Assessment of Early Growth Response 1 in Tumor Suppression of Esophageal Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Real-Time PCR (qRT-PCR)
2.3. Determination of Pathological TNM Classification
2.4. Cell Culture
2.5. siRNA Knockdown
2.6. Western Blot Analysis
2.7. Migration Assay
2.8. Cell Viability Assay
2.9. Statistical Analysis
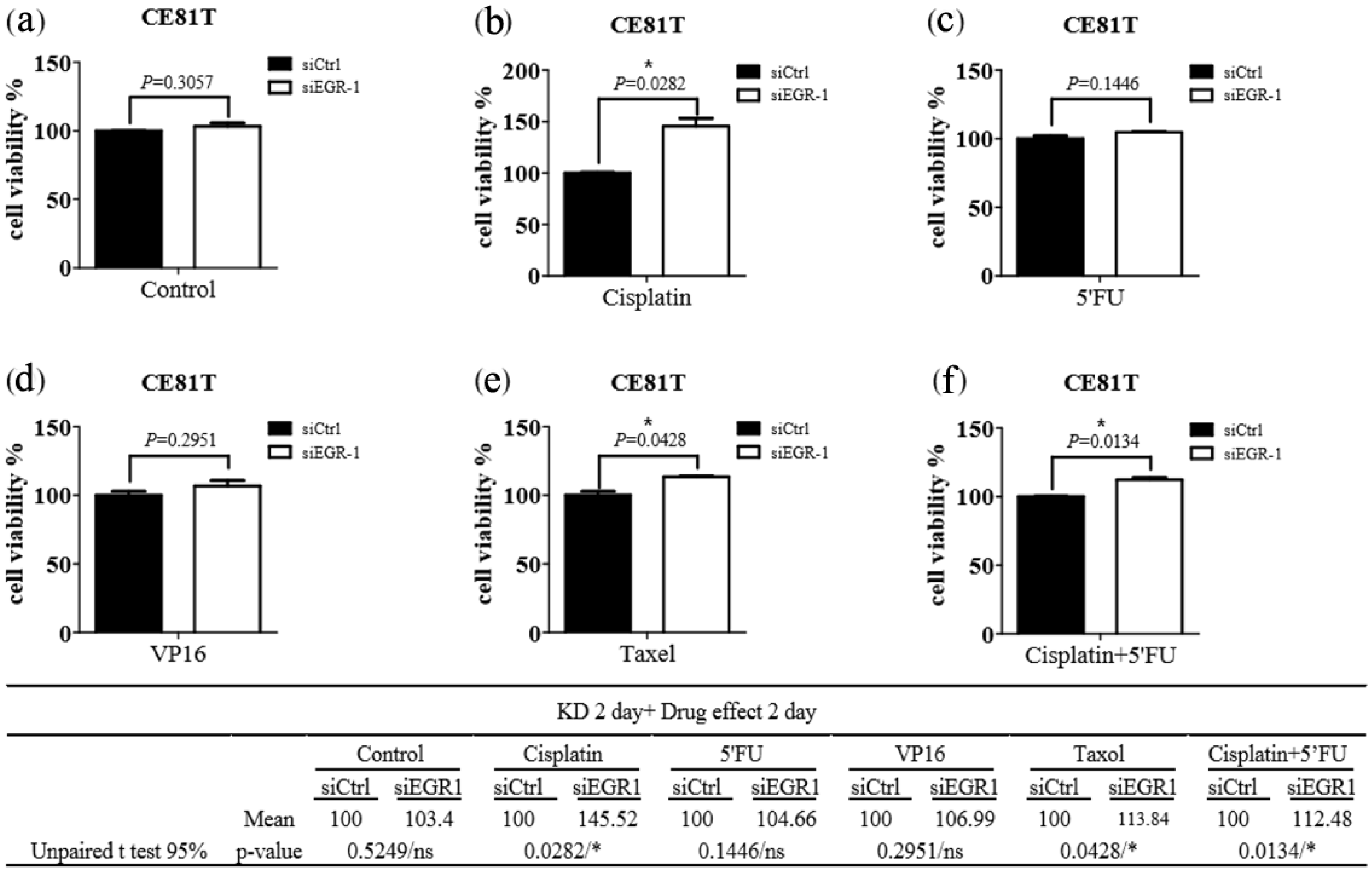
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Uhlenhopp, D.J.; Then, E.O.; Sunkara, T.; Gaduputi, V. Epidemiology of esophageal cancer: Update in global trends, etiology and risk factors. Clin. J. Gastroenterol. 2020, 13, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.K.; Li, A.F.; Wang, Y.C.; Hsieh, C.C.; Huang, M.H.; Hsu, W.H.; Hsu, H.S. Reduced membranous beta-catenin protein expression is associated with metastasis and poor prognosis in squamous cell carcinoma of the esophagus. J. Thorac. Cardiovasc. Surg. 2008, 135, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Krones-Herzig, A.; Mittal, S.; Yule, K.; Liang, H.; English, C.; Urcis, R.; Soni, T.; Adamson, E.D.; Mercola, D. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 2005, 65, 5133–5143. [Google Scholar] [CrossRef]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef]
- Calogero, A.; Arcella, A.; De Gregorio, G.; Porcellini, A.; Mercola, D.; Liu, C.; Lombari, V.; Zani, M.; Giannini, G.; Gagliardi, F.M.; et al. The early growth response gene EGR-1 behaves as a suppressor gene that is down-regulated independent of ARF/Mdm2 but not p53 alterations in fresh human gliomas. Clin. Cancer Res. 2001, 7, 2788–2796. [Google Scholar] [PubMed]
- Huang, R.P.; Fan, Y.; de Belle, I.; Niemeyer, C.; Gottardis, M.M.; Mercola, D.; Adamson, E.D. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int. J. Cancer 1997, 72, 102–109. [Google Scholar] [CrossRef]
- Levin, W.J.; Press, M.F.; Gaynor, R.B.; Sukhatme, V.P.; Boone, T.C.; Reissmann, P.T.; Figlin, R.A.; Holmes, E.C.; Souza, L.M.; Slamon, D.J. Expression patterns of immediate early transcription factors in human non-small cell lung cancer. The Lung Cancer Study Group. Oncogene 1995, 11, 1261–1269. [Google Scholar] [PubMed]
- Saha, S.K.; Islam, S.M.R.; Saha, T.; Nishat, A.; Biswas, P.K.; Gil, M.; Nkenyereye, L.; El-Sappagh, S.; Islam, M.S.; Cho, S.G. Prognostic role of EGR1 in breast cancer: A systematic review. BMB Rep. 2021, 54, 497–504. [Google Scholar] [CrossRef]
- Hsu, H.S.; Lin, M.H.; Jang, Y.H.; Kuo, T.T.; Liu, C.C.; Cheng, T.H. The 4E-BP1/eIF4E ratio is a determinant for rapamycin response in esophageal cancer cells. J. Thorac. Cardiovasc. Surg. 2015, 149, 378–385. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.Y.; Zhuang, C.X.; Yang, H.X.; Liang, Y.R. Expression of Egr-1, c-fos and cyclin D1 in esophageal cancer and its precursors: An immunohistochemical and in situ hybridization study. World J. Gastroenterol. 2004, 10, 476–480. [Google Scholar] [CrossRef]
- Wang, B.; Hendricks, D.T.; Wamunyokoli, F.; Parker, M.I. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer Res. 2006, 66, 3071–3077. [Google Scholar] [CrossRef]
- Peng, W.X.; Pan, F.Y.; Liu, X.J.; Ning, S.; Xu, N.; Meng, F.L.; Wang, Y.Q.; Li, C.J. Hypoxia stabilizes microtubule networks and decreases tumor cell chemosensitivity to anticancer drugs through Egr-1. Anat. Rec. 2010, 293, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xie, Z.; Wang, Z.; Cheng, K.; Liang, K.; Song, Z. Overexpression of miR-191 Predicts Poor Prognosis and Promotes Proliferation and Invasion in Esophageal Squamous Cell Carcinoma. Yonsei Med. J. 2017, 58, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, P.; Wang, S.; Cong, A.; Zhang, Q.; Shen, W.; Li, X.; Zhang, W.; Han, G. miRNA-181a-5p Enhances the Sensitivity of Cells to Cisplatin in Esophageal Adenocarcinoma by Targeting CBLB. Cancer Manag. Res. 2020, 12, 4981–4990. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ma, Q.; Shi, Y.; Li, X.; Wang, M.; Wang, J.; Ge, J.; Chen, Z.; Wang, Z.; Jiang, H. A novel 5-fluorouracil-resistant human esophageal squamous cell carcinoma cell line Eca-109/5-FU with significant drug resistance-related characteristics. Oncol. Rep. 2017, 37, 2942–2954. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Du, X.; Sun, Q.; Wang, Y.; Li, M.; Zang, W.; Liu, K.; Zhao, G. α-solanine enhances the chemosensitivity of esophageal cancer cells by inducing microRNA-138 expression. Oncol. Rep. 2018, 39, 1163–1172. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, X.; Zhang, F.; Mo, S.; Lu, Y.; Wei, W.; Chen, X.; Lan, L.; Lu, B.; Liu, Y. Paclitaxel induces apoptosis of esophageal squamous cell carcinoma cells by downregulating STAT3 phosphorylation at Ser727. Oncol. Rep. 2017, 37, 2237–2244. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, P.; Ma, Q.; Wang, D.; Zhou, T. Cisplatin-based chemoradiotherapy with 5-fluorouracil or pemetrexed in patients with locally advanced, unresectable esophageal squamous cell carcinoma: A retrospective analysis. Mol. Clin. Oncol. 2017, 6, 743–747. [Google Scholar] [CrossRef]
- Saito, T.; Hikita, M.; Kohno, K.; Sato, S.-I.; Takano, H.; Kobayashi, M. Different sensitivities of human esophageal cancer cells to multiple anti-cancer agents and related mechanisms. Cancer 1992, 70, 2402–2409. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, Q.; Liu, Y.; Bai, W.; Yao, Y.; Ding, J.; Lin, L.; Xu, Z.; Cai, Z.; Wang, S.; et al. TCF7L2 and EGR1 synergistic activation of transcription of LCN2 via an ERK1/2-dependent pathway in esophageal squamous cell carcinoma cells. Cell Signal. 2019, 55, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Chang, H.M.; Leung, P.C. Egr-1 mediates epidermal growth factor-induced downregulation of E-cadherin expression via Slug in human ovarian cancer cells. Oncogene 2013, 32, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Eid, M.A.; Kumar, M.V.; Iczkowski, K.A.; Bostwick, D.G.; Tindall, D.J. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998, 58, 2461–2468. [Google Scholar]
- Parra, E.; Ferreira, J. The effect of siRNA-Egr-1 and camptothecin on growth and chemosensitivity of breast cancer cell lines. Oncol. Rep. 2010, 23, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Abdulkadir, S.A.; Carbone, J.M.; Naughton, C.K.; Humphrey, P.A.; Catalona, W.J.; Milbrandt, J. Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum. Pathol. 2001, 32, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ameri, A.H.; Wang, S.; Jansson, K.H.; Casey, O.M.; Yang, Q.; Beshiri, M.L.; Fang, L.; Lake, R.G.; Agarwal, S.; et al. EGR1 regulates angiogenic and osteoclastogenic factors in prostate cancer and promotes metastasis. Oncogene 2019, 38, 6241–6255. [Google Scholar] [CrossRef]
- Kang, H.S.; Ock, J.; Lee, H.J.; Lee, Y.J.; Kwon, B.M.; Hong, S.H. Early growth response protein 1 upregulation and nuclear translocation by 2′-benzoyloxycinnamaldehyde induces prostate cancer cell death. Cancer Lett. 2013, 329, 217–227. [Google Scholar] [CrossRef]
- Zhao, K.; Yu, M.; Zhu, Y.; Liu, D.; Wu, Q.; Hu, Y. EGR-1/ASPP1 inter-regulatory loop promotes apoptosis by inhibiting cyto-protective autophagy. Cell Death Dis. 2017, 8, e2869. [Google Scholar] [CrossRef]
- Lasham, A.; Mehta, S.Y.; Fitzgerald, S.J.; Woolley, A.G.; Hearn, J.I.; Hurley, D.G.; Ruza, I.; Algie, M.; Shelling, A.N.; Braithwaite, A.W.; et al. A novel EGR-1 dependent mechanism for YB-1 modulation of paclitaxel response in a triple negative breast cancer cell line. Int. J. Cancer 2016, 139, 1157–1170. [Google Scholar] [CrossRef]
- Chang, R.; He, H.; Mao, G.; Kong, Z. Upregulating DAB2IP expression via EGR-1 inhibition, a new approach for overcoming fractionated-irradiation-induced cross-tolerance to ionizing radiation and mitomycin C in tumor cells. Int. J. Radiat. Biol. 2017, 93, 386–393. [Google Scholar] [CrossRef]
- Shao, S.; Ju, M.; Lei, J.; Lu, X.; Li, H.; Wang, D.; Xia, C. Egr-1 inhibits colon cancer cell proliferation, migration and invasion via regulating CDKL1 at the transcriptional level. Oncol. Rep. 2021, 46, 169. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.L.; Wu, X.J.; Gong, C.C.; Pei, D.S. Egr-1 suppresses breast cancer cells proliferation by arresting cell cycle progression via down-regulating CyclinDs. Int. J. Clin. Exp. Pathol. 2017, 10, 10212–10222. [Google Scholar] [PubMed]


| Variables | Tumor-Adjacent Normal (n = 144) | Tumor (n = 144) | p Value * | ||
|---|---|---|---|---|---|
| Mean ± SD | Median (Q1~Q3) | Mean ± SD | Median (Q1~Q3) | ||
| EGR-1 | 5.87 ± 10.22 | 2.12 (0.93–7.14) | 3.97 ± 7.58 | 1.30 (0.52–3.45) | 0.0015 |
| Variables | Tumor-Adjacent Normal (n = 3) | Tumor (n = 93) | Metastatic (n = 1) | p Value * | |||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | Mean ± SD | Median | Mean ± SD | Median | ||
| EGR1 | 14.04 ± 0.69 | 14.04 | 12.30 ± 0.13 | 12.27 | 11.56 | - | 0.021 |
| Variables | No (%) | EGR-1 (Continuous) | EGR-1 > 68th Percentile | ||||
|---|---|---|---|---|---|---|---|
| Median (Q1~Q3) | p-Value | Yes (n = 46) | No (n = 98) | p-Value c | |||
| Sex | 0.70 a | 0.65 | |||||
| Male | 139 (96.5) | 1.26 (0.51~3.69) | 95 (96.94) | 44 (95.65) | |||
| Female | 5 (3.5) | 2.42(1.33~2.61) | 3 (3.06) | 2 (4.35) | |||
| Cell Differentiation | 0.16 b | 0.16 | |||||
| Well | 1 (0.7) | 6.37 | 0 (0) | 1 (2.17) | |||
| Moderate | 108 (75.0) | 1.52 (0.55~3.86) | 71 (72.45) | 37 (80.43) | |||
| Poor | 35 (24.3) | 1.05 (0.37~1.86) | 27 (27.55) | 8 (17.39) | |||
| AJCC pathological | 0.84 b | 0.68 | |||||
| I | 15 (10.4) | 0.72 (0.52~7.18) | 9 (9.18) | 6 (13.04) | |||
| II | 72 (50.0) | 1.21 (0.53~2.66) | 52 (53.06) | 20 (43.48) | |||
| III | 53 (36.8) | 1.71 (0.51~4.03) | 34 (34.69) | 19 (41.3) | |||
| IV | 4 (2.8) | 1.41 (0.66~16.39) | 3 (3.06) | 1 (2.17) | |||
| T classification | 0.71 b | 0.81 | |||||
| T1 | 15 (10.4) | 0.91 (0.50~2.53) | 11 (11.22) | 4 (8.7) | |||
| T2 | 30 (20.8) | 1.63 (0.61~3.04) | 20 (20.41) | 10 (21.74) | |||
| T3 | 96 (66.7) | 1.19 (0.50~3.97) | 64 (65.31) | 32 (69.57) | |||
| T4 | 3 (2.1) | 1.26 (0.13~1.97) | 3 (3.06) | 0 (0) | |||
| N classification | 0.47 b | 0.66 | |||||
| N0 | 69 (47.9) | 1.15 (0.54~2.71) | 50 (51.02) | 19 (41.3) | |||
| N1 | 66 (45.8) | 1.68 (0.49~4.03) | 42 (42.86) | 24 (52.17) | |||
| N2 | 8 (5.6) | 1.15 (0.49~4.32) | 5 (5.1) | 3 (6.52) | |||
| N3 | 1 (0.7) | 0.17 | 1 (1.02) | 0 (0) | |||
| M classification | 0.93 a | 1.00 | |||||
| M0 | 140 (97.2) | 1.30 (0.52~3.45) | 95 (96.94) | 45 (97.83) | |||
| M1 | 4 (2.8) | 1.41(0.66~16.39) | 3 (3.06) | 1 (2.17) | |||
| Models a | Univariate Analysis | Multivariate Analysis b | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | aHR (95% CI) | p-Value | |
| EGR-1 (Continuous) | 0.97 (0.93~1.01) | 0.178 | 0.96 (0.92~0.99) | 0.024 |
| EGR-1 > 25th percentile | 0.60 (0.29~1.23) | 0.163 | 0.59 (0.29~1.22) | 0.155 |
| EGR-1 > 50th percentile | 0.95 (0.53~1.70) | 0.867 | 0.76 (0.41~1.39) | 0.372 |
| EGR-1 > 75th percentile | 0.67 (0.33~1.36) | 0.267 | 0.45 (0.20~0.98) | 0.045 |
| EGR-1 > 68th percentile (ROC) | 0.63 (0.31~1.27) | 0.193 | 0.43 (0.20~0.94) | 0.034 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseng, Y.-C.; Shu, C.-W.; Chang, H.-M.; Lin, Y.-H.; Tseng, Y.-H.; Hsu, H.-S.; Goan, Y.-G.; Tseng, C.-J. Assessment of Early Growth Response 1 in Tumor Suppression of Esophageal Squamous Cell Carcinoma. J. Clin. Med. 2022, 11, 5792. https://doi.org/10.3390/jcm11195792
Tseng Y-C, Shu C-W, Chang H-M, Lin Y-H, Tseng Y-H, Hsu H-S, Goan Y-G, Tseng C-J. Assessment of Early Growth Response 1 in Tumor Suppression of Esophageal Squamous Cell Carcinoma. Journal of Clinical Medicine. 2022; 11(19):5792. https://doi.org/10.3390/jcm11195792
Chicago/Turabian StyleTseng, Yen-Chiang, Chih-Wen Shu, Hui-Min Chang, Yi-Hsuan Lin, Yen-Han Tseng, Han-Shui Hsu, Yih-Gang Goan, and Ching-Jiunn Tseng. 2022. "Assessment of Early Growth Response 1 in Tumor Suppression of Esophageal Squamous Cell Carcinoma" Journal of Clinical Medicine 11, no. 19: 5792. https://doi.org/10.3390/jcm11195792





