Exosomal MicroRNA Analyses in Esophageal Squamous Cell Carcinoma Cell Lines
Abstract
:1. Introduction
2. Methods
2.1. Sample Preparation
2.1.1. Cell Culture
2.1.2. Patient Blood Sampling
2.2. Exosome Isolation
2.2.1. ExoDisc Method for Cell Lines
2.2.2. ExoQuick Method for the Plasma
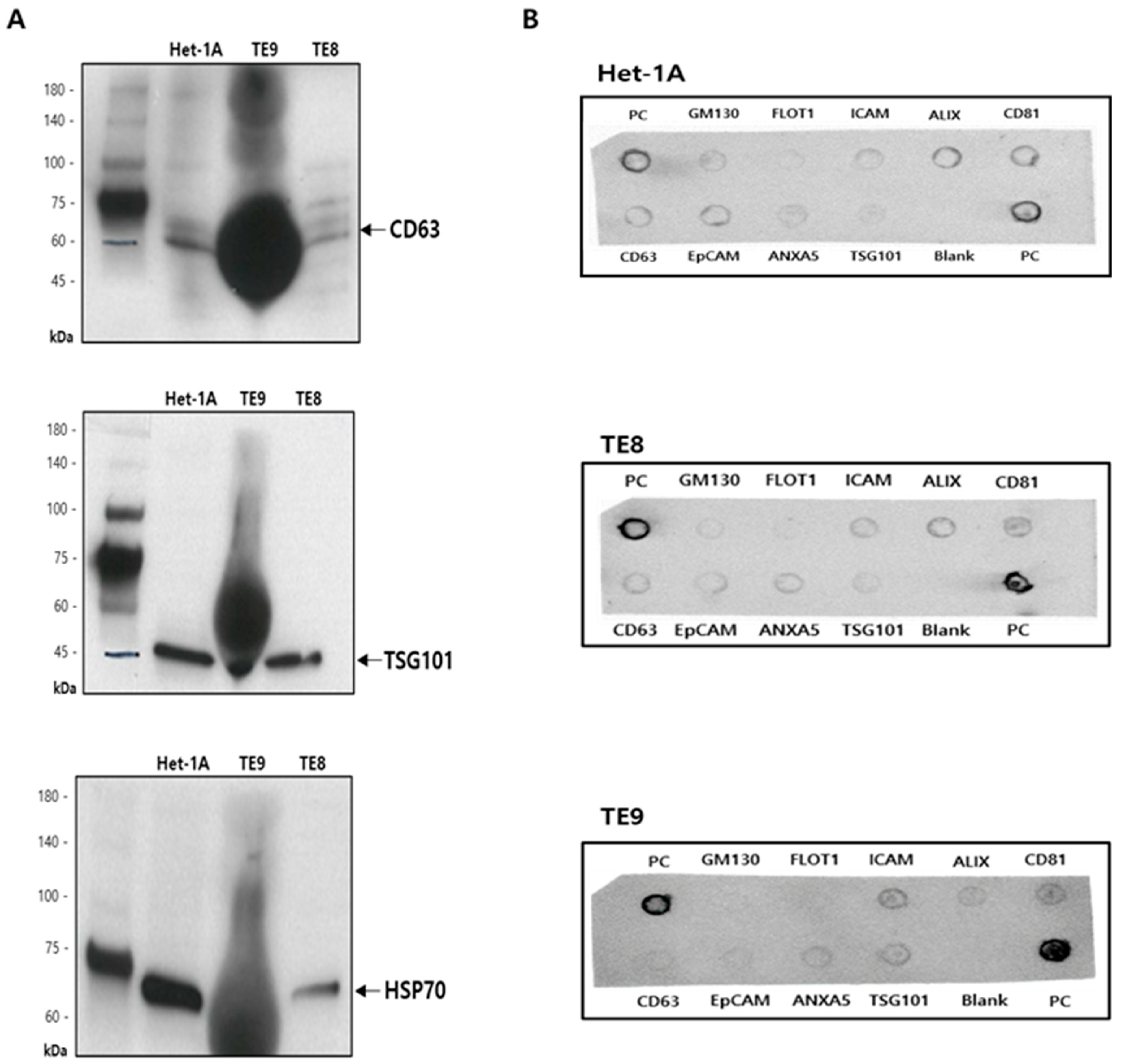
2.3. Characterization of Exosomes
2.3.1. Transmission Electron Microscopy
2.3.2. Western Blotting
2.3.3. Exo-Check Exosome Antibody Array
2.4. RNA Sequencing
2.5. Quantitative Reverse Transcription-Polymerase Chain Reaction
2.6. Bioinformatics Analysis
2.6.1. Differential miRNA Expression
2.6.2. Hierarchical Clustering
2.7. Statistical Analysis
3. Results
3.1. Isolation and Characterization of Exosomes
3.2. Analysis of Diffrentially Expressed Exosomal miRNAs in Esophageal Squamous Cell Carcinoma Cell Lines
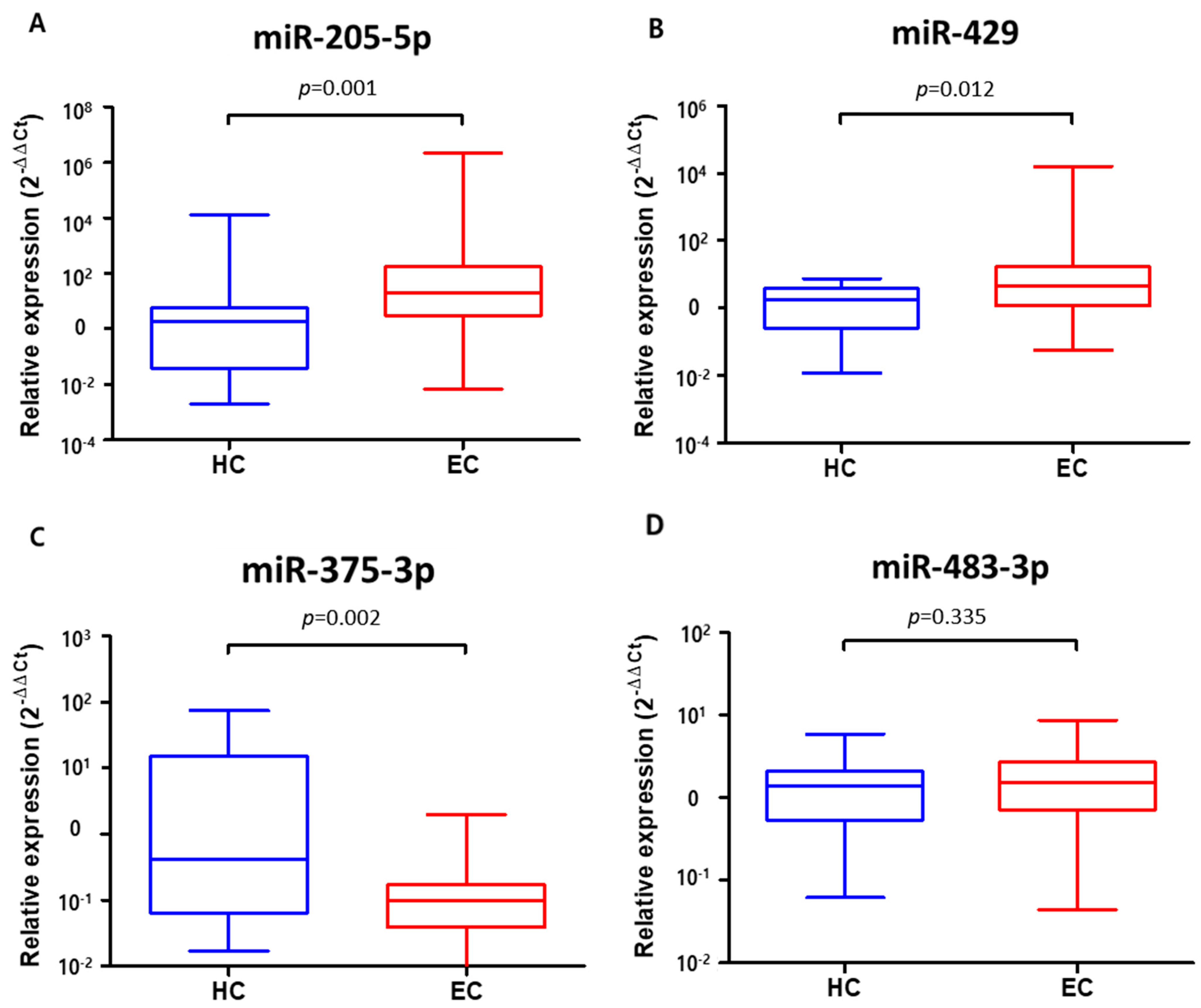
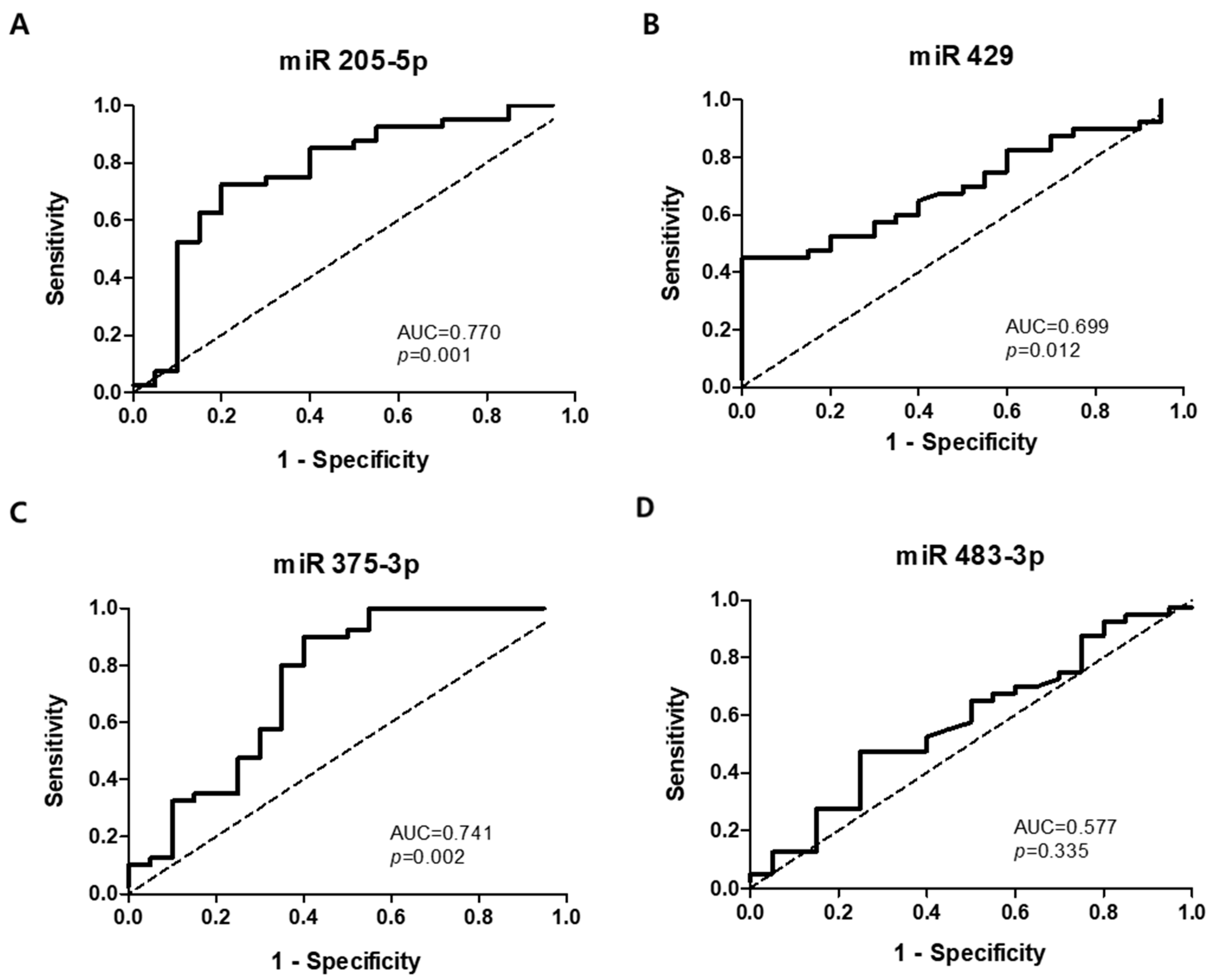
3.3. Verification of Target Exosomal miRNAs in Patients with Esophageal Squamous Cell Carcinoma
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Lin, D.C.; Hao, J.J.; Nagata, Y.; Xu, L.; Shang, L.; Meng, X.; Sato, Y.; Okuno, Y.; Varela, A.M.; Ding, L.W.; et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 467–473. [Google Scholar] [CrossRef]
- Holmes, R.S.; Vaughan, T.L. Epidemiology and pathogenesis of esophageal cancer. Semin. Radiat. Oncol. 2007, 17, 2–9. [Google Scholar] [CrossRef]
- Jung, H.-K. Epidemiology of and risk factors for esophageal cancer in Korea. Korean J. Helicobacter Up. Gastrointest. Res. 2019, 19, 145–148. [Google Scholar] [CrossRef] [Green Version]
- Codipilly, D.C.; Qin, Y.; Dawsey, S.M.; Kisiel, J.; Topazian, M.; Ahlquist, D.; Iyer, P.G. Screening for esophageal squamous cell carcinoma: Recent advances. Gastrointest. Endosc. 2018, 88, 413–426. [Google Scholar] [CrossRef]
- Qi, Y.; Chiu, J.F.; Wang, L.; Kwong, D.L.; He, Q.Y. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics 2005, 5, 2960–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukaszewicz-Zajac, M.; Mroczko, B.; Kozlowski, M.; Niklinski, J.; Laudanski, J.; Szmitkowski, M. Higher importance of interleukin 6 than classic tumor markers (carcinoembryonic antigen and squamous cell cancer antigen) in the diagnosis of esophageal cancer patients. Dis. Esophagus 2012, 25, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.K.; Kim, G.H.; Hoseok, I.; Park, S.J.; Lee, M.W.; Lee, B.E.; Park, D.Y.; Cho, Y.K. Circulating tumor cells detected using fluid-assisted separation technique in esophageal squamous cell carcinoma. J. Gastroenterol. Hepatol. 2019, 34, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Michela, B. Liquid biopsy: A family of possible diagnostic tools. Diagnostics 2021, 11, 1391. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Hur, J.Y.; Choi, Y.L.; Lee, C.H.; Kim, W.S. Current status and future perspectives of liquid biopsy in non-small cell lung cancer. J. Pathol. Transl. Med. 2020, 54, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Bu, H.; He, D.; He, X.; Wang, K. Exosomes: Isolation, analysis, and applications in cancer detection and therapy. Chembiochem 2019, 20, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Zheng, X.; Guo, H.; Xue, X.; Zhang, Y.; Niu, M.; Cui, J.; Liu, H.; Luo, H.; Yang, D.; et al. Serum exosomal miR-941 as a promising oncogenic biomarker for laryngeal squamous cell carcinoma. J. Cancer 2020, 11, 5329–5344. [Google Scholar] [CrossRef] [PubMed]
- Cano, A.; Diaz-Lopez, A.; Moreno-Bueno, G. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag. Res. 2014, 6, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Slack, F.J. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 2011, 11, 849–864. [Google Scholar] [CrossRef] [Green Version]
- Hannafon, B.; Ding, W.-Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-K. Extracellular microRNAs as biomarkers in human disease. Chonnam Med. J. 2015, 51, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Peng, F.; Chen, J. The role of exosomal microRNAs in the tumor microenvironment of breast cancer. Int. J. Mol. Sci. 2019, 20, 3884. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Liu, R.; Yin, L.H.; Pu, Y.P. Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by solexa high-throughput sequencing. Int. J. Mol. Sci. 2014, 15, 15530–15551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamal, N.N.S.B.N.M.; Shahidan, W.N.S. Non-exosomal and exosomal circulatory microRNAs: Which are more valid as biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.Y.; Yuan, T.Z.; Tschannen, M.; Sun, Z.F.; Jacob, H.; Du, M.J.; Liang, M.H.; Dittmar, R.L.; Liu, Y.; Liang, M.Y.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.R.; Sun, D.L.; Ma, L.J.; Deng, Y.R.; Zhang, S.; Dong, L.; Chen, S. Exosomes isolated from CAPS1-overexpressing colorectal cancer cells promote cell migration. Oncol. Rep. 2019, 42, 2528–2536. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. MiR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase C epsilon. Cancer Res. 2009, 69, 2287–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.N.; Hu, K.; Gong, G.H.; Zhu, D.; Wang, Y.X.; Liu, H.L.; Wu, X.Y. Upregulation of miR-205 transcriptionally suppresses SMAD4 and PTEN and contributes to human ovarian cancer progression. Sci. Rep. 2017, 7, 41330. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Zhang, P.; Hu, G.; Xiao, Z.; Xu, F.; Zhong, T.; Huang, F.; Kuang, H.; Zhang, W. Relative expressions of miR-205-5p, miR-205-3p, and miR-21 in tissues and serum of non-small cell lung cancer patients. Mol. Cell Biochem. 2013, 383, 67–75. [Google Scholar] [CrossRef]
- Nagai, H.; Hasegawa, S.; Uchida, F.; Terabe, T.; Kanno, N.I.; Kato, K.; Yamagata, K.; Sakai, S.; Kawashiri, S.; Sato, H.; et al. MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP-2 expression. Int. J. Oncol. 2018, 52, 841–850. [Google Scholar] [CrossRef]
- Ibuki, Y.; Nishiyama, Y.; Tsutani, Y.; Emi, M.; Hamai, Y.; Okada, M.; Tahara, H. Circulating microRNA/isomiRs as novel biomarkers of esophageal squamous cell carcinoma. PLoS ONE 2020, 15, e0231116. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, B.B.; Lu, M.; Zheng, M.J.; Chen, H.; Ding, J.Z.; Xu, A.M.; Xu, Y.H. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. OncoTargets Ther. 2016, 9, 1123–1133. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.T.; Zhao, Q.; Zhou, J.; Shi, R. MiR-429 mediates tumor growth and metastasis in colorectal cancer. Am. J. Cancer Res. 2017, 7, 218. [Google Scholar] [PubMed]
- Wang, H.W.; Hou, L.J.; Li, A.H.; Duan, Y.X.; Gao, H.L.; Song, X.W. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed. Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Zong, M.Z.; Liu, Y.; Zhang, K.; Yi, J.; Chen, L.B. The effects of miR-429 on cell migration and invasion by targeting Slug in esophageal squamous cell carcinoma. Pathol. Res. Pract. 2019, 215, 152526. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, X.J.; Zhou, W.; Chu, Y.X. MicroRNA-429 inhibits the proliferation and migration of esophageal squamous cell carcinoma cells by targeting RAB23 through the NF-kappa B pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.Y.; Wang, H.D.; Yao, X.X.; Zhang, D.; Xie, Y.J.; Cui, R.J.; Zhang, X.W. Circulating microRNAs in cancer: Potential and challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.W.; Lin, J.S.; He, X.X. The emerging role of miR-375 in cancer. Int. J. Cancer 2014, 135, 1011–1018. [Google Scholar] [CrossRef]
- Kong, K.L.; Kwong, D.L.W.; Chan, T.H.-M.; Law, S.Y.-K.; Chen, L.; Li, Y.; Qin, Y.-R.; Guan, X.-Y. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012, 61, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Morimura, R.; Nagata, H.; Kosuga, T.; Iitaka, D.; Konishi, H.; Shiozaki, A.; et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2011, 105, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Yang, W.L.; Ma, J.J.; Zhang, H.W.; Li, Z.; Zhang, L.; Liu, J.Q.; Han, Z.Y.; Wang, H.; Hong, L. Role of miR-483 in digestive tract cancers: From basic research to clinical value. J. Cancer 2018, 9, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Zhao, L.J.; Tan, Y.X.; Ren, H.; Qi, Z.T. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J. Gastroenterol. 2012, 18, 5442–5453. [Google Scholar] [CrossRef]


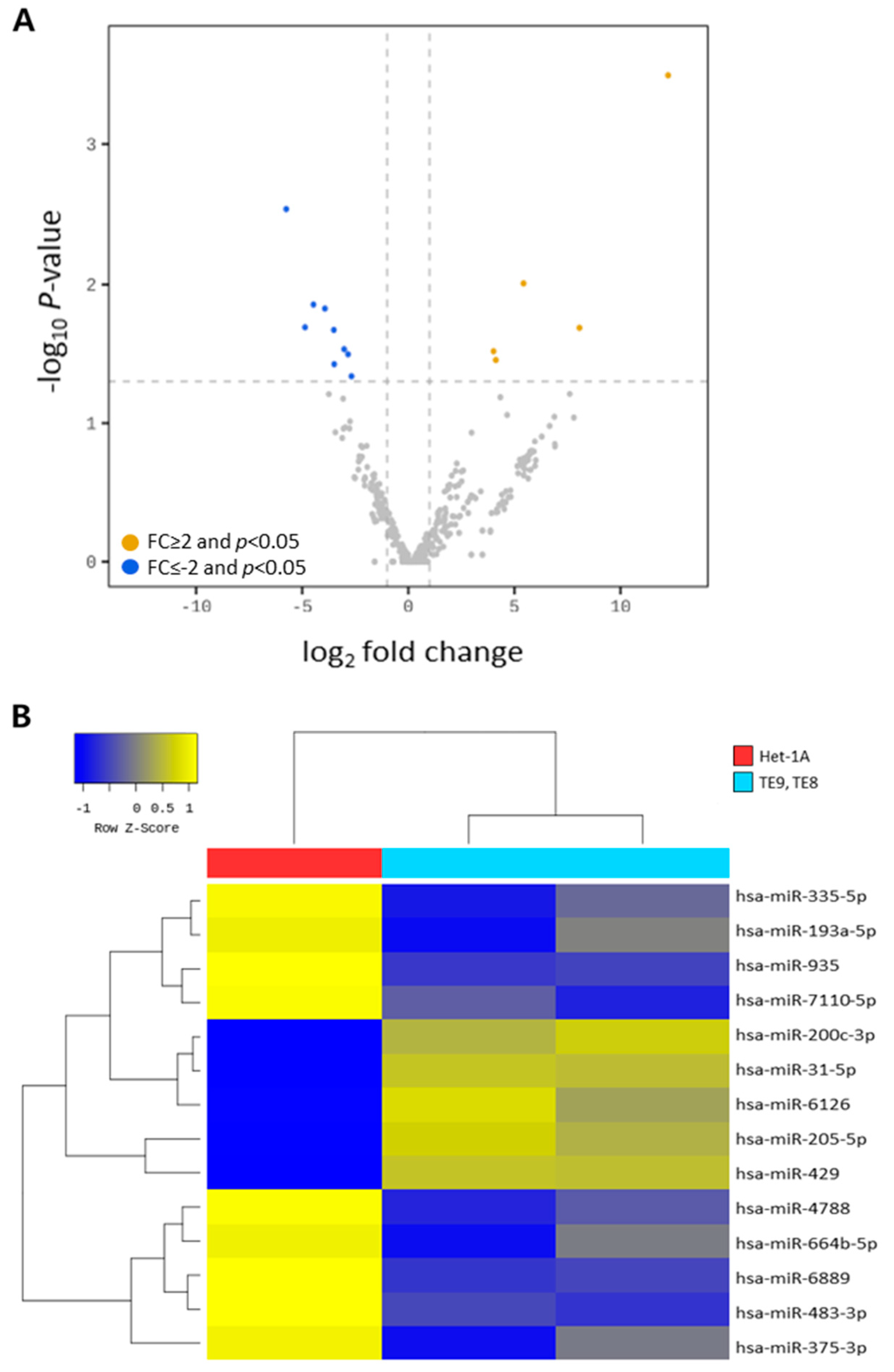
| Mature miRNA | Family | Sequence | Fold Change | p-Value | Expression Level |
|---|---|---|---|---|---|
| hsa-miR-205-5p | hsa-mir-205 | UCCUUCAUUCCACCGGAGUCUG | 4843.079 | <0.001 | Up |
| hsa-miR-429 | hsa-mir-429 | UAAUACUGUCUGGUAAAACCGU | 267.564 | 0.020 | Up |
| hsa-miR-6126 | hsa-mir-6126 | GUGAAGGCCCGGCGGAGA | 43.004 | 0.009 | Up |
| hsa-miR-200c-3p | hsa-mir-200c | UAAUACUGCCGGGUAAUGAUGGA | 17.440 | 0.035 | Up |
| hsa-miR-31-5p | hsa-mir-31 | AGGCAAGAUGCUGGCAUAGCU | 16.186 | 0.030 | Up |
| hsa-miR-193a-5p | hsa-mir-193a | UGGGUCUUUGCGGGCGAGAUGA | −6.395 | 0.045 | Down |
| hsa-miR-335-5p | hsa-mir-335 | UCAAGAGCAAUAACGAAAAAUGU | −7.156 | 0.031 | Down |
| hsa-miR-935 | hsa-mir-935 | CCAGUUACCGCUUCCGCUACCGC | −8.137 | 0.029 | Down |
| hsa-miR-664b-5p | hsa-mir-664b | UGGGCUAAGGGAGAUGAUUGGGUA | −11.285 | 0.037 | Down |
| hsa-miR-7110-5p | hsa-mir-7110 | UGGGGGUGUGGGGAGAGAGAG | −11.416 | 0.021 | Down |
| hsa-miR-4788 | hsa-mir-4788 | UUACGGACCAGCUAAGGGAGGC | −15.234 | 0.014 | Down |
| hsa-miR-6089 | hsa-mir-6089-1 hsa-mir-6089-2 | GGAGGCCGGGGUGGGGCGGGGCGG | −22.213 | 0.013 | Down |
| hsa-miR-375-3p | hsa-mir-375 | UUUGUUCGUUCGGCUCGCGUGA | −29.156 | 0.020 | Down |
| hsa-miR-483-3p | hsa-mir-483 | UCACUCCUCUCCUCCCGUCUU | −53.918 | 0.002 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Kim, G.H.; Park, S.J.; Kwon, C.H.; I, H.; Lee, M.W.; Lee, B.E. Exosomal MicroRNA Analyses in Esophageal Squamous Cell Carcinoma Cell Lines. J. Clin. Med. 2022, 11, 4426. https://doi.org/10.3390/jcm11154426
Kim S, Kim GH, Park SJ, Kwon CH, I H, Lee MW, Lee BE. Exosomal MicroRNA Analyses in Esophageal Squamous Cell Carcinoma Cell Lines. Journal of Clinical Medicine. 2022; 11(15):4426. https://doi.org/10.3390/jcm11154426
Chicago/Turabian StyleKim, Sora, Gwang Ha Kim, Su Jin Park, Chae Hwa Kwon, Hoseok I, Moon Won Lee, and Bong Eun Lee. 2022. "Exosomal MicroRNA Analyses in Esophageal Squamous Cell Carcinoma Cell Lines" Journal of Clinical Medicine 11, no. 15: 4426. https://doi.org/10.3390/jcm11154426
APA StyleKim, S., Kim, G. H., Park, S. J., Kwon, C. H., I, H., Lee, M. W., & Lee, B. E. (2022). Exosomal MicroRNA Analyses in Esophageal Squamous Cell Carcinoma Cell Lines. Journal of Clinical Medicine, 11(15), 4426. https://doi.org/10.3390/jcm11154426







