Randomized Double-Blind Crossover Study for Evaluating a Probiotic Mixture on Gastrointestinal and Behavioral Symptoms of Autistic Children
Abstract
:1. Introduction
2. Methods and ASD Children Enrollment
2.1. Inclusion and Exclusion Criteria
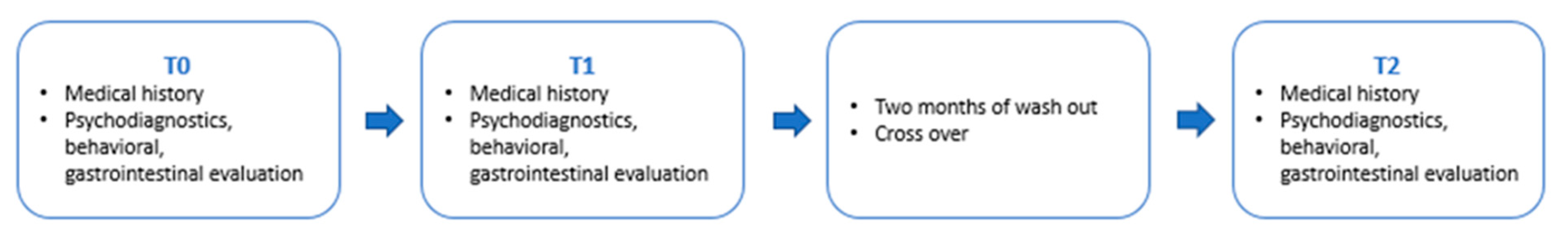
2.2. Study Design
2.3. Data Collection, Clinical Evaluations, and Probiotic/Placebo Administration at the Time Points
2.4. Statistical Method
- -
- The treatment effects;
- -
- The time of treatment administration effect among treatment sequences (T0, T1, and T2);
- -
- The carry-over (sequence) effect computed including an interaction term between time and treatment in the model;
- -
- A random effect term (random intercept) on the patient’s identification code, accounting for correlations among repeated measures of the same patient.
2.5. Fecal Sample DNA Extraction and Metagenome Sequencing
2.6. Bioinformatics Data Analysis
2.7. Data Quality Assessment and Filtering
2.8. Biodiversity Measurements
2.9. Biomarker’s Identification
3. Results
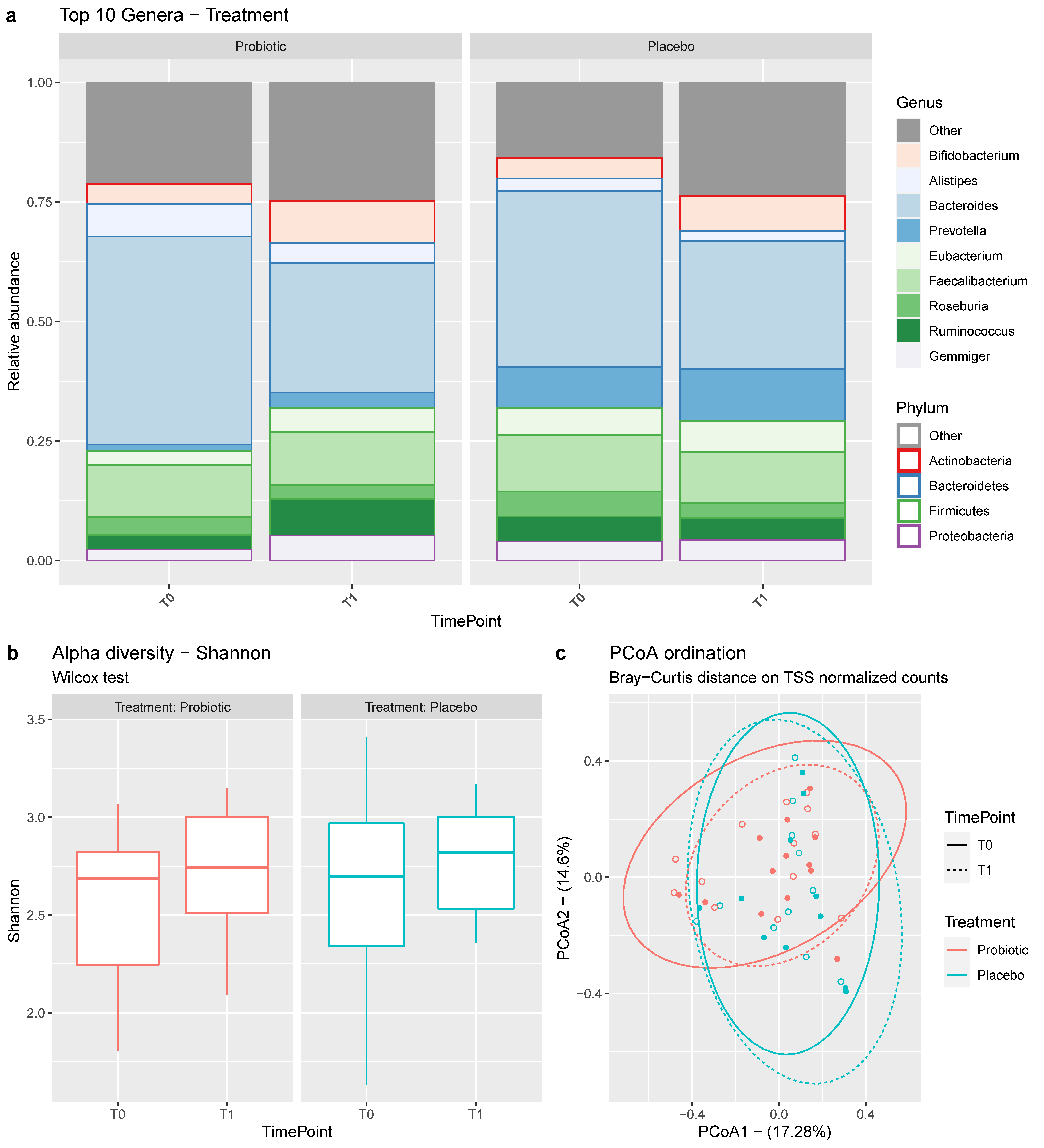
3.1. Microbial Composition of the Samples
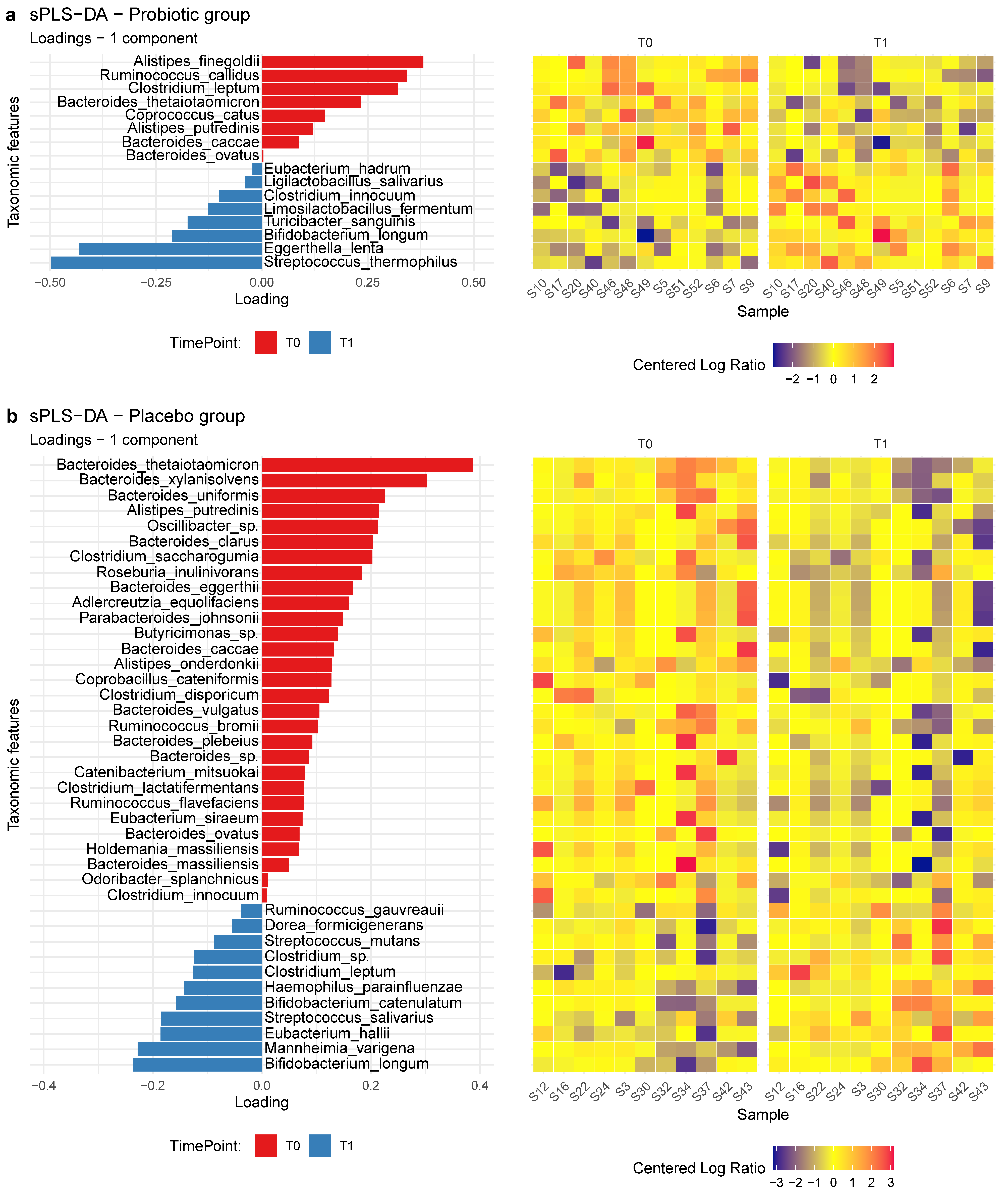
3.2. Biomarker’s Identification
3.3. Clinical Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- American Psychiatric Association. DSM-5 Task Force. In Diagnostic and Statistical Manual of Mental Disorders: DSM-5, 5th ed.; American Psychiatric Association: Arlington, MA, USA, 2013. [Google Scholar]
- Loomers, R.; Hull, L.; Mandy, W.P.L. What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 466–474. [Google Scholar]
- Brugha, T.S.; Spiers, N.; Bankart, J.; Cooper, S.-A.; McManus, S.; Gullon-Scott, F.; Smith, J.; Tyrer, F. epidemiology of autism in adults across age groups and ability levels. Br. J. Psychiatry 2016, 209, 498–503. [Google Scholar] [CrossRef]
- Elsabbagh, M.; Divan, G.; Koh, Y.-J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.J.; Brugha, T.; Erskine, H.; Scheurer, R.; Vos, T.; Scott, J. The epidemiology and global burden of autism spectrum disorders. Psychol. Med. 2015, 45, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Data & Statistics on Autism Spectrum Disorder. January 2019. Available online: https://www.cdc.gov/ncbddd/autism/data.html (accessed on 1 January 2020).
- Tordjman, S.; Somogyi, E.; Coulon, N.; Kermarrec, S.; Cohen, D.; Bronsard, G.; Bonnot, O.; Weismann-Arcache, C.; Botbol, M.; Lauth, B.; et al. Gene × Environment interactions in autism spectrum disorders: Role of epigenetic mechanisms. Front. Psychiatry 2014, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Bölte, S.B.; Girdler, S.; Marschik, P.B. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell. Mol. Life Sci. 2019, 76, 1275–1297. [Google Scholar] [CrossRef] [PubMed]
- Alibek, K.; Farmer, S.; Tskhay, A.; Moldakozhayev, A.; Alibek, K.; Isakov, T. The Role of Infection, Inflammation and Genetic Alterations in ASD Etiopathogenesis: A Review. J. Neurol. Psychiatr. Disord. 2019, 2, 105. [Google Scholar]
- Fakhoury, M. Autistic spectrum disorders: A review of clinical features, theories and diagnosis. Int. J. Dev. Neurosci. 2015, 43, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Jyonouchi, H.; Sun, S.; Itokazu, N. Innate immunity associated with inflammatory responses and cytokine production against common dietary proteins in patients with autism spectrum disorder. Neuropsychobiology 2002, 46, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Neale, B.M.; Kou, Y.; Liu, L.; Ma’ayan, A.; Samocha, K.E.; Sabo, A.; Lin, C.-F.; Stevens, C.; Wang, L.-S.; Makarov, V.; et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012, 485, 242–245. [Google Scholar] [CrossRef]
- Newschaffer, C.J.; Croen, L.A.; Daniels, J.; Giarelli, E.; Grether, J.K.; Levy, S.E.; Mandell, D.S.; Miller, L.A.; Pinto-Martin, J.; Reaven, J.; et al. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health 2007, 28, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, D.A.; Frye, R.E. A review of research trends in physiological abnormalities in autism spectrum disorders: Immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and enviromental toxicant exposures. Mol. Psychiatry 2012, 17, 389–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voung, H.E.; Hsiao, E.Y. Emerging roles for the gut microbiome in autism spectrum disorder. Biol Psychiatry. 2017, 81, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.; Hill, R.J.; Ziviani, J.; Dodrill, P. Features of feeding difficulty in children with Autism Spectrum Disorder. Int. J. Speech. Lang. Pathol. 2014, 16, 151–158. [Google Scholar] [CrossRef]
- Devnani, P.A.; Hegde, A.U. Autism and sleep disorders. J. Pediatr. Neurosci. 2015, 10, 304–307. [Google Scholar] [CrossRef]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Margolis, K.G.; Beversdorf, D.Q.; et al. Association of Rigid-Compulsive Behavior with Functional Constipation in Autism Spectrum Disorder. J. Autism Dev. Disord. 2017, 47, 1673–1681. [Google Scholar] [CrossRef]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Mayer, E.A.; Padua, D.; Tillisch, K. Altered brain-gut axis in autism: Comorbidity or causative machanisms? Bioessays 2014, 36, 933–939. [Google Scholar] [CrossRef]
- Iovene, M.R.; Bombace, F.; Maresca, R.; Sapone, A.; Iardino, P.; Picardi, A.; Marotta, R.; Schiraldi, C.; Siniscalco, D.; Serra, N.; et al. Intestinal Dysbiosis and Yeast isolation in stool of subjects with autism spectrum disorders. Mycopathologia 2017, 182, 349–363. [Google Scholar] [CrossRef]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Caetano, L.; Antunes, M.; Brett Finlay, B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.K.; Melmed, R.D.; Barstow, L.E.; Enriquez, F.J.; Ranger-Moore, J.; Ostrem, J.A. Oral human immunoglobulin for children with autism and gastrointestinal dysfunction: A prospective, open-label study. J. Autism Dev. Disord. 2006, 36, 1053–1064. [Google Scholar] [CrossRef]
- Umbrello, G.; Esposito, S. Microbiota and neurologic diseases: Potential effects of probiotics. J. Transl. Med. 2016, 14, 298. [Google Scholar] [CrossRef]
- Forsythe, P.; Sudo, N.; Dinan, T.; Taylor, V.H.; Bienenstock, J. Mood and gut feelings. Brain Behav. Immun. 2009, 24, 316–323. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Buie, T. Potential etiologic factors of microbiome disruption in autism. Clin. Ther. 2015, 37, 976–983. [Google Scholar] [CrossRef]
- Borre, Y.E.; Moloney, R.D.; Clarke, G.; Dinan, T.G.; Cyran, J.F. The impact of microbiota on brain and behavior: Machanisms &therapeutic potential. Adv. Exp. Med. Biol. 2014, 817, 373–403. [Google Scholar]
- Diaz Heijtz, R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin. Fetal. Neonatal. Med. 2016, 21, 410–417. [Google Scholar] [CrossRef]
- Srikantha, P.; Mohajeri, M.H. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Fond, G.; Boukouaci, W.; Chevalier, G.; Regnault, A.; Eberl, G.; Hamdani, N.; Dickerson, F.; Macgregor, A.; Boyer, L.; Dargel, A.; et al. The “psychomicrobiotic”: Targeting microbiota in major psychiatric disorders: A systematic review. Pathol. Biol. 2015, 63, 35–42. [Google Scholar] [CrossRef]
- Alonso, C.; Vicario, M.; Pigrau, M.; Lobo, B.; Santos, J. Intestinal barrier function and the brain-gut axis. Adv. Exp. Med. Biol. 2014, 817, 73–113. [Google Scholar] [PubMed]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shofey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Pop, M.; Deboy, R.; Eckburg, P.; Turnbaugh, P.; Samuel, B.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Carlson, A.L.; Xia, K.; Azcate-Peril, M.A.; Goldman, B.D.; Ahn, M.; Styner, M.A.; Thompson, A.L.; Geng, X.; Gilmore, J.H.; Knickmeyer, R.C. Infant gut microbiome associated with cognitive development. Biol. Psychiatry 2018, 83, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Borre, Y.; O’Keeffe, G.; Clarke, G.; Stanton, C.; Dinan, T.; Cryan, J. Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends Mol. Med. 2014, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, E.; Orsi, P.; Boso, M.; Broglia, D.; Brondino, N.; Barale, F.; di Nemi, S.U.; Politi, P. Low-grade endotoxemia in patients with severe autism. Neurosci. Lett. 2010, 471, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Kunze, W.A.; Bornstein, J.C.; Furness, J.B. Identification of sensory nerve cells in a peripheral organ (the intestine) of mammal. Neuroscience 1995, 66, 1–4. [Google Scholar] [CrossRef]
- Perez-Burgos, A.; Mao, Y.K.; Bienenstock, J.; Kunze, W.A. The gut-brain axis rewired: Adding a functional vagal nicotinic “sensory synapse”. FASEB J. 2014, 28, 3064–3074. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Summanen, P.H.; Downes, J.; Corbett, K.; Komoriya, T. Detection of Clostridum perfringens toxin genes in the gut microbiota of autistic children. Anaerobe 2017, 45, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Fulceri, F.; Morelli, M.; Santocchi, E.; Cena, H.; Del Bianco, T.; Narzisi, S.; Calderoni, S.; Muratori, F. Gastrointestinal Symptoms and behavioral problems in preschoolers with autism spectrum disorder. Dig. Liver Dis. 2016, 48, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Buie, T.; Campbell, D.B.; Fuchs, G.J., 3rd; Furuta, G.T.; Levy, J.; Vandewater, J.; Whitaker, A.H.; Atkins, D.; Bauman, M.L.; Beaudet, A.L.; et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: A consensus report. Pediatrics 2010, 125 (Suppl. 1), S1–S18. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, X.; Li, J.; Li, F. Association between gut microbiota and autism spectrum disorder: A systematic review and meta-analysis. Front. Psychiatry 2019, 10, 473. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.A.; Savidge, T.C.; Williams, K.C. The Brain-Gut-Microbiome Axis: What Role Does it Play in Autism Spectrum Disorder? Curr. Dev. Disord. Rep. 2016, 3, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Fattorusso, A.; Di Genova, L.; Dell’isola, G.B.; Mencaroni, E.; Esposito, S. Autism spectrum disorders and the gut microbiota. Nutrients 2019, 11, 521. [Google Scholar] [CrossRef]
- Grimaldi, R.; Gibson, G.R.; Vulevic, J.; Giallourou, N.; Castro-Mejía, J.L.; Hansen, L.H.; Leigh Gibson, E.; Nielsen, D.S.; Costabile, A. A prebiotic intervention study in children with autism spectrum disorders (ASDs). Microbiome 2018, 6, 133. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Liu, Y.; Rhoads, J.M. Can probiotics benefit children autism spectrum disorders? World J Gastroenterol. 2016, 22, 10093–10102. [Google Scholar] [CrossRef]
- Van Minnen, L.P.; Timmerman, H.M.; Lutgendorff, F.; Verheem, A.; Harmsen, W.; Konstantinov, S.R.; Smidt, H.; Visser, M.R.; Rijkers, G.T.; Gooszen, H.G.; et al. SurModification of intestinal flora with multispecies probiotics reduces bacterial translocation and improves clinical course in a rat model of acute pancreatitis. Surgery 2007, 141, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Lutgendorff, F.; Akkermans, L.M.; Soderholm, J.D. The role of microbiota and probiotics in stress-induced gastrointestinal damage. Curr. Mol. Med. 2008, 8, 282–298. [Google Scholar] [CrossRef]
- Lutgendorff, F.; Nijmeijer, R.M.; Sandstrom, P.A.; Trulsson, L.M.; Magnusson, K.E.; Timmerman, H.M.; Van Minnen, L.P.; Rijkers, G.T.; Gooszen, H.G.; Akkermans, L.M.; et al. Probiotics prevent intestinal barrier dysfunction in acute pancreatitis in rats via induction of ileal mucosal glutathione biosynthesis. PLoS ONE 2009, 4, e4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caselli, M.; Cassol, F.; Calò, G.; Holton, J.; Zuliani, G.; Gasbarrini, A. Actual concept of “probiotics”: Is it more fuctionals to science or business? World J Gastroenterol. 2013, 19, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Cyran, J.F. Gut instincts: Microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Mental health: Thinking from the gut. Nature 2015, 518, S12–S15. [Google Scholar] [CrossRef] [PubMed]
- Shaaban, S.Y.; El Gendy, Y.G.; Mehanna, N.S.; El Senousy, W.M.; El-Feki, H.A.S.; Saad, K.; El-Asheer, O.M. The role of probiotics in children with autism spectrum disorder: A prospective, open-label study. Nutr. Neurosci. 2018, 21, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, S.S.; Balla, D.A.; Cicchetti, D.V. Vineland Adaptive Behavior Scales. American Guidance Service; Springer: Berlin, Germany, 1984. [Google Scholar]
- Abidin, R.R. Parenting Stress Index—Short Form, 3rd ed.; Psychological Assessment Resources Inc.: Odessa, FL, USA, 1995. [Google Scholar]
- Schopler, E.; Lansing, M.D.; Reichler, R.J.; Marcus, L.M. Pep3 Profilo Psicoeducativo, 3rd ed.; Vannini Editrice: Brescia, Italy, 2006. [Google Scholar]
- Goldstein, S.; Naglieri, J.A. ASRS. Autism Spectrum Rating Scales; Giunti, O.S., Ed.; MHS: Falls Church, VA, USA, 2014. [Google Scholar]
- R Development Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Harrell, F.E.J. rms: Regression Modeling Strategies; R Package Version 4.1-3; Springer Science & Business Media: Nashville, TN, USA, 2014. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar]
- Oksanen, J. Vegan: Community Ecology Package; R Package Version 2.6-2. 2022. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 12 July 2022).
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Costello, M.-E.; Lakis, V.A.; Bartolo, F.; Chua, X.-Y.; Brazeilles, R.; Rondeau, P. MixMC: A multivariate statistical framework to gain insight into microbial communities. PLoS ONE 2016, 11, e0160169. [Google Scholar]
- Higashida, H.; Furuhara, K.; Yamauchi, A.M.; Deguchi, K.; Harashima, A.; Munesue, S.; Lopatina, O.; Gerasimenko, M.; Salmina, A.B.; Zhang, J.S.; et al. Intestinal transepithelial permeability of oxytocin into the blood is dependent on the receptor for advanced glycation end products in mice. Sci. Rep. 2017, 7, 7883. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Lindenberg, A.; Domes, G.; Kirsch, P.; Heinrichs, M. Oxytocin and vasopressin in the human brain: Social neuropeptides for translational medicine. Nat. Rev. Neurosci. 2011, 12, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Shahrestani, S.; Kemp, A.H.; Guastella, A.J. The impact of a single administration of intranasal oxytocin on the recognition of basic emotions in humans: A meta-analysis. Neuropsychopharmacology 2013, 38, 1929–1936. [Google Scholar] [CrossRef]
- Sanctuary, M.R.; Kain, J.N.; Chen, S.Y.; Kalanetra, K.; Lemway, D.G.; Rose, D.R.; Yang, H.T.; Tancredi, D.J.; German, J.B.; Slupsky, C.M.; et al. Pilot study of probiotc/colostrum supplementation on gut function in children with autism and gastrointestinal symptoms. PLoS ONE 2019, 14, E0210064. [Google Scholar] [CrossRef] [PubMed]
- Kaluzna-Czaplinska, J.; Blaszczyk, S. The level of anabinitol in autistic children after probiotic therapy. Nutrition 2012, 28, 124–126. [Google Scholar] [CrossRef] [PubMed]
- West, R.; Roberts, E.; Sichel, L.S.; Sichel, J. Improvements in gastrointestinal symptoms among children with autism spectrum disorder receiving the Delpro Probiotic and immunomodulator formulation. J. Prob. Health 2013, 1, 102. [Google Scholar]
- Santocchi, E.; Guiducci, L.; Prosperi, M.; Calderoni, S.; Gaggini, M.; Apicella, F.; Tancredi, R.; Billeci, L.; Mastromarino, P.; Grossi, E.; et al. Effects of probiotic supplementation on gatrointestinal, sensory and core symptoms in autism spectrum disorders: A randomized controlled trial. Front. Psychiatry 2020, 11, 550593. [Google Scholar] [CrossRef] [PubMed]
- Siemann, J.K.; Muller, C.L.; Forsberg, C.G.; Blakely, R.D.; Veenstra-VanderWeele, J.; Wallace, M.T. An autism-associated serotonin transporter variant disrupts multisensory processing. Transl. Psychiatri. 2017, 7, e1067. [Google Scholar] [CrossRef]
- Parracho, H.M.; Gibson, G.R.; Knott, F.; Bosscher, D.; Kleerebezem, M.; McCartney, A.L. A double-blind, placebo-controlled, crossover-designed probiotic feeding study in children diagnosed with autistic spectrum disorders. Int. J. Probiotics Prebiotics 2010, 5, 69–74. [Google Scholar]
- Slykerman, R.F.; Kang, J.; Van Zyl, N.; Barthow, C.; Wickens, K.; Stanley, T.; Coomarasamy, C.; Purdie, G.; Murphy, R.; Crane, J.; et al. Effect of early probiotic supplementation on childhood cognition, behavior and mood a randomized, placebo-controlled trial. Acta Paediatr. 2018, 107, 2173–2178. [Google Scholar] [CrossRef]
- Jones, R.M.; Carberry, C.; Hamo, A.; Lord, C. Placebo-like response in absence of treatment in children with autism. Autism Res. 2017, 10, 1567–1572. [Google Scholar] [CrossRef] [PubMed]

| Variable | N | Probiotic Group | Placebo Group | Overall | p Value |
|---|---|---|---|---|---|
| - | (N = 30) | (N = 31) | (N = 61) | ||
| Age | 61 | 40.2/49.5/71.5 | 37.5/47.0/58.0 | 39.0/48.0/65.0 | 0.44 |
| Gender: 0 | 61 | 20% (6) | 16% (5) | 18% (11) | 0.694 |
| 1 | - | 80% (24) | 84% (26) | 82% (50) | |
| Childbirth: 0 | 61 | 70% (21) | 74% (23) | 72% (44) | 0.715 |
| 1 | - | 30% (9) | 26% (8) | 28% (17) | |
| Breastfeeding: 0 | 60 | 40% (12) | 70% (21) | 55% (33) | 0.063 |
| 1 | - | 23% (7) | 10% (3) | 17% (10) | |
| 2 | - | 37% (11) | 20% (6) | 28% (17) | |
| Intolerances: 0 | 61 | 90% (27) | 87% (27) | 89% (54) | 0.722 |
| 1 | - | 10% (3) | 13% (4) | 11% (7) | |
| Rehabilitation. therapy: 0 | 61 | 10% (3) | 6% (2) | 8% (5) | 0.614 |
| 1 | - | 90% (27) | 94% (29) | 92% (56) | |
| Ages. Diagnosis | 60 | 30.2/37.0/50.5 41.0+/−15.9 | 30.2/37.5/48.8 40.4+/−12.5 | 30.0/37.5/49.2 40.7+/−14.2 | 0.942 |
| Karyotype: 1 | 7 | 100% (5) | 100% (2) | 100% (7) | |
| 0 | - | 0% (0) | 0% (0) | 0% (0) | |
| X. fragile: 0 | 39 | 100% (17) | 95% (21) | 97% (38) | 0.373 |
| 1 | - | 0% (0) | 5% (1) | 3% (1) | |
| CGH. array: 0 | 45 | 90% (19) | 79% (19) | 84% (38) | 0.296 |
| 1 | - | 10% (2) | 21% (5) | 16% (7) | |
| Epilessia: 0 | 55 | 96% (25) | 100% (29) | 98% (54) | 0.286 |
| 1 | - | 4% (1) | 0% (0) | 2% (1) | |
| MRI: 0 | 45 | 12% (3) | 24% (5) | 18% (8) | 0.322 |
| 1 | - | 88% (21) | 76% (16) | 82% (37) | |
| AA.plasmatic.acids.organic.urinary: 0 | 48 | 5% (1) | 12% (3) | 8% (4) | 0.382 |
| 1 | - | 95% (21) | 88% (23) | 92% (44) | |
| Verbal: 0 | 61 | 53% (16) | 52% (16) | 52% (32) | 0.893 |
| 1 | - | 47% (14) | 48% (15) | 48% (29) | |
| Other diagnosis in. brothers sisters: 0 | 60 | 80% (24) | 83% (25) | 82% (49) | 0.739 |
| 1 | - | 20% (6) | 17% (5) | 18% (11) | |
| Pharmacological therapy: 0 | 61 | 90% (27) | 97% (30) | 93% (57) | 0.285 |
| 1 | - | 10% (3) | 3% (1) | 7% (4) |
| Panel A Child Absolute Value | |||
|---|---|---|---|
| Estimate (Treatment vs. Placebo) | Standard Error | p Value | |
| Diarrhea | −0.15 | 0.07 | 0.05 |
| Receptive language | 18 | 8.44 | 0.03 |
| Panel B Parent absolute value | |||
| Estimate (Treatment vs. placebo) | Standard Error | pvalue | |
| PSSI.mother | −18.9 | 8.75 | 0.03 |
| PSSI.father.DC | −15.42 | 7.21 | 0.04 |
| PSSI.father.total Stress | −18.9 | 7.68 | 0.01 |
| Panel C Child variation | |||
| Estimate (Treatment vs. placebo) | Standard Error | pvalue | |
| Abdominal pain | −0.28 | 0.15 | 0.05 |
| Communication (Vineland) | 11.3 | 4.6 | 0.01 |
| Disadaptive behaviors (PEP3) | 8.59 | 4.31 | 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guidetti, C.; Salvini, E.; Viri, M.; Deidda, F.; Amoruso, A.; Visciglia, A.; Drago, L.; Calgaro, M.; Vitulo, N.; Pane, M.; et al. Randomized Double-Blind Crossover Study for Evaluating a Probiotic Mixture on Gastrointestinal and Behavioral Symptoms of Autistic Children. J. Clin. Med. 2022, 11, 5263. https://doi.org/10.3390/jcm11185263
Guidetti C, Salvini E, Viri M, Deidda F, Amoruso A, Visciglia A, Drago L, Calgaro M, Vitulo N, Pane M, et al. Randomized Double-Blind Crossover Study for Evaluating a Probiotic Mixture on Gastrointestinal and Behavioral Symptoms of Autistic Children. Journal of Clinical Medicine. 2022; 11(18):5263. https://doi.org/10.3390/jcm11185263
Chicago/Turabian StyleGuidetti, Cristina, Elena Salvini, Maurizio Viri, Francesca Deidda, Angela Amoruso, Annalisa Visciglia, Lorenzo Drago, Matteo Calgaro, Nicola Vitulo, Marco Pane, and et al. 2022. "Randomized Double-Blind Crossover Study for Evaluating a Probiotic Mixture on Gastrointestinal and Behavioral Symptoms of Autistic Children" Journal of Clinical Medicine 11, no. 18: 5263. https://doi.org/10.3390/jcm11185263





