The Role of TSHR, PTEN and RASSF1A Promoters’ Methylation Status for Non-Invasive Detection of Papillary Thyroid Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. DNA Samples
2.3. DNA Extraction
2.4. Bisulfite Conversion
2.5. Quantitative Methylation-Specific PCR
2.6. Statistical Analysis
3. Results
3.1. Study Population
3.2. DNA Methylation in Thyroid Cancer Tissue and Adjacent Normal Tissue Groups
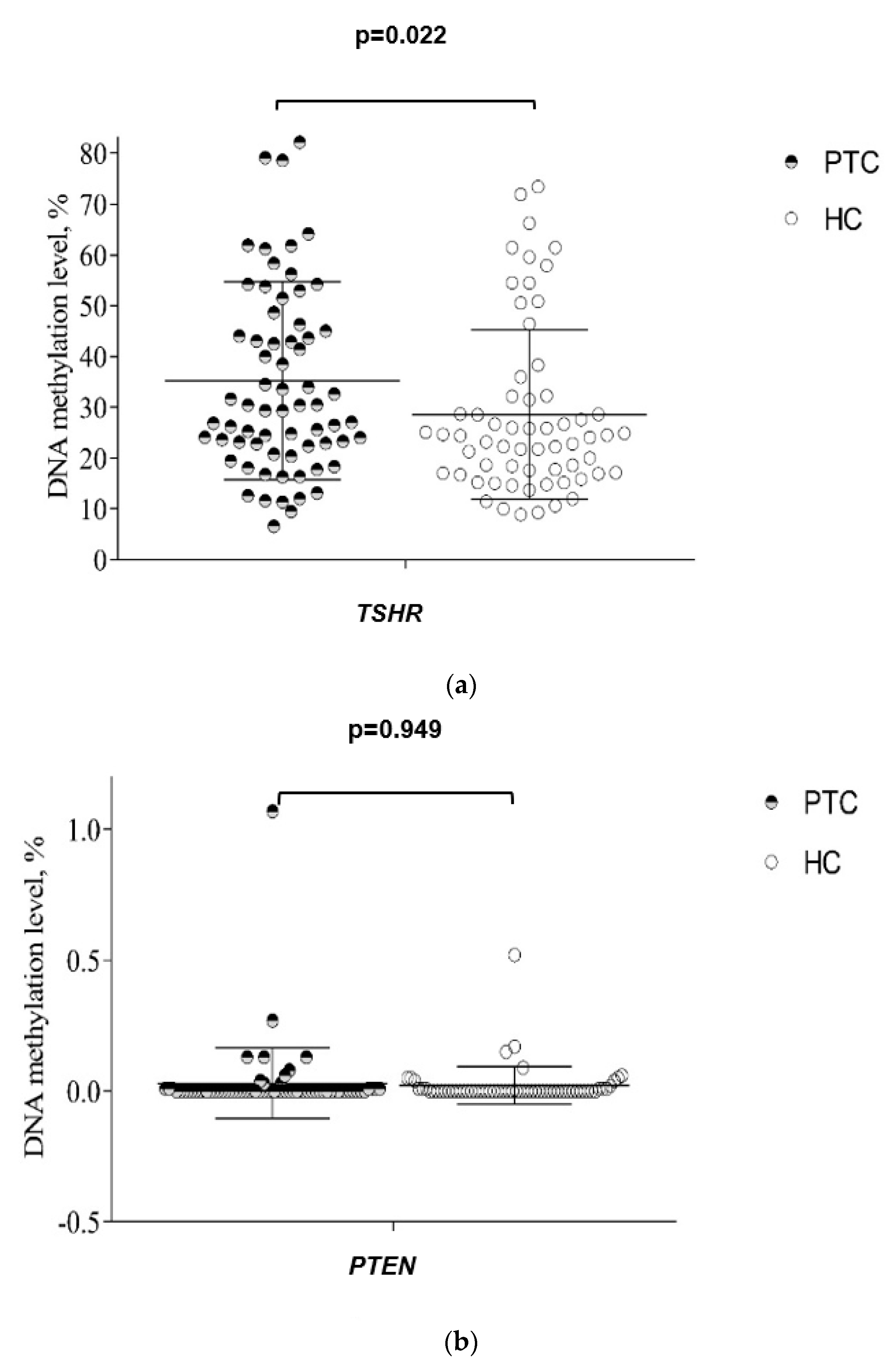
3.3. DNA Methylation in Plasma from PTC and HC Groups
3.4. DNA Methylation in Plasma PTC Patients before and after Surgery
3.5. Association of DNA Methylation Level in Plasma with Clinicopathological Features of PTC
3.6. The Diagnostic Value of Plasma TSHR, PTEN, and RASSF1A Methylation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Limaiem, F.; Rehman, A.; Mazzoni, T. Papillary Thyroid Carcinoma; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Fugazzola, L.; Elisei, R.; Fuhrer, D.; Jarzab, B.; Leboulleux, S.; Newbold, K.; Smit, J. 2019 European Thyroid Association Guidelines for the Treatment and Follow-Up of Advanced Radioiodine-Refractory Thyroid Cancer. Eur. Thyroid J. 2019, 8, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Miyauchi, A.; Kihara, M.; Fukushima, M.; Higashiyama, T.; Miya, A. Overall Survival of Papillary Thyroid Carcinoma Patients: A Single-Institution Long-Term Follow-Up of 5897 Patients. World J. Surg. 2018, 42, 615–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medas, F.; Canu, G.L.; Boi, F.; Lai, M.L.; Erdas, E.; Calo, P.G. Predictive Factors of Recurrence in Patients with Differentiated Thyroid Carcinoma: A Retrospective Analysis on 579 Patients. Cancers 2019, 11, 1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalska, A.; Walczyk, A.; Kowalik, A.; Palyga, I.; Trybek, T.; Kopczynski, J.; Kajor, M.; Chrapek, M.; Pieciak, L.; Chlopek, M.; et al. Increase in Papillary Thyroid Cancer Incidence Is Accompanied by Changes in the Frequency of the BRAF V600E Mutation: A Single-Institution Study. Thyroid 2016, 26, 543–551. [Google Scholar] [CrossRef]
- Lewinski, A.; Adamczewski, Z. Papillary thyroid carcinoma: A cancer with an extremely diverse genetic background and prognosis. Pol. Arch. Intern. Med. 2017, 127, 388–389. [Google Scholar] [CrossRef] [Green Version]
- Ward, L.S.; Kloos, R.T. Molecular markers in the diagnosis of thyroid nodules. Arq. Bras. Endocrinol. Metabol. 2013, 57, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Alsina, J.; Alsina, R.; Gulec, S. A Concise Atlas of Thyroid Cancer Next-Generation Sequencing Panel ThyroSeq v.2. Mol. Imaging Radionucl. Ther. 2017, 26, 102–117. [Google Scholar] [CrossRef]
- Mon, S.Y.; Riedlinger, G.; Abbott, C.E.; Seethala, R.; Ohori, N.P.; Nikiforova, M.N.; Nikiforov, Y.E.; Hodak, S.P. Cancer risk and clinicopathological characteristics of thyroid nodules harboring thyroid-stimulating hormone receptor gene mutations. Diagn. Cytopathol. 2018, 46, 369–377. [Google Scholar] [CrossRef]
- Chu, Y.D.; Yeh, C.T. The Molecular Function and Clinical Role of Thyroid Stimulating Hormone Receptor in Cancer Cells. Cells 2020, 9, 1730. [Google Scholar] [CrossRef]
- Kartal, K.; Onder, S.; Kosemehmetoglu, K.; Kilickap, S.; Tezel, Y.G.; Kaynaroglu, V. Methylation status of TSHr in well-differentiated thyroid cancer by using cytologic material. BMC Cancer 2015, 15, 824. [Google Scholar] [CrossRef] [Green Version]
- Qu, M.; Wan, S.; Ren, B.; Wu, H.; Liu, L.; Shen, H. Association between TSHR gene methylation and papillary thyroid cancer: A meta-analysis. Endocrine 2020, 69, 508–515. [Google Scholar] [CrossRef]
- Beg, S.; Siraj, A.K.; Jehan, Z.; Prabakaran, S.; Al-Sobhi, S.S.; Al-Dawish, M.; Al-Dayel, F.; Al-Kuraya, K.S. PTEN loss is associated with follicular variant of Middle Eastern papillary thyroid carcinoma. Br. J. Cancer 2015, 112, 1938–1943. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.M.; Cheng, F.; Teng, L.S. The association between phosphatase and tensin homolog hypermethylation and patients with breast cancer, a meta-analysis and literature review. Sci. Rep. 2016, 6, 32723. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Lee, M.H.; Kim, D.W.; Lee, S.; Huang, S.; Ryu, M.J.; Kim, Y.K.; Kim, S.J.; Kim, S.J.; Hwang, J.H. Cross-regulation between oncogenic BRAFV600E kinase and the MST1 pathway in papillary thyroid Carcinoma. PLoS ONE 2011, 6, e16180. [Google Scholar] [CrossRef] [Green Version]
- Khatami, F.; Larijani, B.; Heshmat, R.; Nasiri, S.; Haddadi-Aghdam, M.; Teimoori-Toolabi, L.; Tavangar, S.M. Hypermethylated RASSF1 and SLC5A8 promoters alongside BRAF(V600E) mutation as biomarkers for papillary thyroid carcinoma. J. Cell. Physiol. 2020, 235, 6954–6968. [Google Scholar] [CrossRef]
- Shou, F.; Xu, F.; Li, G.; Zhao, Z.; Mao, Y.; Yang, F.; Wang, H.; Guo, H. RASSF1A promoter methylation is associated with increased risk of thyroid cancer: A meta-analysis. Onco Targets Ther. 2017, 10, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Tuttle, R.M.; Haugen, B.; Perrier, N.D. Updated American Joint Committee on Cancer/Tumor-Node-Metastasis Staging System for Differentiated and Anaplastic Thyroid Cancer (Eighth Edition): What Changed and Why? Thyroid 2017, 27, 751–756. [Google Scholar] [CrossRef]
- Hou, P.; Ji, M.; Xing, M. Association of PTEN gene methylation with genetic alterations in the phosphatidylinositol 3-kinase/AKT signaling pathway in thyroid tumors. Cancer 2008, 113, 2440–2447. [Google Scholar] [CrossRef]
- Rogeri, C.D.; Silveira, H.C.S.; Causin, R.L.; Villa, L.L.; Stein, M.D.; de Carvalho, A.C.; Arantes, L.M.R.B.; Scapulatempo-Neto, C.; Possati-Resende, J.C.; Antoniazzi, M. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol. Oncol. 2018, 150, 545–551. [Google Scholar] [CrossRef]
- Lehmann, U.; Langer, F.; Feist, H.; Glockner, S.; Hasemeier, B.; Kreipe, H. Quantitative assessment of promoter hypermethylation during breast cancer development. Am. J. Pathol. 2002, 160, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.A.; Fan, C.Y.; Zou, C.; Bodenner, D.; Kokoska, M.S. Methylation status of genes in papillary thyroid carcinoma. Arch. Otolaryngol. Head Neck Surg. 2007, 133, 1006–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Li, T.; Yang, W.; Chai, X.; Chen, K.; Wei, L.; Duan, S.; Li, B.; Qin, Y. Analysis of DNA methylation in plasma for monitoring hepatocarcinogenesis. Genet. Test. Mol. Biomark. 2015, 19, 295–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schotten, L.M.; Darwiche, K.; Seweryn, M.; Yildiz, V.; Kneuertz, P.J.; Eberhardt, W.E.E.; Eisenmann, S.; Welter, S.; Sisson, B.E.; Pietrzak, M.; et al. DNA methylation of PTGER4 in peripheral blood plasma helps to distinguish between lung cancer, benign pulmonary nodules and chronic obstructive pulmonary disease patients. Eur. J. Cancer 2021, 147, 142–150. [Google Scholar] [CrossRef]
- Miller, B.F.; Petrykowska, H.M.; Elnitski, L. Assessing ZNF154 methylation in patient plasma as a multicancer marker in liquid biopsies from colon, liver, ovarian and pancreatic cancer patients. Sci. Rep. 2021, 11, 221. [Google Scholar] [CrossRef]
- Ceolin, L.; Goularte, A.P.P.; Ferreira, C.V.; Romitti, M.; Maia, A.L. Global DNA methylation profile in medullary thyroid cancer patients. Exp. Mol. Pathol. 2018, 105, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wu, Y.; Wang, Z.; Li, Y.; Wang, J.; Shao, G.; Yang, Y.; Shi, B. Diagnostic significance of DNA methylation of PTEN and DAPK in thyroid tumors. Clin. Endocrinol. 2020, 93, 187–195. [Google Scholar] [CrossRef]
- Stephen, J.K.; Chen, K.M.; Merritt, J.; Chitale, D.; Divine, G.; Worsham, M.J. Methylation markers differentiate thyroid cancer from benign nodules. J. Endocrinol. Investig. 2018, 41, 163–170. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, K. Relationship between methylation of TSHR and NIS gene promoter regions and clinicopathological characteristics in thyroid papillary carcinoma. Shandong Med. J. 2017, 57, 83–85. [Google Scholar] [CrossRef]
- Zheng, C.; Lu, X.; Ling, Z.; Ge, M. Methylation of TSHR gene promoter in papillary thyroid carcinoma and its clinical significance. J. Chin. Oncol. 2017, 23, 257–261. [Google Scholar] [CrossRef]
- Liu, T.; Men, Q.; Su, X.; Chen, W.; Zou, L.; Li, Q.; Song, M.; Ouyang, D.; Chen, Y.; Li, Z.; et al. Downregulated expression of TSHR is associated with distant metastasis in thyroid cancer. Oncol. Lett. 2017, 14, 7506–7512. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Cai, D.; Chen, H.; Zhang, H.; Zhang, Z.; Li, J. The relevance between the promoter hypermethylation of tshr and p16 gene and clinicopathological parameters in human papillary thyroid carcinoma. J. Cap. Med. Univ. 2012, 33, 361–365. [Google Scholar]
- Mohammadi-asl, J.; Larijani, B.; Khorgami, Z.; Tavangar, S.M.; Haghpanah, V.; Kheirollahi, M.; Mehdipour, P. Qualitative and quantitative promoter hypermethylation patterns of the P16, TSHR, RASSF1A and RARβ2 genes in papillary thyroid carcinoma. Med. Oncol. 2011, 28, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, C.; Liu, J.; Tang, X.; Li, Z. DNA methylation alterations as therapeutic prospects in thyroid cancer. J. Endocrinol. Investig. 2019, 42, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Yang, J.; Yang, K.; Huang, Y. The relationship between RASSF1A promoter methylation and thyroid carcinoma: A meta-analysis of 14 articles and a bioinformatics of 2 databases (PRISMA). Medicine 2017, 96, e8630. [Google Scholar] [CrossRef]
- Brait, M.; Loyo, M.; Rosenbaum, E.; Ostrow, K.L.; Markova, A.; Papagerakis, S.; Zahurak, M.; Goodman, S.M.; Zeiger, M.; Sidransky, D.; et al. Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics 2012, 7, 710–719. [Google Scholar] [CrossRef] [Green Version]
- Qu, F.; Xue, W. RASSF1A methylation and its clinical roles in papillary thyroid carcinoma. J. Nantong Univ. (Med. Sci.) 2012, 32, 490–492. [Google Scholar]





| Characteristic | PTC n = 68 | HC n = 86 | p-Value |
|---|---|---|---|
| Gender | |||
| Male | 8 (11.8%) | 11 (12.79%) | |
| Female | 60 (88.2%) | 75 (87.2%) | |
| Age at initial surgery (years) | 48.19 (14.9) | 45.30 (12.07) | p = 0.221 |
| T (TNM), n (%) | - | - | |
| pT1a | 27 (39.7) | ||
| pT1b | 7 (10.3) | ||
| pT2 | 4 (5.9) | ||
| pT3a | 19 (27.9) | ||
| pT3b | 11 (16.2) | ||
| Tumor size (cm) | - | - | |
| ≤2 | 48 (70.6) | ||
| >2 | 20 (29.4) | ||
| Lymph node metastases at initial surgery | - | - | |
| Yes | 16 (23.5) | ||
| No | 52 (76.5) | ||
| Variant of PTC, n (%) | - | - | |
| The classical variant | 29 (42.6) | ||
| The follicular variant | 18 (26.5) | ||
| The diffuse sclerosing variant | 17 (25.0) | ||
| The tall cell carcinoma | 4 (5.9) | ||
| Extrathyroidal extension | - | - | |
| Yes | 30 (44.1) | ||
| No | 38 (55.9) | ||
| Lymphovascular invasion | - | - | |
| Yes | 36 (52.9) | ||
| No | 32 (47.1) | ||
| Multifocality | - | - | |
| Yes | 16 (23.5) | ||
| No | 52 (76.5) |
| Marker | Methylation Level: MEAN (SD) | p-Value | |
|---|---|---|---|
| Pre-Operative PTC (n = 68) | Post-Operative PTC (n = 62) | ||
| TSHR | 34.105 (17.790) | 26.901 (11.617) | 0.003 * |
| PTEN | 0.029 (0.134) | 0.003 (0.014) | 0.031 * |
| RASSF1A | 1.010 (3.248) | 0.816 (4.072) | 0.903 |
| Methylation Level: MEAN (Minimum–Maximum) | |||
|---|---|---|---|
| Characteristic | TSHR | PTEN | RASSF1A |
| 35.24 (6.60–100.00) | 0.02 (0.00–1.07) | 1.18 (0.00–36.74) | |
| % | % | % | |
| p-Value | p-Value | p-Value | |
| p-ValueGender | 0.505 | 0.456 | 0.059 |
| Male | 33.06 (11.35–78.60) | 0.01 (0.00–0.06) | 0.94 (0.00–6.66) |
| Female | 35.54 (6.60–100.00) | 0.03 (0.00–1.07) | 1.02 (0.00–21.74) |
| Age at initial surgery (years) | 0.599 | 0.204 | 0.061 |
| ≤55 years | 35.95 (11.63–100.00) | 0.04 (0.00–1.07) | 0.79 (0.00–21.74) |
| >55 years | 33.95 (6.60–79.13) | 0.01 (0.00–0.13) | 1.41 (0.00–9.36) |
| pT (TNM) | 0.422 | 0.148 | 0.627 |
| pT1 | 37.15 (6.60–100.00) | 0.04 (0.00–1.07) | 1.35 (0.00–21.74) |
| pT2–3 | 33.22 (11.35–79.13) | 0.01 (0.00–0.13) | 0.65 (0.00–7.10) |
| Tumor size (cm) | <0.001 * | 0.743 | 0.164 |
| ≤ 2 | 28.84 (6.60–58.32) | 0.04 (0.00–1.07) | 1.28 (0.00–21.74) |
| > 2 | 50.62 (12.04–100.00) | 0.01 (0.00–0.13) | 0.36 (0.00–7.22) |
| Lymph node metastases at initial surgery | 0.010 * | 0.368 | 0.487 |
| Yes | 47.01 (12.62–82.19) | 0.04 (0.00–0.27) | 1.04 (0.00–9.36) |
| No | 31.62 (6.60–100.00) | 0.03 (0.00–1.07) | 1.00 (0.00–21.74) |
| Variant of PTC | 0.300 | 0.791 | 0.824 |
| Aggressive histology of PTC | 31.32 (12.62–64.15) | 0.01 (0.00–0.08) | 0.95 (0.00–9.36) |
| Non-aggressive subtypes of PTC | 37.12 (6.60–100.00) | 0.04 (0.00–1.07) | 1.04 (0.00–21.74) |
| Extrathyroidal extension | 0.621 | 0.608 | 0.501 |
| Yes | 38.12 (13.13–100.00) | 0.05 (0.00–1.07) | 0.65 (0.00–7.22) |
| No | 32.97 (6.60–78.60) | 0.01 (0.00–0.27) | 1.24 (0.00–21.74) |
| Lymphovascular invasion | 0.020 * | 0.726 | 0.578 |
| Yes | 40.96 (12.62–100.00) | 0.04 (0.00–1.07) | 0.72 (0.00–7.22) |
| No | 28.81 (6.60–61.79) | 0.02 (0.00–0.27) | 0.42 (0.00–21.74) |
| Multifocality | 0.013 * | 0.094 | 0.437 |
| Yes | 47.19 (13.13–100.00) | 0.08 (0.00–0.27) | 0.86 (0.00–7.10) |
| No | 31.57 (6.60–82.19) | 0.01 (0.00–1.07) | 1.06 (0.00–21.74) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimaitė, R.; Kazokaitė, M.; Kondrotienė, A.; Daukšienė, D.; Sabaliauskaitė, R.; Žukauskaitė, K.; Žilaitienė, B.; Jarmalaitė, S.; Daukša, A. The Role of TSHR, PTEN and RASSF1A Promoters’ Methylation Status for Non-Invasive Detection of Papillary Thyroid Carcinoma. J. Clin. Med. 2022, 11, 4917. https://doi.org/10.3390/jcm11164917
Klimaitė R, Kazokaitė M, Kondrotienė A, Daukšienė D, Sabaliauskaitė R, Žukauskaitė K, Žilaitienė B, Jarmalaitė S, Daukša A. The Role of TSHR, PTEN and RASSF1A Promoters’ Methylation Status for Non-Invasive Detection of Papillary Thyroid Carcinoma. Journal of Clinical Medicine. 2022; 11(16):4917. https://doi.org/10.3390/jcm11164917
Chicago/Turabian StyleKlimaitė, Raimonda, Mintautė Kazokaitė, Aistė Kondrotienė, Dalia Daukšienė, Rasa Sabaliauskaitė, Kristina Žukauskaitė, Birutė Žilaitienė, Sonata Jarmalaitė, and Albertas Daukša. 2022. "The Role of TSHR, PTEN and RASSF1A Promoters’ Methylation Status for Non-Invasive Detection of Papillary Thyroid Carcinoma" Journal of Clinical Medicine 11, no. 16: 4917. https://doi.org/10.3390/jcm11164917
APA StyleKlimaitė, R., Kazokaitė, M., Kondrotienė, A., Daukšienė, D., Sabaliauskaitė, R., Žukauskaitė, K., Žilaitienė, B., Jarmalaitė, S., & Daukša, A. (2022). The Role of TSHR, PTEN and RASSF1A Promoters’ Methylation Status for Non-Invasive Detection of Papillary Thyroid Carcinoma. Journal of Clinical Medicine, 11(16), 4917. https://doi.org/10.3390/jcm11164917






