The Impact of Modifying Sunitinib Treatment Scheduling on Renal Cancer Tumor Biology and Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Mouse Models
2.2. Sunitinib Administration Protocols
2.3. Tumor Growth Assessment
2.4. Histological Assessment
2.5. RNA Isolation and Quantification
2.6. Next Generation Sequencing Analysis of mRNA Transcripts
2.7. Statistical and Bioinformatics Analysis
3. Results
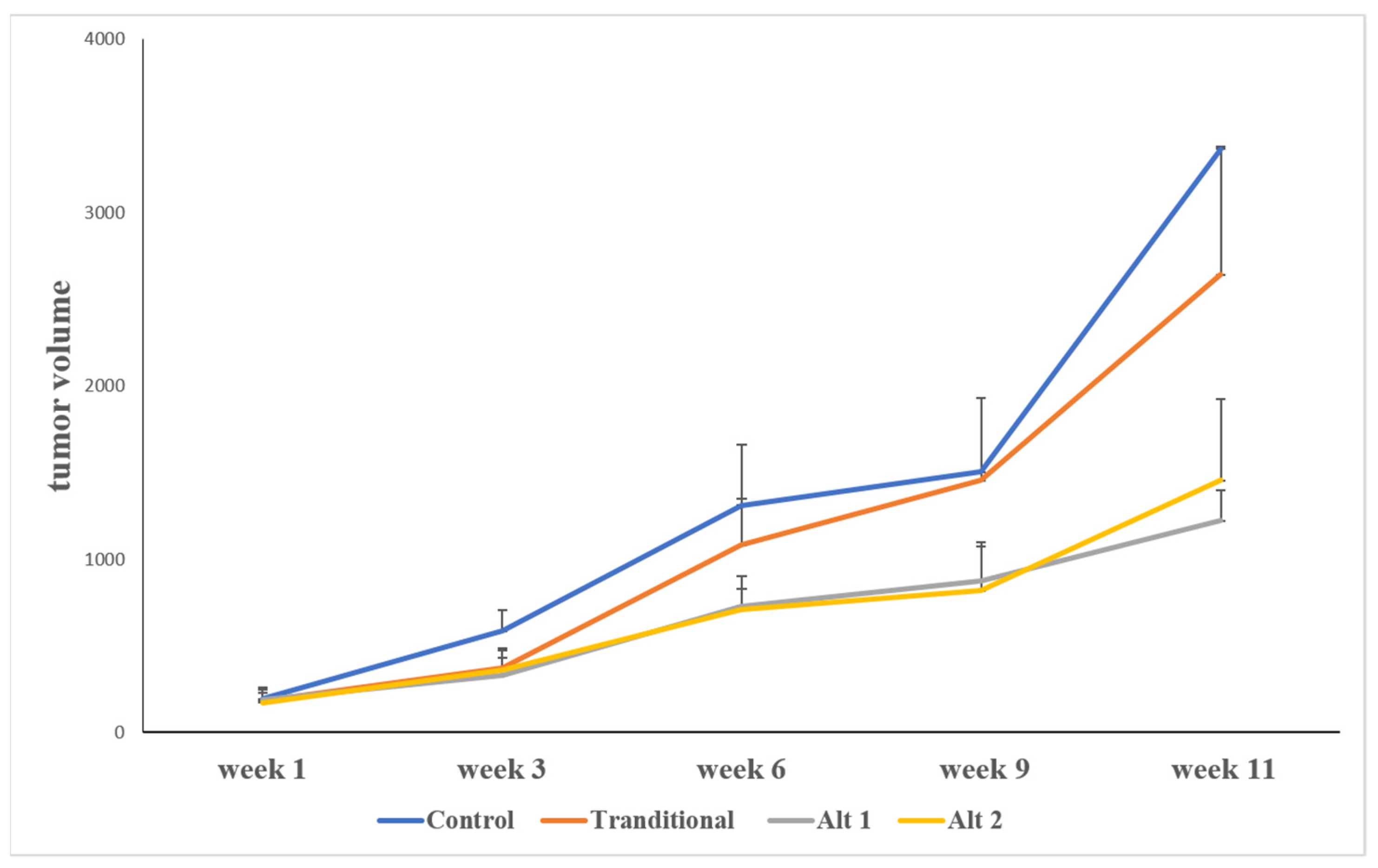
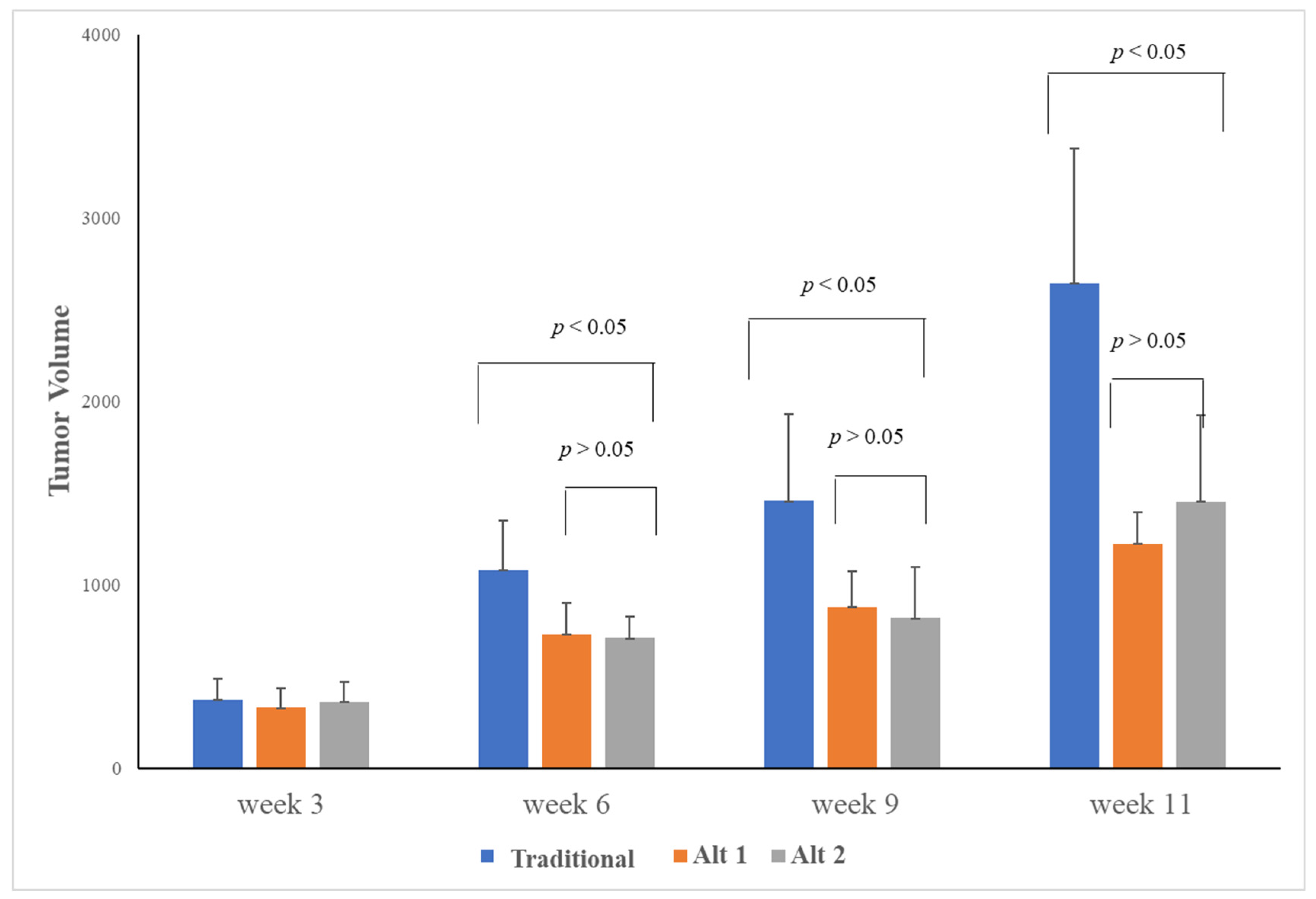
3.1. Alternative Treatment Scheduling Delayed Tumor Growth and Drug Resistance
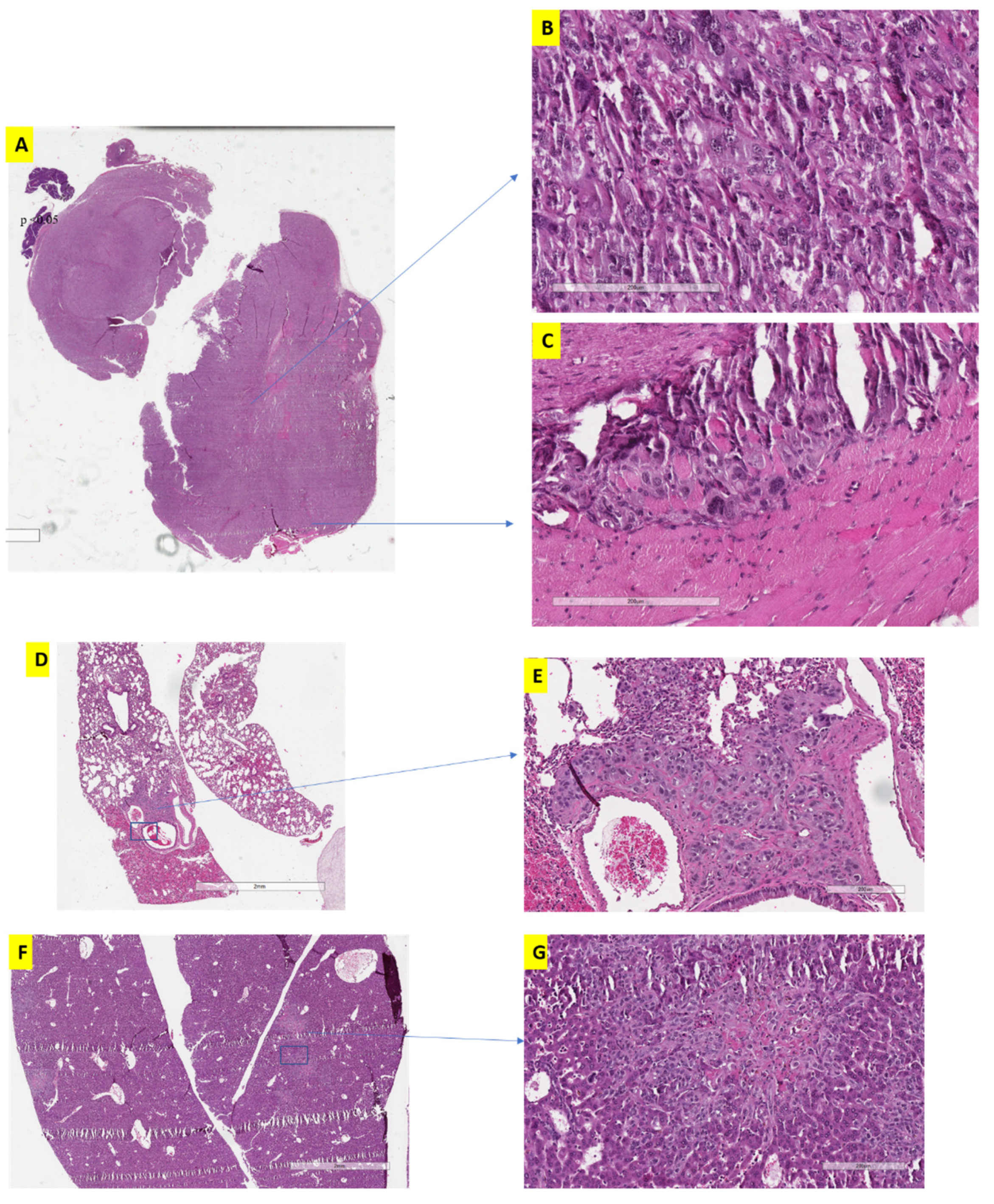
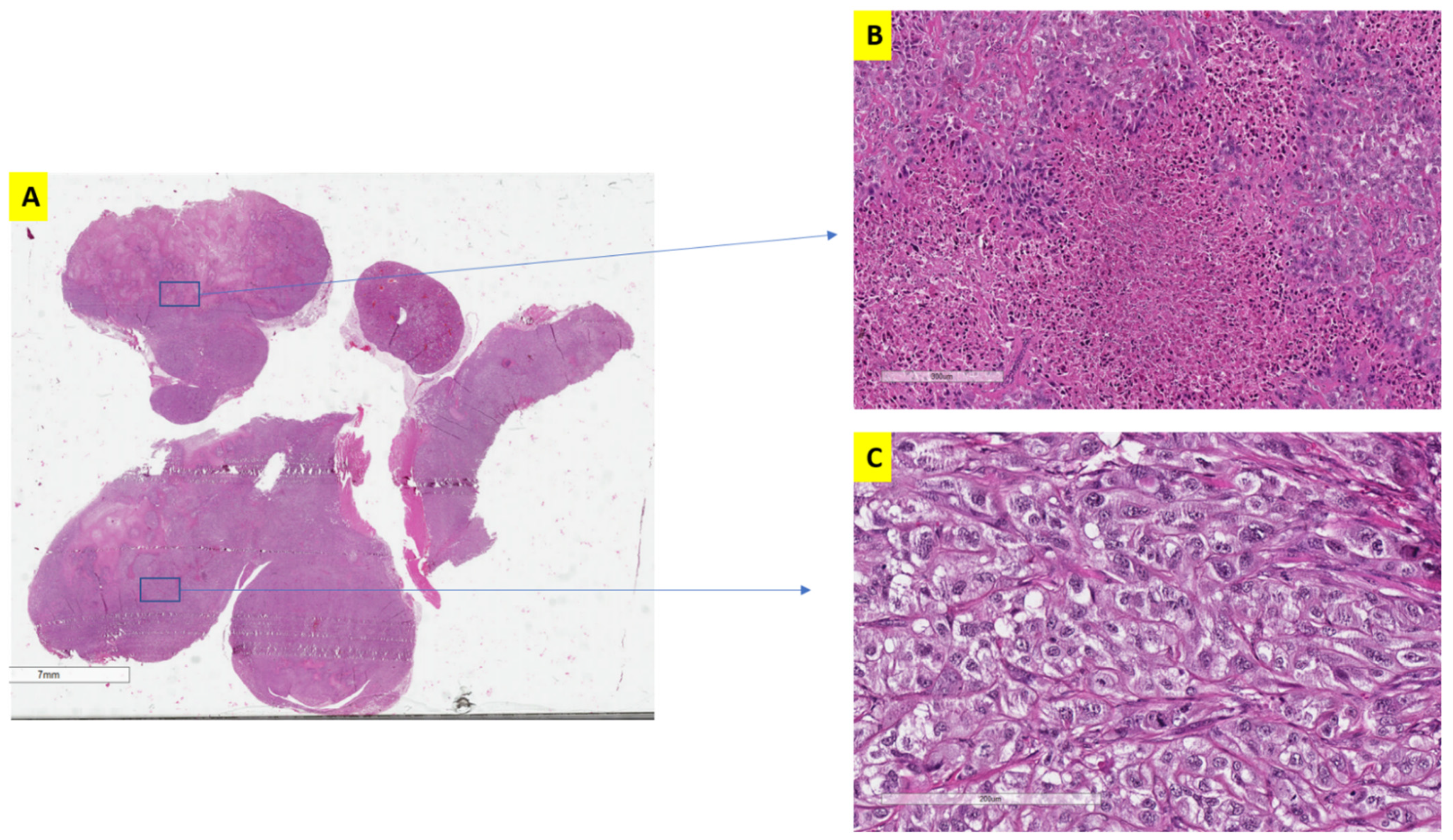
3.2. The Effect of Alternative Treatment Scheduling on Tumor Morphology and Behaviour
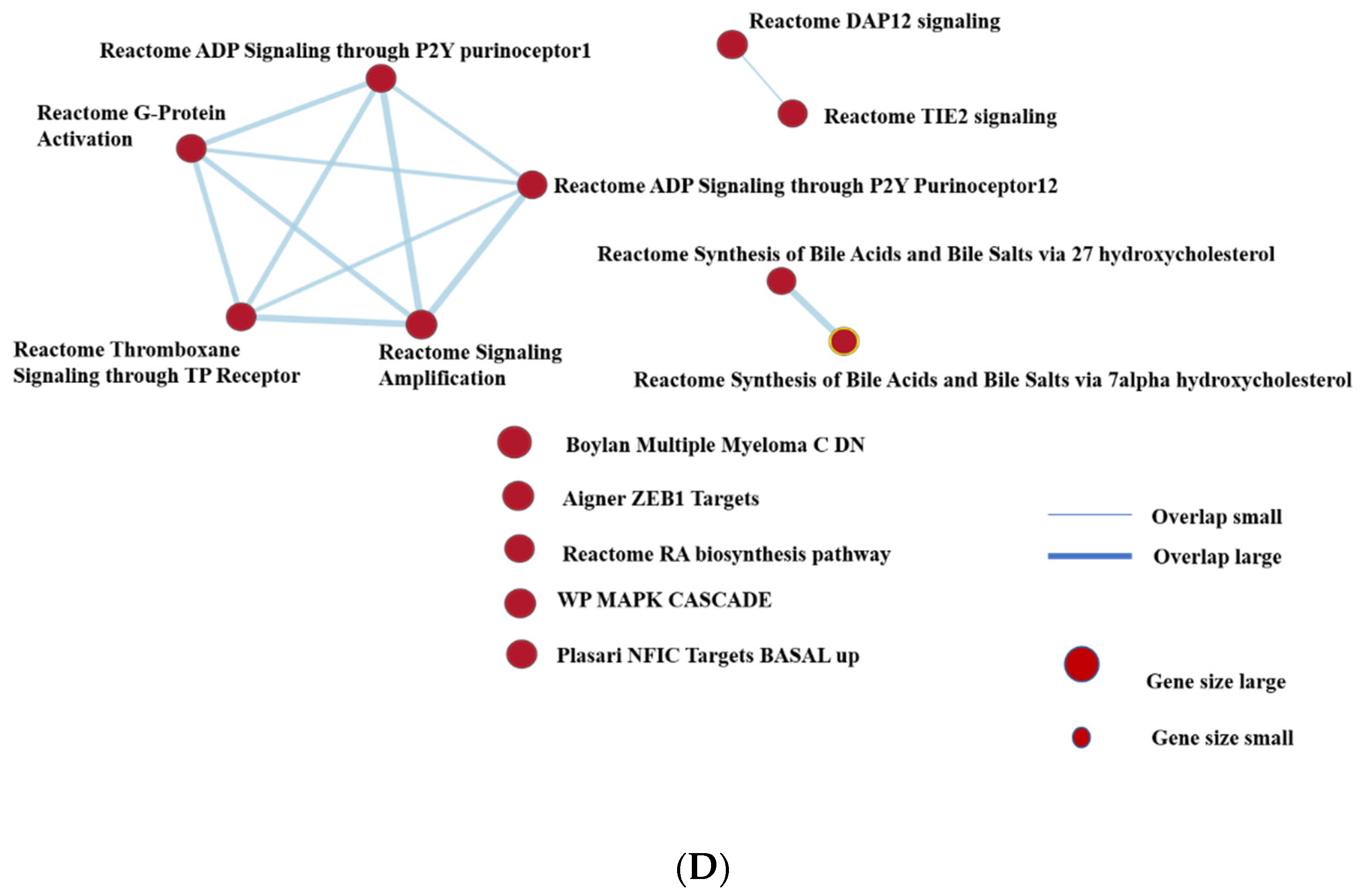
3.3. Transcriptomic Profile and Behavior Were Different between Traditional and Alternative Treatment Groups
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Mahasin, S.Z.; Aloudah, N.; Al-Surimi, K.; Alkhateeb, S.S. Epidemiology profile of renal cell carcinoma: A 10-year patients’ experience at King Abdulaziz Medical City, National Guard Health Affairs, Saudi Arabia. Urol. Ann. 2018, 10, 59–64. [Google Scholar] [CrossRef]
- Arsanious, A.; Bjarnason, G.A.; Yousef, G.M. From bench to bedside: Current and future applications of molecular profiling in renal cell carcinoma. Mol. Cancer 2009, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Lichner, Z.; Saleeb, R.; Butz, H.; Ding, Q.; Nofech-Mozes, R.; Riad, S.; Farag, M.; Varkouhi, A.K.; Dos Santos, C.C.; Kapus, A.; et al. Sunitinib induces early histomolecular changes in a subset of renal cancer cells that contribute to resistance. FASEB J. 2019, 33, 1347–1359. [Google Scholar] [CrossRef]
- Youssef, Y.M.; White, N.M.; Grigull, J.; Krizova, A.; Samy, C.; Mejia-Guerrero, S.; Evans, A.; Yousef, G.M. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur. Urol. 2011, 59, 721–730. [Google Scholar] [CrossRef]
- Inamura, K. Renal Cell Tumors: Understanding Their Molecular Pathological Epidemiology and the 2016 WHO Classification. Int. J. Mol. Sci. 2017, 18, 2195. [Google Scholar] [CrossRef]
- Barata, P.C.; Rini, B.I. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Arén Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Powles, T.; Burotto, M.; Escudier, B.; Bourlon, M.T.; Zurawski, B.; Oyervides Juárez, V.M.; Hsieh, J.J.; Basso, U.; Shah, A.Y.; et al. Nivolumab plus Cabozantinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2021, 384, 829–841. [Google Scholar] [CrossRef]
- Motzer, R.; Alekseev, B.; Rha, S.Y.; Porta, C.; Eto, M.; Powles, T.; Grünwald, V.; Hutson, T.E.; Kopyltsov, E.; Méndez-Vidal, M.J.; et al. Lenvatinib plus Pembrolizumab or Everolimus for Advanced Renal Cell Carcinoma. N. Engl. J. Med. 2021, 384, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Hutson, T.E.; Figlin, R.A.; Lechuga, M.J.; Valota, O.; Serfass, L.; Rosbrook, B.; Motzer, R.J. Sunitinib in Patients with Metastatic Renal Cell Carcinoma: Clinical Outcome According to International Metastatic Renal Cell Carcinoma Database Consortium Risk Group. Clin. Genitourin. Cancer 2018, 16, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, A. Activity of SU11248, a multitargeted inhibitor of vascular endothelial growth factor receptor and platelet-derived growth factor receptor, in patients with metastatic renal cell carcinoma and various other solid tumors. J. Steroid Biochem. Mol. Biol. 2008, 108, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, G.A.; Knox, J.J.; Kollmannsberger, C.K.; Soulieres, D.; Ernst, D.S.; Zalewski, P.; Canil, C.M.; Winquist, E.; Hotte, S.J.; North, S.A.; et al. The efficacy and safety of sunitinib given on an individualised schedule as first-line therapy for metastatic renal cell carcinoma: A phase 2 clinical trial. Eur. J. Cancer 2019, 108, 69–77. [Google Scholar] [CrossRef]
- Butz, H.; Ding, Q.; Nofech-Mozes, R.; Lichner, Z.; Ni, H.; Yousef, G.M. Elucidating mechanisms of sunitinib resistance in renal cancer: An integrated pathological-molecular analysis. Oncotarget 2018, 9, 4661–4674. [Google Scholar] [CrossRef] [PubMed]
- Bracarda, S.; Iacovelli, R.; Boni, L.; Rizzo, M.; Derosa, L.; Rossi, M.; Galli, L.; Procopio, G.; Sisani, M.; Longo, F.; et al. Sunitinib administered on 2/1 schedule in patients with metastatic renal cell carcinoma: The RAINBOW analysis. Ann. Oncol. 2015, 26, 2107–2113. [Google Scholar] [CrossRef]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Lichner, Z.; Saleh, C.; Subramaniam, V.; Seivwright, A.; Prud’homme, G.J.; Yousef, G.M. miR-17 inhibition enhances the formation of kidney cancer spheres with stem cell/ tumor initiating cell properties. Oncotarget 2015, 6, 5567–5581. [Google Scholar] [CrossRef]
- Carlini, M.J.; Recouvreux, M.S.; Simian, M.; Nagai, M.A. Gene expression profile and cancer-associated pathways linked to progesterone receptor isoform a (PRA) predominance in transgenic mouse mammary glands. BMC Cancer 2018, 18, 682. [Google Scholar] [CrossRef]
- Hammers, H.J.; Verheul, H.M.; Salumbides, B.; Sharma, R.; Rudek, M.; Jaspers, J.; Shah, P.; Ellis, L.; Shen, L.; Paesante, S.; et al. Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: Evidence from a xenograft study. Mol. Cancer Ther. 2010, 9, 1525–1535. [Google Scholar] [CrossRef]
- Khella, H.W.Z.; Butz, H.; Ding, Q.; Rotondo, F.; Evans, K.R.; Kupchak, P.; Dharsee, M.; Latif, A.; Pasic, M.D.; Lianidou, E.; et al. miR-221/222 Are Involved in Response to Sunitinib Treatment in Metastatic Renal Cell Carcinoma. Mol. Ther. J. Am. Soc. Gene Ther. 2015, 23, 1748–1758. [Google Scholar] [CrossRef]
- Lichner, Z.; Butz, H.; Mozes, R.; Riad, S.; Kapus, A.; Yousef, G.M. Sunitinib Treatment Induces the Formation of Stem Cell-Like Cancer Cells in Clear Cell Renal Cell Carcinoma. Lab. Investig. A J. Tech. Methods Pathol. 2016, 96 (Suppl. 1), 246A. [Google Scholar]
- Sahores, A.; Carozzo, A.; May, M.; Gómez, N.; Di Siervi, N.; De Sousa Serro, M.; Yaneff, A.; Rodríguez-González, A.; Abba, M.; Shayo, C.; et al. Multidrug transporter MRP4/ABCC4 as a key determinant of pancreatic cancer aggressiveness. Sci. Rep. 2020, 10, 14217. [Google Scholar] [CrossRef]
- Colavita, J.P.M.; Todaro, J.S.; de Sousa, M.; May, M.; Gómez, N.; Yaneff, A.; Di Siervi, N.; Aguirre, M.V.; Guijas, C.; Ferrini, L.; et al. Multidrug resistance protein 4 (MRP4/ABCC4) is overexpressed in clear cell renal cell carcinoma (ccRCC) and is essential to regulate cell proliferation. Int. J. Biol. Macromol. 2020, 161, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Fargeas, C.A.; Corbeil, D.; Huttner, W.B. AC133 antigen, CD133, prominin-1, prominin-2, etc.: Prominin family gene products in need of a rational nomenclature. Stem. Cells 2003, 21, 506–508. [Google Scholar] [CrossRef]
- Ding, Q.; Miyazaki, Y.; Tsukasa, K.; Matsubara, S.; Yoshimitsu, M.; Takao, S. CD133 facilitates epithelial-mesenchymal transition through interaction with the ERK pathway in pancreatic cancer metastasis. Mol. Cancer 2014, 13, 15. [Google Scholar] [CrossRef]
- Takao, S.; Ding, Q.; Matsubara, S. Pancreatic cancer stem cells: Regulatory networks in the tumor microenvironment and targeted therapy. J. Hepatobiliary Pancreat Sci. 2012, 19, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Fujiwara, T.; Sato, H.; Terui, A.; Hisaka, A. Investigation of Metabolomic Changes in Sunitinib-Resistant Human Renal Carcinoma 786-O Cells by Capillary Electrophoresis-Time of Flight Mass Spectrometry. Biol. Pharm. Bull. 2018, 41, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Hartsough, E.J.; Weiss, M.B.; Heilman, S.A.; Purwin, T.J.; Kugel, C.H., 3rd; Rosenbaum, S.R.; Erkes, D.A.; Tiago, M.; HooKim, K.; Chervoneva, I.; et al. CADM1 is a TWIST1-regulated suppressor of invasion and survival. Cell Death Dis. 2019, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Ahmat Amin, M.K.B.; Shimizu, A.; Zankov, D.P.; Sato, A.; Kurita, S.; Ito, M.; Maeda, T.; Yoshida, T.; Sakaue, T.; Higashiyama, S.; et al. Epithelial membrane protein 1 promotes tumor metastasis by enhancing cell migration via copine-III and Rac1. Oncogene 2018, 37, 5416–5434. [Google Scholar] [CrossRef] [PubMed]
- SAMD1. Available online: https://www.proteinatlas.org/ENSG00000141858-SAMD1/pathology/renal+cancer (accessed on 15 July 2021).
- Swan, R.; Alnabulsi, A.; Cash, B.; Alnabulsi, A.; Murray, G.I. Characterisation of the oxysterol metabolising enzyme pathway in mismatch repair proficient and deficient colorectal cancer. Oncotarget 2016, 7, 46509–46527. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Gene Set Name | Size | Enrichment Score | Normalized Enrichment Score | NOM p-val | FDR q-val |
|---|---|---|---|---|---|
| HALLMARK_PROTEIN_SECRETION | 96 | 0.644088 | 1.524993 | 0.0001 | 0.00576 |
| HALLMARK_ANDROGEN_RESPONSE | 99 | 0.622935 | 1.48529 | 0.0002 | 0.00625 |
| HALLMARK_HEME_METABOLISM | 195 | 0.54383 | 1.322556 | 0.0005 | 0.1018 |
| HALLMARK_MITOTIC_SPINDLE | 198 | 0.510461 | 1.245078 | 0.001 | 0.171488 |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 200 | 0.513705 | 1.252967 | 0.002 | 0.212878 |
| HALLMARK_MTORC1_SIGNALING | 197 | 0.516115 | 1.25293 | 0.002 | 0.177398 |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 198 | 0.513156 | 1.244218 | 0.006 | 0.15162 |
| HALLMARK_MYC_TARGETS_V1 | 196 | 0.499329 | 1.21534 | 0.01 | 0.171292 |
| HALLMARK_BILE_ACID_METABOLISM | 112 | 0.516231 | 1.230885 | 0.019076 | 0.162642 |
| HALLMARK_UV_RESPONSE_DN | 142 | 0.506569 | 1.219168 | 0.023 | 0.17801 |
| HALLMARK_G2M_CHECKPOINT | 196 | 0.480435 | 1.162778 | 0.034 | 0.296556 |
| _PI3K_AKT_MTOR_SIGNALING | 104 | 0.50546 | 1.209761 | 0.04911 | 0.160687 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.S.; Ding, Q.; Lichner, Z.; Kim, S.S.; Saleeb, R.; Farag, M.; Di Meo, A.; Plant, P.; Kaldas, M.; Bjarnason, G.A.; et al. The Impact of Modifying Sunitinib Treatment Scheduling on Renal Cancer Tumor Biology and Resistance. J. Clin. Med. 2022, 11, 369. https://doi.org/10.3390/jcm11020369
Lin HS, Ding Q, Lichner Z, Kim SS, Saleeb R, Farag M, Di Meo A, Plant P, Kaldas M, Bjarnason GA, et al. The Impact of Modifying Sunitinib Treatment Scheduling on Renal Cancer Tumor Biology and Resistance. Journal of Clinical Medicine. 2022; 11(2):369. https://doi.org/10.3390/jcm11020369
Chicago/Turabian StyleLin, Harrison Sicheng, Qiang Ding, Zsuzsanna Lichner, Sung Sun Kim, Rola Saleeb, Mina Farag, Ashley Di Meo, Pamela Plant, Mirit Kaldas, Georg Arnold Bjarnason, and et al. 2022. "The Impact of Modifying Sunitinib Treatment Scheduling on Renal Cancer Tumor Biology and Resistance" Journal of Clinical Medicine 11, no. 2: 369. https://doi.org/10.3390/jcm11020369
APA StyleLin, H. S., Ding, Q., Lichner, Z., Kim, S. S., Saleeb, R., Farag, M., Di Meo, A., Plant, P., Kaldas, M., Bjarnason, G. A., & Yousef, G. M. (2022). The Impact of Modifying Sunitinib Treatment Scheduling on Renal Cancer Tumor Biology and Resistance. Journal of Clinical Medicine, 11(2), 369. https://doi.org/10.3390/jcm11020369






