; Pepi, M.; Caiani, E.G. Automated left and right ventricular chamber segmentation in cardiac magnetic resonance images using dense fully convolutional neural network. Comput. Methods. Programs Biomed. 2021, 204, 106059. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Fedorov, V.V.; Fu, X.; Cheng, E.; Macleod, R.; Zhao, J. Fully Automatic Left Atrium Segmentation From Late Gadolinium Enhanced Magnetic Resonance Imaging Using a Dual Fully Convolutional Neural Network. IEEE Trans. Med. Imaging 2019, 38, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhuang, X.; Khan, H.; Haldar, S.; Nyktari, E.; Li, L.; Wage, R.; Ye, X.; Slabaugh, G.G.; Mohiaddin, R.; et al. Fully automatic segmentation and objective assessment of atrial scars for long-standing persistent atrial fibrillation patients using late gadolinium-enhanced MRI. Med. Phys. 2018, 45, 1562–1576. [Google Scholar] [CrossRef] [PubMed]
- Zabihollahy, F.; White, J.A.; Ukwatta, E. Convolutional neural network-based approach for segmentation of left ventricle myocardial scar from 3D late gadolinium enhancement MR images. Med. Phys. 2019, 46, 1740–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moccia, S.; Banali, R.; Martini, C.; Muscogiuri, G.; Pontone, G.; Pepi, M.; Caiani, E.G. Development and testing of a deep learning-based strategy for scar segmentation on CMR-LGE images. Magma 2019, 32, 187–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Xu, L.; Gao, Z.; Zhao, S.; Zhang, H.; Zhang, Y.; Du, X.; Zhao, S.; Ghista, D.; Liu, H.; et al. Direct delineation of myocardial infarction without contrast agents using a joint motion feature learning architecture. Med. Image Anal. 2018, 50, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Wolterink, J.M.L.T.; Viergever, M.A.; Išgum, I. Automatic segmentation and disease classification using cardiac cine MR images. Lect. Notes Comput. Sci. 2018, 10663, 101–110. [Google Scholar]
- Snaauw, G.; Gong, D.; Maicas, G.; Hengel, A.V.D.; Niessen, W.J.; Verjans, J.; Carneiro, G. End-to-end diagnosis and segmentation learning from cardiac magnetic resonance imaging. Proc. Int. Symp. Biomed. Imaging 2019, 2019, 802–805. [Google Scholar]
- Wang, S.; Chauhan, D.; Patel, H.; Amir-Khalili, A.; da Silva, I.F.; Sojoudi, A.; Friedrich, S.; Singh, A.; Landeras, L.; Miller, T.; et al. Assessment of right ventricular size and function from cardiovascular magnetic resonance images using artificial intelligence. J. Cardiovasc. Magn. Reson. 2022, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Alandejani, F.; Alabed, S.; Garg, P.; Goh, Z.M.; Karunasaagarar, K.; Sharkey, M.; Salehi, M.; Aldabbagh, Z.; Dwivedi, K.; Mamalakis, M.; et al. Training and clinical testing of artificial intelligence derived right atrial cardiovascular magnetic resonance measurements. J. Cardiovasc. Magn. Reson. 2022, 24, 25. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, A.S.; Rausch, J.; Neisius, U.; Chan, R.H.; Maron, M.S.; Appelbaum, E.; Menze, B.; Nezafat, R. Automated Cardiac MR Scar Quantification in Hypertrophic Cardiomyopathy Using Deep Convolutional Neural Networks. JACC Cardiovasc. Imaging 2018, 11, 1917–1918. [Google Scholar] [CrossRef] [PubMed]
- Kamesh Iyer, S.; Tasdizen, T.; Burgon, N.; Kholmovski, E.; Marrouche, N.; Adluru, G.; Di Bella, E. Compressed sensing for rapid late gadolinium enhanced imaging of the left atrium: A preliminary study. Magn. Reson. Imaging 2016, 34, 846–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahmy, A.S.; El-Rewaidy, H.; Nezafat, M.; Nakamori, S.; Nezafat, R. Automated analysis of cardiovascular magnetic resonance myocardial native T(1) mapping images using fully convolutional neural networks. J. Cardiovasc. Magn. Reson. 2019, 21, 7. [Google Scholar] [CrossRef] [PubMed]
- Farrag, N.A.; Lochbihler, A.; White, J.A.; Ukwatta, E. Evaluation of fully automated myocardial segmentation techniques in native and contrast-enhanced T1-mapping cardiovascular magnetic resonance images using fully convolutional neural networks. Med. Phys. 2021, 48, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Hann, E.; Ferreira, V.M.; Neubauer, S.; Piechnik, S.K. Deep learning for fully automatic contouring of the left ventricle in cardiac T1 mapping. In Proceedings of the CMR 2018–A Joint EuroCMR/SCMR Meeting Abstract Supplement, Barcelona, Spain, 31 January–3 February 2018; pp. 401–402. [Google Scholar]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallières, M.; Zwanenburg, A.; Badic, B.; Cheze Le Rest, C.; Visvikis, D.; Hatt, M. Responsible Radiomics Research for Faster Clinical Translation. J. Nucl. Med. 2018, 59, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Tourassi, G.D. Journey toward computer-aided diagnosis: Role of image texture analysis. Radiology 1999, 213, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Baessler, B.; Mannil, M.; Oebel, S.; Maintz, D.; Alkadhi, H.; Manka, R. Subacute and Chronic Left Ventricular Myocardial Scar: Accuracy of Texture Analysis on Nonenhanced Cine MR Images. Radiology 2018, 286, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schofield, R.; Ganeshan, B.; Fontana, M.; Nasis, A.; Castelletti, S.; Rosmini, S.; Treibel, T.; Manisty, C.; Endozo, R.; Groves, A.; et al. Texture analysis of cardiovascular magnetic resonance cine images differentiates aetiologies of left ventricular hypertrophy. Clin. Radiol. 2019, 74, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Engan, K.; Eftestol, T.; Orn, S.; Kvaloy, J.T.; Woie, L. Exploratory data analysis of image texture and statistical features on myocardium and infarction areas in cardiac magnetic resonance images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2010, 2010, 5728–5731. [Google Scholar] [PubMed]
- Kotu, L.P.; Engan, K.; Skretting, K.; Måløy, F.; Orn, S.; Woie, L.; Eftestøl, T. Probability mapping of scarred myocardium using texture and intensity features in CMR images. Biomed. Eng. Online 2013, 12, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larroza, A.; Materka, A.; López-Lereu, M.P.; Monmeneu, J.V.; Bodí, V.; Moratal, D. Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur. J. Radiol. 2017, 92, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, R.E.C.M.; Dwivedi, G.; Dennie, C.; Fuller, L.; Dick, A.; Ruddy, T.; Peña, E. Quantitative texture features as objective metrics of enhancement heterogeneity in hypertrophic cardiomyopathy. J. Cardiovasc. Magn. Reson. 2014, 16, P351. [Google Scholar] [CrossRef] [Green Version]
- Neisius, U.; El-Rewaidy, H.; Nakamori, S.; Rodriguez, J.; Manning, W.J.; Nezafat, R. Radiomic Analysis of Myocardial Native T(1) Imaging Discriminates Between Hypertensive Heart Disease and Hypertrophic Cardiomyopathy. JACC Cardiovasc. Imaging 2019, 12, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Baessler, B.; Luecke, C.; Lurz, J.; Klingel, K.; von Roeder, M.; de Waha, S.; Besler, C.; Maintz, D.; Gutberlet, M.; Thiele, H.; et al. Cardiac MRI Texture Analysis of T1 and T2 Maps in Patients with Infarctlike Acute Myocarditis. Radiology 2018, 289, 357–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baessler, B.; Luecke, C.; Lurz, J.; Klingel, K.; Das, A.; von Roeder, M.; De Waha-Thiele, S.; Besler, C.; Rommel, K.-P.; Maintz, D.; et al. Cardiac MRI and Texture Analysis of Myocardial T1 and T2 Maps in Myocarditis with Acute versus Chronic Symptoms of Heart Failure. Radiology 2019, 292, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Khened, V.M.A.; Krishnamurthi, G. Densely connected fully convolutional network for short-axis cardiac cine MR image segmentation and heart diagnosis using random forest. Lect. Notes Comput. Sci. 2018, 10663, 140–151. [Google Scholar]
- Ammar, A.; Bouattane, O.; Youssfi, M. Automatic cardiac cine MRI segmentation and heart disease classification. Comput. Med. Imaging. Graph. 2021, 88, 101864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, G.; Gao, Z.; Xu, C.; Zhang, Y.; Shi, R.; Keegan, J.; Xu, L.; Zhang, H.; Fan, Z.; et al. Deep Learning for Diagnosis of Chronic Myocardial Infarction on Nonenhanced Cardiac Cine MRI. Radiology 2019, 291, 606–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalakrishnan, V.; Menon, P.G.; Madan, S. cMRI-BED: A novel informatics framework for cardiac MRI biomarker extraction and discovery applied to pediatric cardiomyopathy classification. Biomed. Eng. Online 2015, 14, S7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantilla, J.G.M.; Bellanger, J.J.; Paredes, J.L. Machine learning techniques for LV wall motion classification based on Spatio-temporal profiles from cardiac cine MRI. In Proceedings of the 12th International Conference on Machine Learning Appl ICMLA, Miami, FL, USA, 4–7 December 2013; pp. 167–172. [Google Scholar]
- Lekadir, K.; Leiner, T.; Young, A.A.; Petersen, S.E. Editorial: Current and Future Role of Artificial Intelligence in Cardiac Imaging. Front. Cardiovasc. Med. 2020, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Rodriguez, J.; Martinez, F. Regional Multiscale Motion Representation for Cardiac Disease Prediction. In Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA). In Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA), Bucaramanga, Colombia, 24–26 April 2019; pp. 1–5. [Google Scholar]
- Afshin, M.; Ben Ayed, I.; Punithakumar, K.; Law, M.; Islam, A.; Goela, A.; Peters, T.; Li, S. Regional assessment of cardiac left ventricular myocardial function via MRI statistical features. IEEE Trans. Med. Imaging 2014, 33, 481–494. [Google Scholar] [CrossRef]
- Baessato, F.; Guglielmo, M.; Muscogiuri, G.; Baggiano, A.; Fusini, L.; Scafuri, S.; Babbaro, M.; Mollace, R.; Collevecchio, A.; Guaricci, A.I.; et al. Stress CMR in Known or Suspected CAD: Diagnostic and Prognostic Role. Biomed Res. Int. 2021, 2021, 6678029. [Google Scholar] [CrossRef] [PubMed]
- Baggiano, A.; Del Torto, A.; Guglielmo, M.; Muscogiuri, G.; Fusini, L.; Babbaro, M.; Collevecchio, A.; Mollace, R.; Scafuri, S.; Mushtaq, S.; et al. Role of CMR Mapping Techniques in Cardiac Hypertrophic Phenotype. Diagnostics 2020, 10, 770. [Google Scholar] [CrossRef] [PubMed]
- Peretto, G.; Barison, A.; Forleo, C.; Di Resta, C.; Esposito, A.; Aquaro, G.D.; Scardapane, A.; Palmisano, A.; Emdin, M.; Resta, N.; et al. Late gadolinium enhancement role in arrhythmic risk stratification of patients with LMNA cardiomyopathy: Results from a long-term follow-up multicentre study. Europace 2020, 22, 1864–1872. [Google Scholar] [CrossRef] [PubMed]
- Pontone, G.; Andreini, D.; Guaricci, A.I.; Guglielmo, M.; Baggiano, A.; Muscogiuri, G.; Fusini, L.; Fazzari, F.; Berzovini, C.; Pasquini, A.; et al. Association between Haptoglobin Phenotype and Microvascular Obstruction in Patients with STEMI: A Cardiac Magnetic Resonance Study. JACC Cardiovasc. Imaging 2019, 12, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Guaricci, A.I.; Carità, P.; Lorenzoni, V.; Casavecchia, G.; Rabbat, M.; Ieva, R.; Brunetti, N.D.; Andreini, D.; Di Biase, M.; Marenzi, G.; et al. QT-interval evaluation in primary percutaneous coronary intervention of ST-segment elevation myocardial infarction for prediction of myocardial salvage index. PLoS ONE 2018, 13, e0192220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontone, G.; Guaricci, A.I.; Andreini, D.; Ferro, G.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Muscogiuri, G.; Lorenzoni, V.; Mushtaq, S.; et al. Prognostic Stratification of Patients With ST-Segment-Elevation Myocardial Infarction (PROSPECT): A Cardiac Magnetic Resonance Study. Circ. Cardiovasc. Imaging 2017, 10, e006428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontone, G.; Andreini, D.; Guaricci, A.I.; Rota, C.; Guglielmo, M.; Mushtaq, S.; Baggiano, A.; Beltrama, V.; Fusini, L.; Solbiati, A.; et al. The STRATEGY Study (Stress Cardiac Magnetic Resonance Versus Computed Tomography Coronary Angiography for the Management of Symptomatic Revascularized Patients): Resources and Outcomes Impact. Circ. Cardiovasc. Imaging 2016, 9, e005171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontone, G.; Andreini, D.; Bertella, E.; Loguercio, M.; Guglielmo, M.; Baggiano, A.; Aquaro, G.D.; Mushtaq, S.; Salerni, S.; Gripari, P.; et al. Prognostic value of dipyridamole stress cardiac magnetic resonance in patients with known or suspected coronary artery disease: A mid-term follow-up study. Eur. Radiol. 2016, 26, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Fang, M.; Cui, C.; Chen, X.; Yin, G.; Prasad, S.K.; Dong, D.; Tian, J.; Zhao, S. LGE-CMR-derived texture features reflect poor prognosis in hypertrophic cardiomyopathy patients with systolic dysfunction: Preliminary results. Eur. Radiol. 2018, 28, 4615–4624. [Google Scholar] [CrossRef] [PubMed]
- Ambale-Venkatesh, B.; Yang, X.; Wu, C.O.; Liu, K.; Hundley, W.G.; McClelland, R.; Gomes, A.S.; Folsom, A.R.; Shea, S.; Guallar, E.; et al. Cardiovascular Event Prediction by Machine Learning: The Multi-Ethnic Study of Atherosclerosis. Circ. Res. 2017, 121, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Kotu, L.P.; Engan, K.; Borhani, R.; Katsaggelos, A.K.; Ørn, S.; Woie, L.; Eftestøl, T. Cardiac magnetic resonance image-based classification of the risk of arrhythmias in post-myocardial infarction patients. Artif. Intell. Med. 2015, 64, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Peressutti, D.; Sinclair, M.; Bai, W.; Jackson, T.; Ruijsink, J.; Nordsletten, D.; Asner, L.; Hadjicharalambous, M.; Rinaldi, C.; Rueckert, D.; et al. A framework for combining a motion atlas with non-motion information to learn clinically useful biomarkers: Application to cardiac resynchronisation therapy response prediction. Med. Image Anal. 2017, 35, 669–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tat, E.; Bhatt, D.L.; Rabbat, M.G. Addressing bias: Artificial intelligence in cardiovascular medicine. Lancet Digit. Health. 2020, 2, e635–e636. [Google Scholar] [CrossRef] [PubMed]







| Method | Image Substrate | Application | |
|---|---|---|---|
| Muscogiuri et al. (2021) [32] | DL | 2D multisegment late gadolinium enhancement | Noise reduction |
| Forman et al. (2015) [33] | CS | Free-breathing whole-heart coronary MRA | Reduction of respiratory motion artifacts |
| Forman et al. (2014) [34] | CS | High-resolution 3D whole-heart coronary MRA | Shortening of acquisition time |
| Schemper et al. (2018) [27] | CNN | Cine | Automatic reconstruction |
| Frick et al. (2011) [36] | ML | CMR imaging | Automatic view planning |
| Yokoyama et al. (2015) [37] | ML | CMR imaging | Automatic slice alignment method |
| Nitta et al. (2013) [38] | ML | CMR imaging | Automatic slice alignment method |
| Oktay et al. (2017) [39] | ML | Cine | Localization of anatomical landmarks |
| Lu et al. (2011) [40] | ML | CMR imaging | Automatic view planning |
| Blansit et al. (2019) [41] | DL | CMR imaging | Localizaion of anatomical landmarks |
| Lebet et al. (2020) [42] | CNN | CMR imaging | Improvement of image quality |
| Van Der Velde et al. (2021) [43] | DL | LGE | Improvement of image quality |
| Hauptmann et al. (2019) [44] | CNN | CMR imaging | Shortening of reconstruction time and improvement of image quality |
| Sandino et al. (2021) [45] | DL | Cine | Shortening of reconstruction time and improvement of image quality |
| Kustner et al. (2020) [46] | DL | Cine | Shortening of reconstruction time and improvement of image quality |
| Method | Image Substrate | Application | |
|---|---|---|---|
| Romaguera et al. (2018) [53] | CNN | CMR imaging | Ventricular segmentation |
| Bernard et al. (2018) [54] | DL | CMR imaging | Ventricular segmentation |
| Bai et al. (2018) [55] | DL | CMR imaging | Ventricular segmentation |
| Penso et al. (2021) [56] | DL | CMR imaging | Ventricular segmentation |
| Xiong et al. (2019) [57] | CNN | LGE | Atrial segmentation |
| Yang et al. (2018) [58] | DL | LGE | Atrial scar segmentation |
| Zabihollahy et al. (2019) [59] | DL | LGE | Myocardial scar segmentation |
| Moccia et al. (2019) [60] | DL | LGE | Myocardial scar segmentation |
| Xu et al. (2018) [61] | CNN | Cine | Myocardial infarction area segmentation |
| Author | Method | Image Substrate | Application |
|---|---|---|---|
| Fahmy et al. (2018) [66] | CNN | LGE | Segmentation and quantification of scar volume in patients with HCM |
| Hann et al. (2018) [70] | DL | T1 mapping | Automated LV segmentation of T1 maps in order to speed up LGE quantification based on T1 mapping |
| Thornhill et al. (2014) [79] | Radiomics and TA | LGE | Detection of myocardial fibrosis in patients with HCM |
| Schofield et al. (2019) [75] | Radiomics and TA | Cine | Differentiation among several causes of myocardial hypertrophy (HCM, amyloid, and aortic stenosis) and healthy controls |
| Engan et al. (2010) [76] | Radiomics and TA | LGE | Discrimination of patients with low and high risk of arrhythmias |
| Kotu et al. (2013) [77] | Radiomics and TA | LGE | Automated segmentation of scarred tissue areas |
| Larroza et al. (2017) [78] | Radiomics and TA | LGE, Cine | Differential diagnosis between acute and chronic infarction |
| Neisius et al. (2019) [80] | Radiomics and TA | Native T1 mapping | Discrimination between hypertrophic cardiomyopathy and hypertensive heart disease |
| Baessler, et al. (Radiology 2018 Nov) [81] | Radiomics and TA | Native T1–T2 mapping | Diagnostic accuracy in acute infarct-like myocarditis |
| Baessler, et al. (Radiology 2018 Jan) [82] | Radiomics and TA | Native T1–T2 mapping | Diagnostic accuracy in chronic myocardial inflammation/myocarditis |
| Author | Method | Image Substrate | Myocardial Disease |
|---|---|---|---|
| Khened et al. (2018) [83] | CNN | Cine | HCM, DCM, MI, and ARVC |
| Ammar et al. (2021) [84] | CNN | Cine | HCM, DCM, MI, and ARVC |
| Neisius et al. (2019) [80] | Radiomics and TA | Native T1 maps | Discrimination between HCM and hypertensive heart disease |
| Baessler et al. (Radiology 2018 Jan) [74] | Radiomics and TA | Cine | Differentiation of chronic from subacute MI |
| Zhang et al. (2019) [85] | DL | Cine | Chronic MI |
| Gopalakrishnan et al. (2015) [86] | ML | HCM, DCM, ARVC, LVNC, and myocarditis | |
| Wolterink et al. (2018) [62] | RF | Cine | Healthy, HCM, DCM, ARVC, and MI |
| Snaauw et al. (2019) [63] | CNN | Healthy, HCM, DCM, ARVC, and MI | |
| Baessler et al. (Radiology 2019) [82] | Radiomics and TA | Native T1–T2 mapping | Acute or chronic heart failure-like myocarditis |
| Mantilla et al. (2013) [87] | ML | Cine | Abnormal wall motion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Argentiero, A.; Muscogiuri, G.; Rabbat, M.G.; Martini, C.; Soldato, N.; Basile, P.; Baggiano, A.; Mushtaq, S.; Fusini, L.; Mancini, M.E.; et al. The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review. J. Clin. Med. 2022, 11, 2866. https://doi.org/10.3390/jcm11102866
Argentiero A, Muscogiuri G, Rabbat MG, Martini C, Soldato N, Basile P, Baggiano A, Mushtaq S, Fusini L, Mancini ME, et al. The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review. Journal of Clinical Medicine. 2022; 11(10):2866. https://doi.org/10.3390/jcm11102866
Chicago/Turabian StyleArgentiero, Adriana, Giuseppe Muscogiuri, Mark G. Rabbat, Chiara Martini, Nicolò Soldato, Paolo Basile, Andrea Baggiano, Saima Mushtaq, Laura Fusini, Maria Elisabetta Mancini, and et al. 2022. "The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review" Journal of Clinical Medicine 11, no. 10: 2866. https://doi.org/10.3390/jcm11102866
APA StyleArgentiero, A., Muscogiuri, G., Rabbat, M. G., Martini, C., Soldato, N., Basile, P., Baggiano, A., Mushtaq, S., Fusini, L., Mancini, M. E., Gaibazzi, N., Santobuono, V. E., Sironi, S., Pontone, G., & Guaricci, A. I. (2022). The Applications of Artificial Intelligence in Cardiovascular Magnetic Resonance—A Comprehensive Review. Journal of Clinical Medicine, 11(10), 2866. https://doi.org/10.3390/jcm11102866













