Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer
Abstract
:1. Introduction
2. Materials and Methods
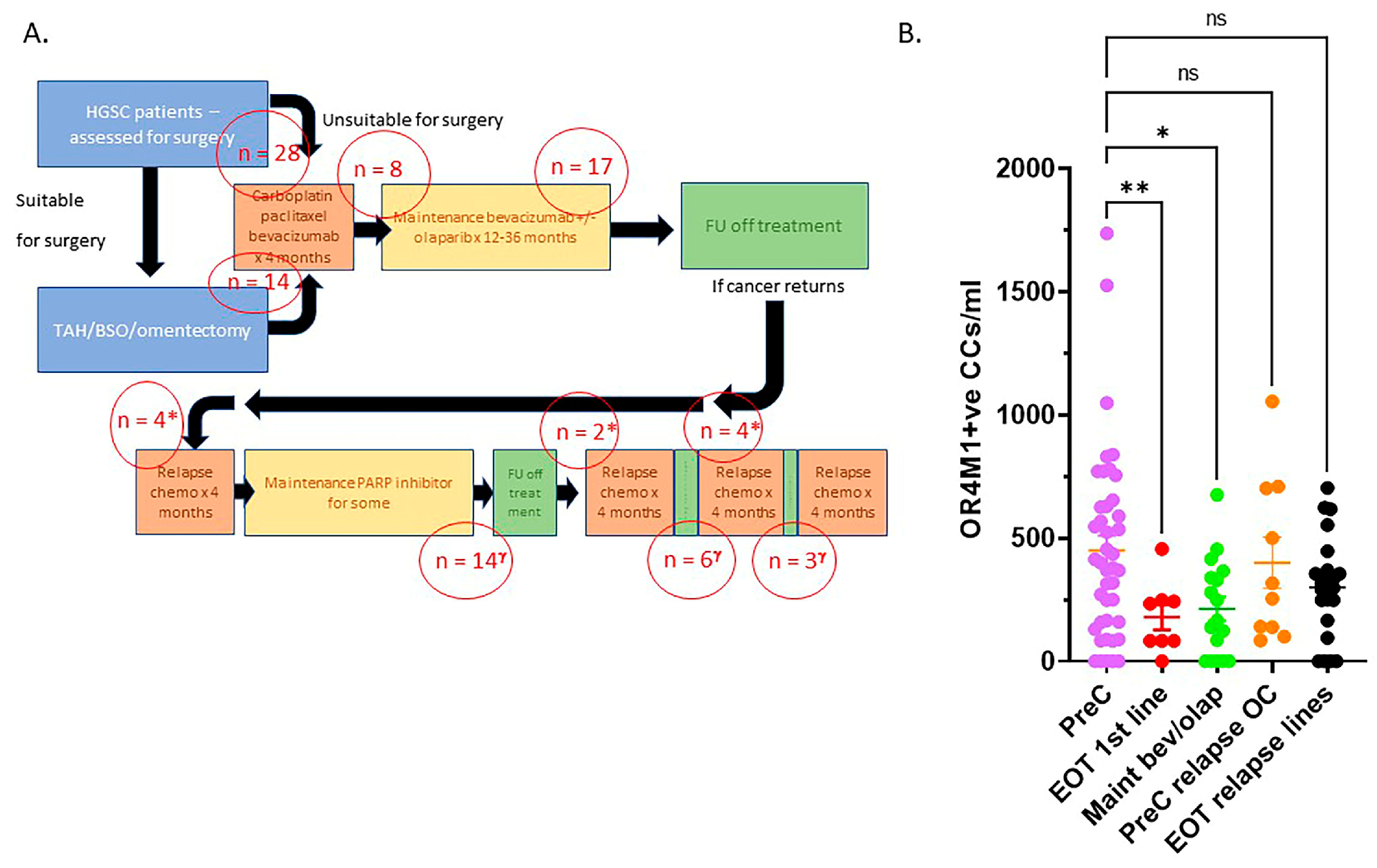
2.1. Blood Samples and ImageStream Mark II Analysis
2.2. Tissue Culture
2.3. RNA Extraction and cDNA Synthesis
2.4. RT-qPCR Analysis
2.5. RNA Sequencing
2.6. RNA Sequencing Statistical Analysis
2.7. Functional Annotation
2.8. Western Blot
2.9. Immunohistochemical Staining
3. Results
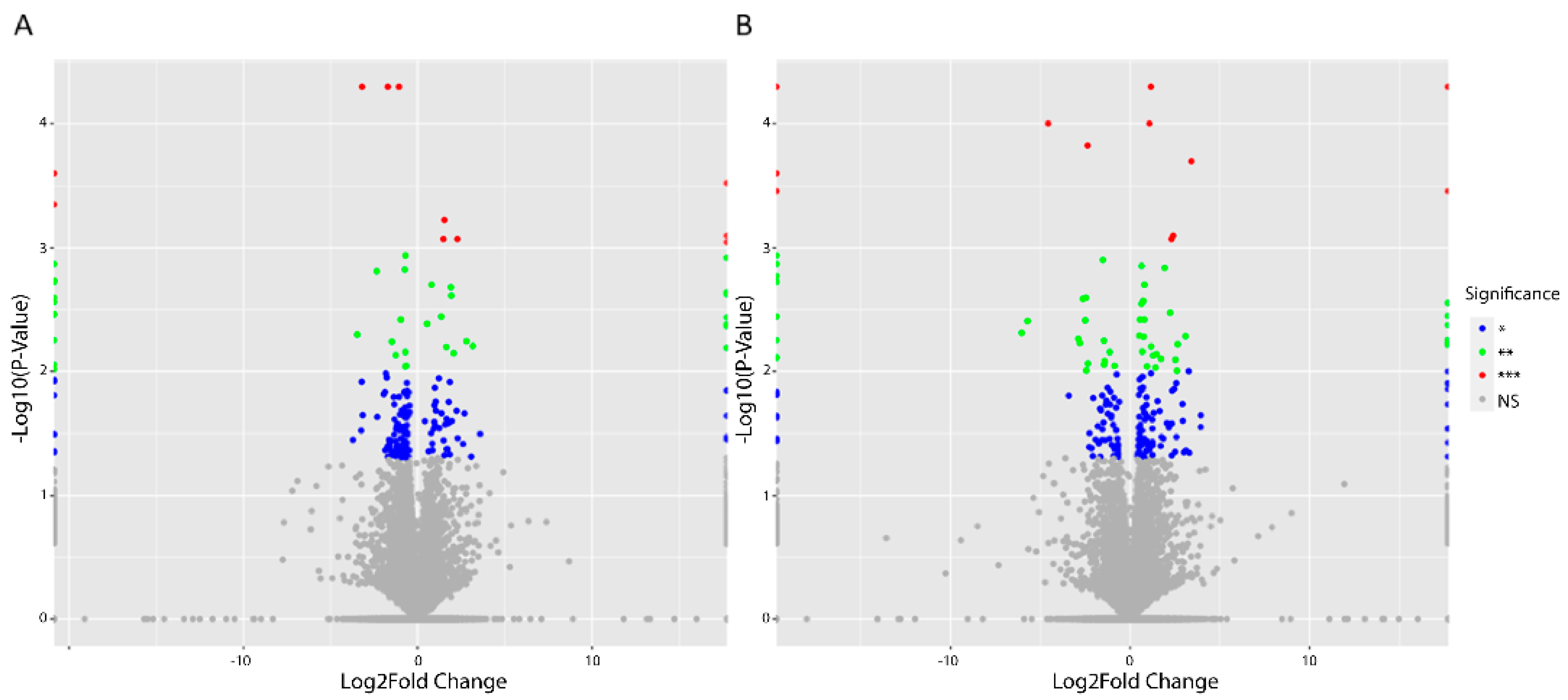
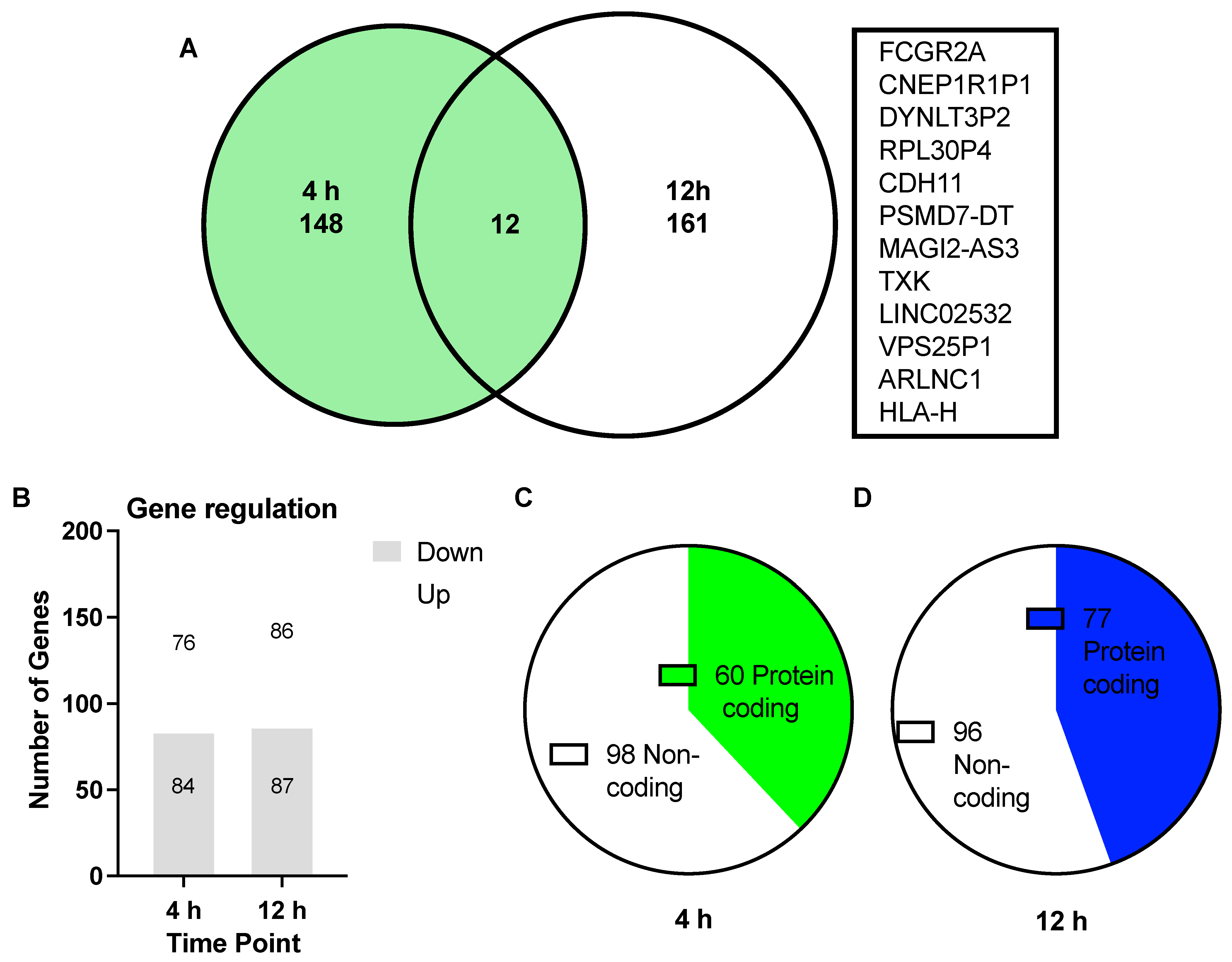
3.1. Differentially Expressed Genes (DEGs) upon Treatment with Asprosin
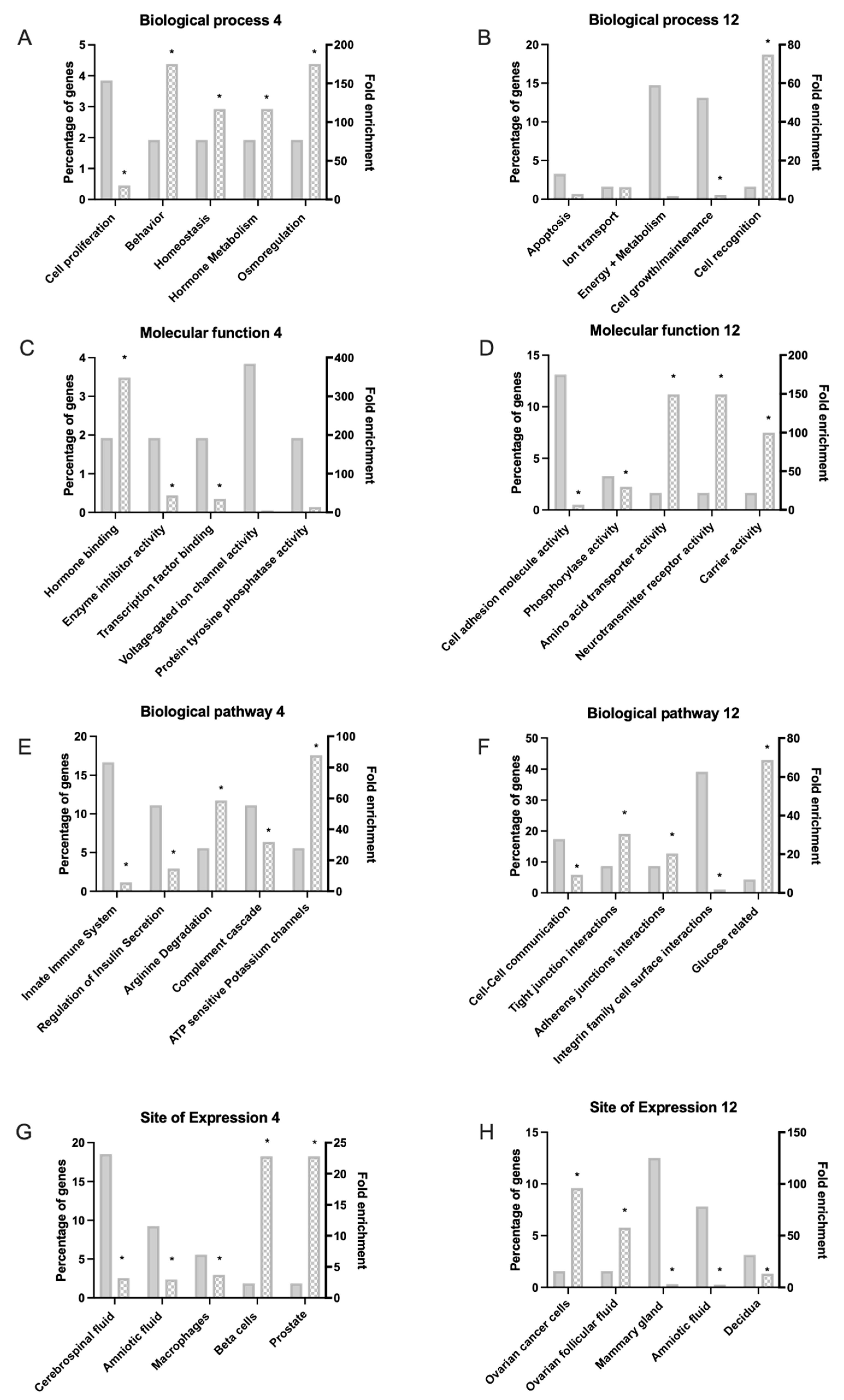
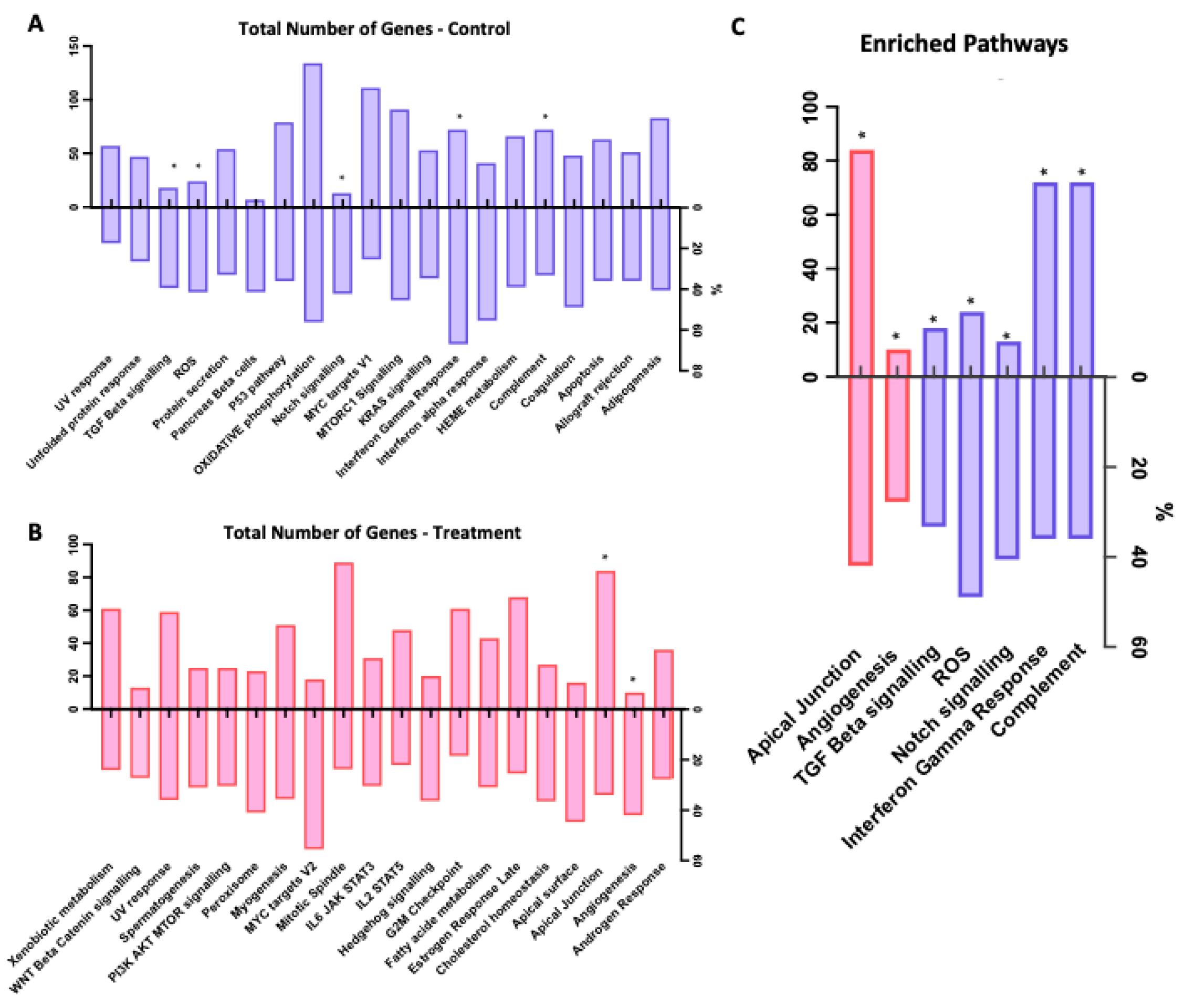
3.2. Functional Analysis of DEGs
3.3. Validation of Differentially Expressed Genes
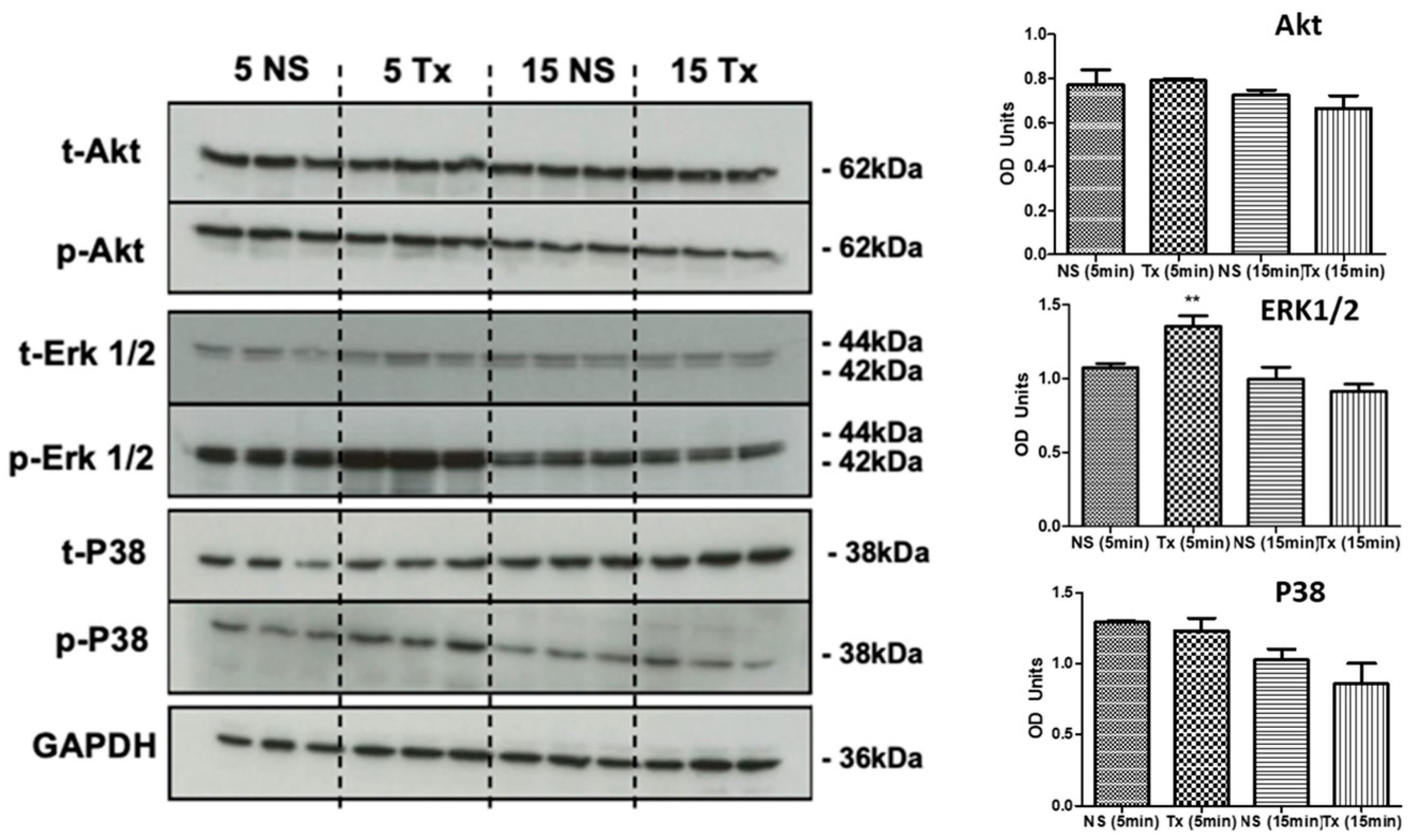
3.4. Asprosin Induces Phosphorylation of ERK1/2
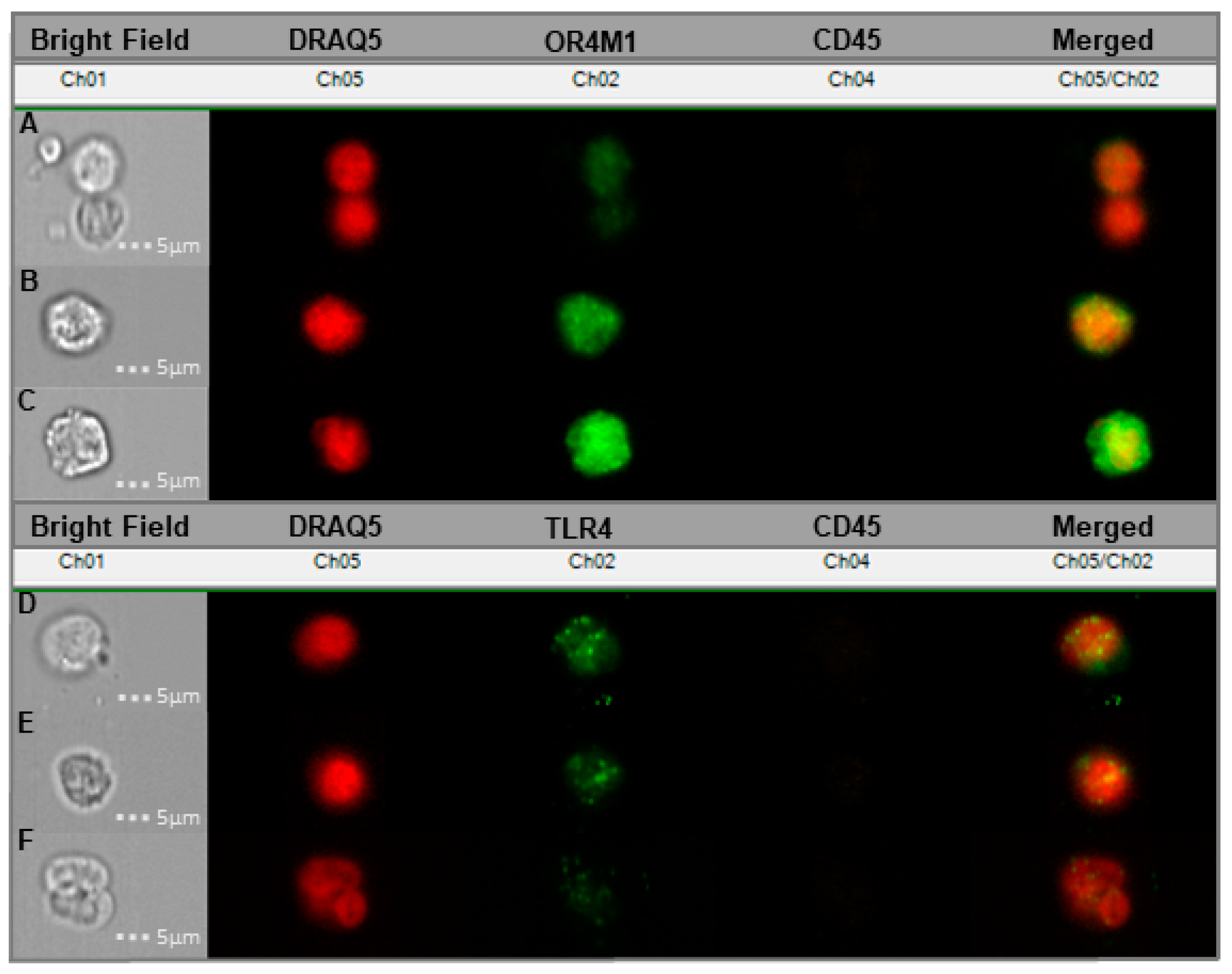
3.5. Cancer-Associated Circulating Cells Express Receptors for Asprosin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Romere, C.; Duerrschmid, C.; Bournat, J.; Constable, P.; Jain, M.; Xia, F.; Saha, P.K.; Del Solar, M.; Zhu, B.; York, B.; et al. Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 2016, 165, 566–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, E.; Shan, H.; Chen, L.; Long, A.; Zhang, Y.; Liu, Y.; Jia, L.; Wei, F.; Han, J.; Li, T.; et al. OLFR734 Mediates Glucose Metabolism as a Receptor of Asprosin. Cell Metab. 2019, 30, 319–328.e8. [Google Scholar] [CrossRef] [PubMed]
- Duerrschmid, C.; He, Y.; Wang, C.; Li, C.; Bournat, J.C.; Romere, C.; Saha, P.K.; Lee, M.E.; Phillips, K.J.; Jain, M.; et al. Asprosin is a centrally acting orexigenic hormone. Nat. Med. 2017, 23, 1444–1453. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-Y.; Lin, T.-A.; Liu, K.-H.; Liao, C.-H.; Liu, Y.-Y.; Wu, V.C.-C.; Wen, M.-S.; Yeh, T.-S. Serum asprosin levels and bariatric surgery outcomes in obese adults. Int. J. Obes. 2018, 43, 1019–1025. [Google Scholar] [CrossRef]
- Wang, M.; Yin, C.; Wang, L.; Liu, Y.; Li, H.; Li, M.; Yi, X.; Xiao, Y. Serum Asprosin Concentrations Are Increased and Associated with Insulin Resistance in Children with Obesity. Ann. Nutr. Metab. 2019, 75, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qu, H.; Xiong, X.; Qiu, Y.; Liao, Y.; Chen, Y.; Zheng, Y.; Zheng, H. Plasma Asprosin Concentrations Are Increased in Individuals with Glucose Dysregulation and Correlated with Insulin Resistance and First-Phase Insulin Secretion. Mediat. Inflamm. 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiecek, M.; Szymura, J.; Maciejczyk, M.; Kantorowicz, M.; Szygula, Z. Acute Anaerobic Exercise Affects the Secretion of Asprosin, Irisin, and Other Cytokines—A Comparison Between Sexes. Front. Physiol. 2018, 9, 1782. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.N.; Shill, A.L.; Thackray, A.E.; Stensel, D.J.; Bishop, N.C. Fasted plasma asprosin concentrations are associated with menstrual cycle phase, oral contraceptive use and training status in healthy women. Eur. J. Appl. Physiol. 2020, 121, 793–801. [Google Scholar] [CrossRef]
- Zhong, L.; Long, Y.; Wang, S.; Lian, R.; Deng, L.; Ye, Z.; Wang, Z.; Liu, B. Continuous elevation of plasma asprosin in pregnant women complicated with gestational diabetes mellitus: A nested case-control study. Placenta 2020, 93, 17–22. [Google Scholar] [CrossRef]
- Baykus, Y.; Yavuzkir, S.; Ustebay, S.; Ugur, K.; Deniz, R.; Aydin, S. Asprosin in umbilical cord of newborns and maternal blood of gestational diabetes, preeclampsia, severe preeclampsia, intrauterine growth retardation and macrosemic fetus. Peptides 2019, 120, 170132. [Google Scholar] [CrossRef]
- Chang, C.L.; Huang, S.Y.; Hsu, Y.C.; Chin, T.H.; Soong, Y.K. The serum level of irisin, but not asprosin, is abnormal in polycystic ovary syndrome patients. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alan, M.; Gürlek, B.; Yilmaz, A.; Aksit, M.; Aslanipour, B.; Gulhan, I.; Mehmet, C.; Taner, C.E. Asprosin: A novel peptide hormone related to insulin resistance in women with polycystic ovary syndrome. Gynecol. Endocrinol. 2018, 35, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Long, A.; Wang, Y. The Asprosin-OLFR734 hormonal signaling axis modulates male fertility. Cell Discov. 2019, 5, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maylem, E.R.S.; Spicer, L.J.; Batalha, I.; Schutz, L.F. Discovery of a possible role of asprosin in ovarian follicular function. J. Mol. Endocrinol. 2021, 66, 35–44. [Google Scholar] [CrossRef]
- Kerslake, R.; Hall, M.; Vagnarelli, P.; Jeyaneethi, J.; Randeva, H.S.; Pados, G.; Kyrou, I.; Karteris, E. A pancancer overview of FBN1, asprosin and its cognate receptor OR4M1 with detailed expression profiling in ovarian cancer. Oncol. Lett. 2021, 22, 1–14. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- La Vecchia, C. Ovarian cancer: Epidemiology and risk factors. Eur. J. Cancer Prev. 2017, 26, 55–62. [Google Scholar] [CrossRef]
- Slatnik, C.L.; Duff, E. Ovarian cancer. Nurse Pr. 2015, 40, 47–54. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Wu, H.; Dai, C.; Pan, Q.; Ding, Z.; Hu, D.; Ji, B.; Luo, Y.; Hu, X. Beyond Warburg effect–dual metabolic nature of cancer cells. Sci. Rep. 2014, 4, 4927. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocaman, N.; Yuksel, E.I.; Demir, B.; Calik, I.; Cicek, D. Two Novel Biomarker Candidates for Differentiating Basal Cell Carcinoma from Trichoblastoma; Asprosin and Meteorine Like Peptide. Tissue Cell 2022, 76, 101752. [Google Scholar] [CrossRef] [PubMed]
- Kang, B. OUP accepted manuscript. JNCI J. Natl. Cancer Inst. 2021. [Google Scholar] [CrossRef]
- Akkus, G.; Koyuturk, L.C.; Yilmaz, M.; Hancer, S.; Ozercan, I.H.; Kuloglu, T. Asprosin and meteorin-like protein immunoreactivity in invasive ductal breast carcinoma stages. Tissue Cell 2022, 77, 1855. [Google Scholar] [CrossRef]
- Kumar, J.; Chudasama, D.; Roberts, C.; Kubista, M.; Sjöback, R.; Chatterjee, J.; Anikin, V.; Karteris, E.; Hall, M. Detection of Abundant Non-Haematopoietic Circulating Cancer-Related Cells in Patients with Advanced Epithelial Ovarian Cancer. Cells 2019, 8, 732. [Google Scholar] [CrossRef] [Green Version]
- Mutch, D.G.; Prat, J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol. Oncol. 2014, 133, 401–404. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, D792–D799. [Google Scholar] [CrossRef] [Green Version]
- Zahra, A.; Kerslake, R.; Kyrou, I.; Randeva, H.S.; Sisu, C.; Karteris, E. Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary. Int. J. Mol. Sci. 2022, 23, 5334. [Google Scholar] [CrossRef]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2020, 433, 166747. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Rogers-Broadway, K.-R.; Chudasama, D.; Pados, G.; Tsolakidis, D.; Goumenou, A.; Hall, M.; Karteris, E. Differential effects of rapalogues, dual kinase inhibitors on human ovarian carcinoma cells in vitro. Int. J. Oncol. 2016, 49, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerslake, R.; Randeva, H.S.; Jonigk, D.; Werlein, C.; Robertus, J.L.; Katopodis, P.; Jasker, P.; Spandidos, D.A.; Kyrou, I.; Karteris, E. Protein expression of transmembrane protease serine 4 in the gastrointestinal tract and in healthy, cancer, and SARS-CoV-2 infected lungs. Mol. Med. Rep. 2022, 25, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Somers, E.B.; Ross, E.N.; Kline, J.B.; O’Shannessy, D.J.; Schweizer, C.; Weil, S.; Grasso, L.; Nicolaides, N.C. FCGR2A and FCGR3A Genotypes Correlate with Farletuzumab Response in Patients with First-Relapsed Ovarian Cancer Exhibiting Low CA125. Cytogenet. Genome Res. 2017, 152, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA MAGI2-AS3 is a New Player with a Tumor Suppressive Role in High Grade Serous Ovarian Carcinoma. Cancers 2019, 11, 2008. [Google Scholar] [CrossRef] [Green Version]
- Bi, J.; Huang, Y.; Liu, Y. Effect of NOP2 knockdown on colon cancer cell proliferation, migration, and invasion. Transl Cancer Res. 2019, 8, 6. [Google Scholar] [CrossRef]
- Mucaki, E.J.; Zhao, J.Z.L.; Lizotte, D.J.; Rogan, P.K. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduct. Target. Ther. 2019, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Madarampalli, B.; Watts, G.; Panipinto, P.M.; Nguygen, H.N.; Brenner, M.B.; Noss, E.H. Interactions between cadherin-11 and platelet-derived growth factor receptor-alpha signaling link cell adhesion and proliferation. Biochim. et Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 1516–1524. [Google Scholar] [CrossRef]
- Passanha, F.R.; Divinagracia, M.L.; LaPointe, V.L.S. Cadherin-11 Regulates Cell Proliferation via the PDGFRβ-ERK1/2 Signaling Pathway in Human Mesenchymal Stem Cells. Stem Cells 2022, 40, 165–174. [Google Scholar] [CrossRef]
- Von Bülow, C.; Oliveira-Ferrer, L.; Löning, T.; Trillsch, F.; Mahner, S.; Milde-Langosch, K. Cadherin-11 mRNA and protein expression in ovarian tumors of different malignancy: No evidence of oncogenic or tumor-suppressive function. Mol. Clin. Oncol. 2015, 3, 1067–1072. [Google Scholar] [CrossRef] [Green Version]
- Qian, J.; Gong, Z.-C.; Zhang, Y.-N.; Wu, H.-H.; Zhao, J.; Wang, L.-T.; Ye, L.-J.; Liu, D.; Wang, W.; Kang, X.; et al. Lactic acid promotes metastatic niche formation in bone metastasis of colorectal cancer. Cell Commun. Signal. 2021, 19, 1–15. [Google Scholar] [CrossRef]
- Yang, Z.; Yan, C.; Yu, Z.; He, C.; Li, J.; Li, C.; Yan, M.; Liu, B.; Wu, Y.; Zhu, Z. Downregulation of CDH11 Promotes Metastasis and Resistance to Paclitaxel in Gastric Cancer Cells. J. Cancer 2021, 12, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Nara, K.; Yoshikawa, H.; Suzuki, N. Txk, a member of the non-receptor tyrosine kinase of the Tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1α and regulates interferon-γ gene transcription in Th1 cells. Clin. Exp. Immunol. 2006, 147, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wester, L.; He, J.; Geiger, T.; Moerkens, M.; Siddappa, R.; Helmijr, J.A.; Timmermans, M.M.; Look, M.P.; Van Deurzen, C.H.M.; et al. IGF1R signaling drives antiestrogen resistance through PAK2/PIX activation in luminal breast cancer. Oncogene 2018, 37, 1869–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Zeng, B.; Li, Y.; Wang, H.; Zhang, X. LINC02532 Contributes to Radiosensitivity in Clear Cell Renal Cell Carcinoma through the miR-654-5p/YY1 Axis. Molecules 2021, 26, 7040. [Google Scholar] [CrossRef]
- Zhang, Y.; Pitchiaya, S.; Cieślik, M.; Niknafs, Y.S.; Tien, J.C.-Y.; Hosono, Y.; Iyer, M.K.; Yazdani, S.; Subramaniam, S.; Shukla, S.; et al. Analysis of the androgen receptor–regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 2018, 50, 814–824. [Google Scholar] [CrossRef]
- Chai, Y.; Wang, L.; Qu, Y.; Hu, Z. LncRNA MAGI2-As3 Suppresses the Proliferation and Invasion of Cervical Cancer by Sponging MiR-15b. J. Heal. Eng. 2022, 2022, 1–9. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, X.; Li, B.; Meng, X. MAGI2-AS3 suppresses MYC signaling to inhibit cell proliferation and migration in ovarian cancer through targeting miR-525-5p/MXD1 axis. Cancer Med. 2020, 9, 6377–6386. [Google Scholar] [CrossRef]
- Xiang, Z.; Song, S.; Zhu, Z.; Sun, W.; Gifts, J.E.; Sun, S.; Li, Q.S.; Yu, Y.; Li, K.K. LncRNAs GIHCG and SPINT1-AS1 Are Crucial Factors for Pan-Cancer Cells Sensitivity to Lapatinib. Front. Genet. 2019, 10, 25. [Google Scholar] [CrossRef]
- Roychowdhury, A.; Samadder, S.; Das, P.; Mazumder, D.I.; Chatterjee, A.; Addya, S.; Mondal, R.; Roy, A.; Roychoudhury, S.; Panda, C.K. Deregulation of H19 is associated with cervical carcinoma. Genomics 2019, 112, 961–970. [Google Scholar] [CrossRef]
- Xiao-Jie, L.; Ai-Mei, G.; Li-Juan, J.; Jiang, X. Pseudogene in cancer: Real functions and promising signature. J. Med. Genet. 2014, 52, 17–24. [Google Scholar] [CrossRef]
- Katopodis, P.; Dong, Q.; Halai, H.; Fratila, C.I.; Polychronis, A.; Anikin, V.; Sisu, C.; Karteris, E. In Silico and In Vitro Analysis of lncRNA XIST Reveals a Panel of Possible Lung Cancer Regulators and a Five-Gene Diagnostic Signature. Cancers 2020, 12, 3499. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liang, X.; Zhang, X.; Ni, Y.; Zhou, X.; Zhao, X. Metabolic cross-talk between ovarian cancer and the tumor microenvironment—Providing potential targets for cancer therapy. Front. Biosci. 2022, 27, 139. [Google Scholar] [CrossRef] [PubMed]
- Mishra, I.; Xie, W.R.; Bournat, J.C.; He, Y.; Wang, C.; Silva, E.S.; Liu, H.; Ku, Z.; Chen, Y.; Erokwu, B.O.; et al. Protein tyrosine phosphatase receptor δ serves as the orexigenic asprosin receptor. Cell Metab. 2022, 34, 549–563.e8. [Google Scholar] [CrossRef]
- Pizzuti, L.; Sergi, D.; Mandoj, C.; Antoniani, B.; Sperati, F.; Chirico, A.; Di Lauro, L.; Valle, M.; Garofalo, A.; Vizza, E.; et al. GLUT 1 receptor expression and circulating levels of fasting glucose in high grade serous ovarian cancer. J. Cell. Physiol. 2017, 233, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Rouaud, F.; Sluysmans, S.; Flinois, A.; Shah, J.; Vasileva, E.; Citi, S. Scaffolding proteins of vertebrate apical junctions: Structure, functions and biophysics. Biochim. et Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183399. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-H.; Zhang, Y.-Z.; Wang, Y.-S.; Ma, X.-X. Identification of novel cell glycolysis related gene signature predicting survival in patients with endometrial cancer. Cancer Cell Int. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Kang, H.; Wang, N.; Wang, X.; Zhang, Y.; Lin, S.; Mao, G.; Liu, D.; Dang, C.; Zhou, Z. A glycolysis-related gene signature predicts prognosis of patients with esophageal adenocarcinoma. Aging 2020, 12, 25828–25844. [Google Scholar] [CrossRef]
- Halimi, S.A.; Maeda, D.; Shinozaki-Ushiku, A.; Koso, T.; Matsusaka, K.; Tanaka, M.; Arimoto, T.; Oda, K.; Kawana, K.; Yano, T.; et al. Claudin-18 overexpression in intestinal-type mucinous borderline tumour of the ovary. Histopathology 2013, 63, 534–544. [Google Scholar] [CrossRef]
- Cuylan, Z.F.; Karabuk, E.; Oz, M.; Turan, A.T.; Meydanli, M.M.; Taşkın, S.; Sari, M.E.; Sahin, H.; Ulukent, S.C.; Akbayir, O.; et al. Comparison of stage III mucinous and serous ovarian cancer: A case-control study. J. Ovarian Res. 2018, 11, 91. [Google Scholar] [CrossRef]
- Sahin, U.; Koslowski, M.; Dhaene, K.; Usener, D.; Brandenburg, G.; Seitz, G.; Huber, C.; Türeci, O. Claudin-18 Splice Variant 2 Is a Pan-Cancer Target Suitable for Therapeutic Antibody Development. Clin. Cancer Res. 2008, 14, 7624–7634. [Google Scholar] [CrossRef] [Green Version]
- Hall, M.; Bertelli, G.; Li, L.; Green, C.; Chan, S.; Yeoh, C.C.; Hasan, J.; Jones, R.; Ograbek, A.; Perren, T. Role of front-line bevacizumab in advanced ovarian cancer: The OSCAR study. Int. J. Gynecol. Cancer 2019, 30, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-M.; Hwang, K.-A.; Choi, K.-C. Potential roles of reactive oxygen species derived from chemical substances involved in cancer development in the female reproductive system. BMB Rep. 2018, 51, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Perez-Fidalgo, J.A.; Ortega, B.; Simon, S.; Samartzis, E.P.; Boussios, S. NOTCH signalling in ovarian cancer angiogenesis. Ann. Transl. Med. 2020, 8, 1705. [Google Scholar] [CrossRef]
- Ceccarelli, S.; Megiorni, F.; Bellavia, D.; Marchese, C.; Screpanti, I.; Checquolo, S. Notch3 Targeting: A Novel Weapon against Ovarian Cancer Stem Cells. Stem Cells Int. 2019, 2019, 1–8. [Google Scholar] [CrossRef]
- Alsina-Sanchis, E.; Figueras, A.; Lahiguera, Á.; Vidal, A.; Casanovas, O.; Graupera, M.; Villanueva, A.; Viñals, F. The TGFβ pathway stimulates ovarian cancer cell proliferation by increasing IGF1R levels. Int. J. Cancer 2016, 139, 1894–1903. [Google Scholar] [CrossRef] [Green Version]
- Chaudhry, S.S.; Cain, S.A.; Morgan, A.; Dallas, S.L.; Shuttleworth, C.A.; Kielty, C.M. Fibrillin-1 regulates the bioavailability of TGFβ1. J. Cell Biol. 2007, 176, 355–367. [Google Scholar] [CrossRef]
- Muthu, M.L.; Reinhardt, D.P. Fibrillin-1 and fibrillin-1-derived asprosin in adipose tissue function and metabolic disorders. J. Cell Commun. Signal. 2020, 14, 159–173. [Google Scholar] [CrossRef]
- Davis, M.R.; Summers, K.M. Structure and function of the mammalian fibrillin gene family: Implications for human connective tissue diseases. Mol. Genet. Metab. 2012, 107, 635–647. [Google Scholar] [CrossRef]
- Katopodis, P.; Kerslake, R.; Zikopoulos, A.; Beri, N.; Anikin, V. p38β-MAPK11 and its role in female cancers. J. Ovarian Res. 2021, 14, 1–12. [Google Scholar] [CrossRef]
- Steinmetz, R.; Wagoner, H.A.; Zeng, P.; Hammond, J.R.; Hannon, T.S.; Meyers, J.L.; Pescovitz, O.H. Mechanisms Regulating the Constitutive Activation of the Extracellular Signal-Regulated Kinase (ERK) Signaling Pathway in Ovarian Cancer and the Effect of Ribonucleic Acid Interference for ERK1/2 on Cancer Cell Proliferation. Mol. Endocrinol. 2004, 18, 2570–2582. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Yun, S.; Jeong, J.H.; Jung, T.W. Asprosin impairs insulin secretion in response to glucose and viability through TLR4/JNK-mediated inflammation. Mol. Cell. Endocrinol. 2019, 486, 96–104. [Google Scholar] [CrossRef]
- Wang, A.-C.; Ma, Y.-B.; Wu, F.-X.; Ma, Z.-F.; Liu, N.-F.; Gao, R.; Gao, Y.-S.; Sheng, X.-G. TLR4 induces tumor growth and inhibits paclitaxel activity in MyD88-positive human ovarian carcinoma in vitro. Oncol. Lett. 2013, 7, 871–877. [Google Scholar] [CrossRef]


| Gene | Strand F to R |
|---|---|
| TXK | ACGGAGGCTGCCATAAAACAT |
| GGATTGATTGAAAGGCGTGTCT | |
| FCGR2A | CCTGAGAGCGACTCCATTCAG |
| GTCTGTAAACAGATTTCATCCGTCCT | |
| MAGI2-AS3 | GCTCTCATAGGCCACCTTGC |
| CTCCATCCTCATTCTCTCACCAC | |
| GAPDH | GGAGAAGGCTGGGGC |
| GATGGCATGGACTGTGG |
| Condition | Average Total Reads |
|---|---|
| Control (4 h) | 70,027,291 |
| 100 nM Asprosin (4 h) | 69,128,541 |
| Control (12 h) | 68,811,451 |
| 100 nM Asprosin (12 h) | 74,371,331 |
| Antibodies | Dilution | Source |
|---|---|---|
| GAPDH | 1:1000 | Cell Signalling |
| ERK 1/2 | 1:1000 | Thermo Fisher Scientific |
| P38 | 1:1000 | Cell Signalling |
| Akt | 1:1000 | Cell Signalling |
| Phospho-ERK 1/2 | 1:1000 | Thermo Fisher Scientific |
| Phospho-P38 | 1:1000 | Cell Signalling |
| Phospho-Akt | 1:1000 | Cell Signalling |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerslake, R.; Sisu, C.; Panfilov, S.; Hall, M.; Khan, N.; Jeyaneethi, J.; Randeva, H.; Kyrou, I.; Karteris, E. Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. J. Clin. Med. 2022, 11, 5942. https://doi.org/10.3390/jcm11195942
Kerslake R, Sisu C, Panfilov S, Hall M, Khan N, Jeyaneethi J, Randeva H, Kyrou I, Karteris E. Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. Journal of Clinical Medicine. 2022; 11(19):5942. https://doi.org/10.3390/jcm11195942
Chicago/Turabian StyleKerslake, Rachel, Cristina Sisu, Suzana Panfilov, Marcia Hall, Nabeel Khan, Jeyarooban Jeyaneethi, Harpal Randeva, Ioannis Kyrou, and Emmanouil Karteris. 2022. "Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer" Journal of Clinical Medicine 11, no. 19: 5942. https://doi.org/10.3390/jcm11195942
APA StyleKerslake, R., Sisu, C., Panfilov, S., Hall, M., Khan, N., Jeyaneethi, J., Randeva, H., Kyrou, I., & Karteris, E. (2022). Differential Regulation of Genes by the Glucogenic Hormone Asprosin in Ovarian Cancer. Journal of Clinical Medicine, 11(19), 5942. https://doi.org/10.3390/jcm11195942










