Applying an Improved Stacking Ensemble Model to Predict the Mortality of ICU Patients with Heart Failure
Abstract
:1. Introduction
- We propose an accurate and medically intuitive framework for predicting mortality in the ICU based on a comprehensive list of key characteristics of patients with HF in the ICU. The model is based on ensemble learning theory and stacking methods, and constructs a heterogeneous ensemble learning model to improve the generalization and prediction performance of the model.
- For our model we adopted the most popular and most diverse classifiers in current literature, including six different ML techniques. The generated classifier lists are used to construct the proposed stacking models. The different meta classifiers were tested and the best performing estimators were selected. The predictive capabilities of our stacking model outperform the results of a single classifier and standard ensemble techniques, achieving encouraging accuracy and strong generalization performance.
- In addition, feature importance provides a specific score for each feature, and these scores indicate the impact of each feature on model performance. The importance score represents the degree to which each input variable adds value to the decision in the constructed model. We also obtained the clinical characteristics of patients with HF in the ICU that had the greatest impact on model prediction.
2. Materials and Methods
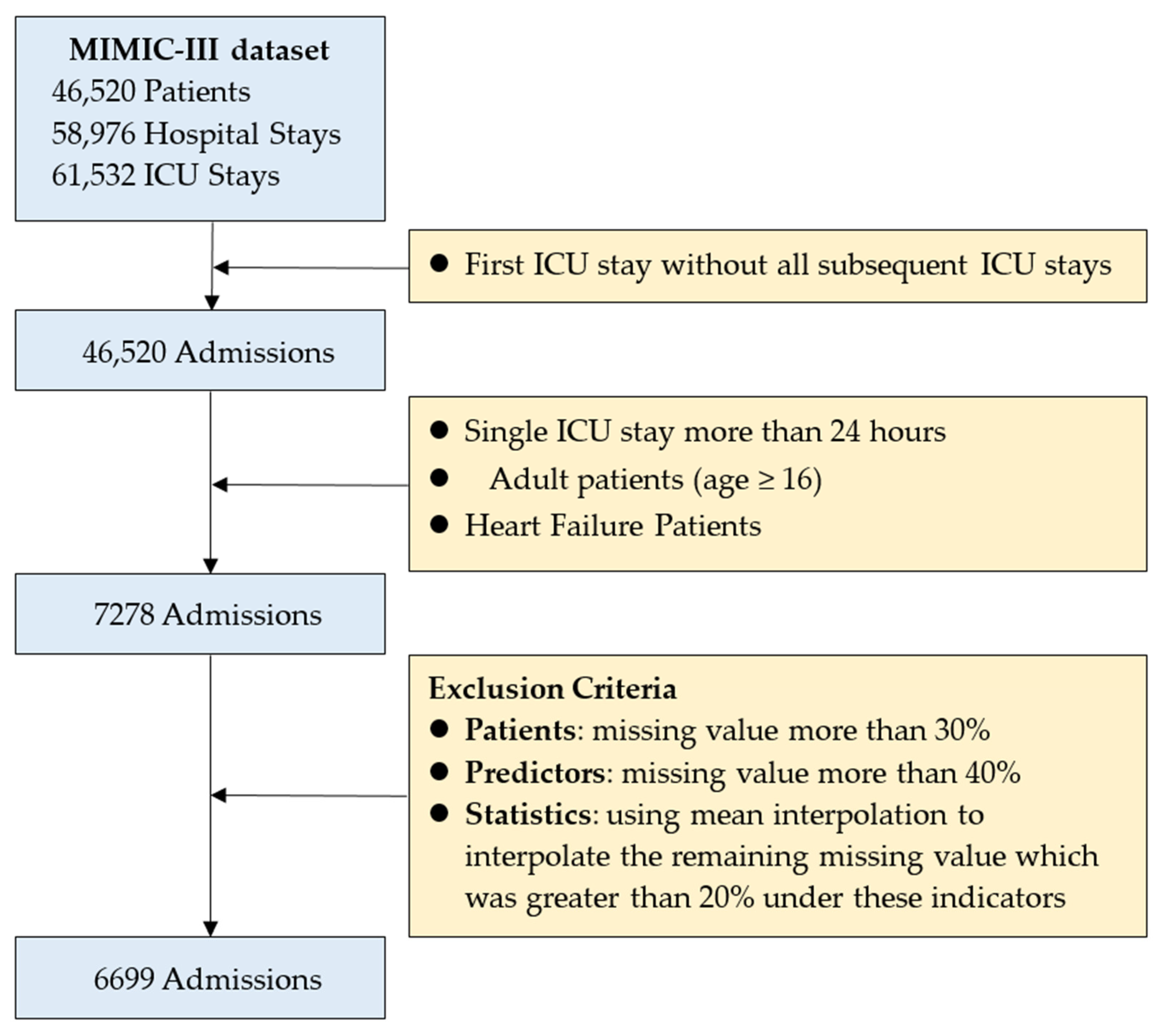
2.1. Patient Selection and Variable Selection
2.2. Proposed Framework
- Random Forest (RF) is an ensemble supervised ML algorithm. It uses decision trees as the basic classifier. RF generates many classifiers and combines their results by majority voting [53]. In the regression model, the output categories are numerical and the mean or average of the predicted outputs is used. The random forest algorithm is well suited to handle datasets with missing values. It also performs well on large datasets and can sort the features by importance. The advantage of using RF is that the algorithm provides higher accuracy compared to a single decision tree, it can handle datasets with a large number of predictive variables, and it can be used for variable selection [54].
- Support Vector Classifier (SVC) performs classification and regression analysis on linear and non-linear data. SVC aims to identify classes by creating decision hyperplanes in a non-linear manner in a higher eigenspace [55]. SVC is a robust tool to address data bias and variance and leads to accurate prediction of binary or multiclass classifications. In addition, SVC is robust to overfitting and has significant generalization capabilities [56].
- The K-Nearest Neighbors (KNN) algorithm does not require training. It is used to predict binary or sequential outputs. The data is divided into clusters and the number of nearest neighbors is specified by declaring the value of “K”, a constant. KNN is an algorithm [53] that stores all available instances and classifies new instances based on a similarity measure (e.g., distance function). Due to its simple implementation and excellent performance, it has been widely used in classification and regression prediction problems [57].
- Light Gradient Boosting Machine (LGBM) is an ensemble approach that combines predictions from multiple decision trees to make well-generalized final predictions. LGBM divides the consecutive eigenvalues into K intervals and selects the demarcation points from the K values. This process greatly accelerates the prediction speed and reduces storage space required without degrading the prediction accuracy [58,59]. LGBM is a gradient boosting decision tree learning algorithm that has been widely used for feature selection, classification and regression [60].
- The Bootstrap aggregating (Bagging) algorithm, also known as bagging algorithm, is an ensemble learning algorithm in the field of machine learning. It was originally proposed by Leo Breiman in 1994. Bagging algorithm can be combined with other classification and regression algorithms to improve its accuracy, stability, and avoid overfitting by reducing the variance of the results. Bagging is an ensemble method, i.e., a method of combining multiple predictors. It helps to avoid overfitting and variance reduction of the model to the data and has been used in a series of microarray studies [61,62]. We implemented Bagging using python’s sklearn library. We chose an ensemble of 500 DecisionTreeClassifier classifiers with a maximum sample set of 100 for each classifier, sampled each time using self-sampling, and trained all other hyperparameters by applying the sklearn default values.
- The self-adaptive nature of the Adaptive Boosting (AdaBoost) method is that the wrong samples of the previous classifier are used to train the next classifier, therefore, the AdaBoost method is sensitive to noisy data and anomalous data. It trains a basic classifier and assigns higher weights to the misclassified samples. After that, it is applied to the next process. The iterative process continues until the stopping condition is reached or the error rate becomes small enough [63,64]. We implemented AdaBoost using python’s sklearn library, choosing a maximum number of iterations of 50 for our hyperparameters and using the default values in sklearn for the rest of the training model.
2.3. Stacking Ensemble Technique
- The original dataset S is randomly divided into K sub-datasets {S1, S2, ⋯, Sn}. Taking base learner 1 as an example, each sub-dataset Si (i = 1, 2, ⋯, K) is verified separately, and the remaining K − 1 sub-datasets are used as training sets to obtain K prediction results. Merge into set D1, which has the same length as S.
- Perform the same operation on other n − 1 base learners to obtain the set D2, D3, ⋯, Dn. Combining the prediction results of n base learners, a new dataset D = {D1, D2, ⋯, Dn} is obtained, which constitutes the input data of the second-layer meta-learner.
- The second-layer prediction model can detect and correct errors in the first-layer prediction model in time, and improve the accuracy of the prediction model.
2.4. Synthetic Minority Oversampling Technique (SMOTE)
2.5. Evaluation Criteria
3. Results
3.1. Baseline Characteristics
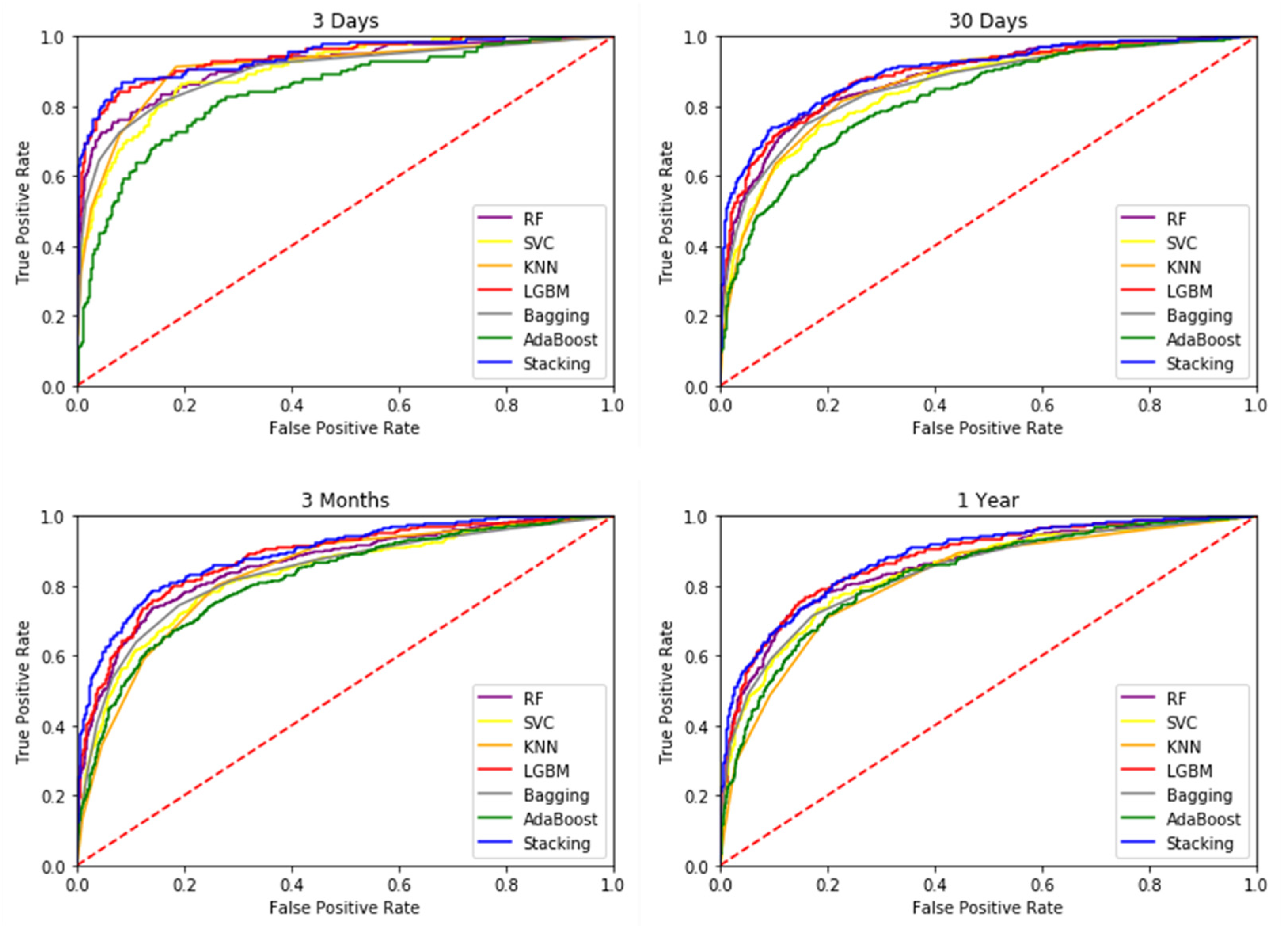
3.2. Mortality Prediction Results of Different Models
3.3. Interpretation of Variable Importance
4. Discussion
5. Conclusions
- Compared to structured data that have been used for clinical outcome prediction, the information available in diagnostic records and test reports in unstructured data is still underutilized by medical research. These diagnostic data are important references for clinical decision-making because they record multifaceted information about the patient’s visit, such as the focus of care, preliminary medical assessment, and the generation of different recommendations for the final diagnosis. Future research suggests that structured data and unstructured data can be integrated for more detailed classification and study [84].
- The MIMIC-III data is relatively rich and complete, and this study only modeled and predicted the mortality of patients; subsequent studies can be conducted to evaluate the readmission, length of stay, medication use, and complications of patients with reference to the framework of this study. This type of study can be made more objective and complete if it can be extended to conduct more comprehensive evaluation and analysis.
- The evolution of variables over time can be collected from patient EHR data in an attempt to obtain better predictive effects. In terms of research methods, future research can attempt different ML methods as well as deep learning methods that have recently been applied to solve time-series data more effectively. For example, long short-term memory, recurrent neural network, CNN models [85,86] are common deep learning models.
- With the popularity and increasing prevalence of AI, telemedicine and robotics, which have emerged in response to the recent COVID-19, imaging AI and speech AI can be incorporated. Combining existing clinical data, diagnostic reports, medical image images, etc., can improve medical culture and quality of care, which will be an important issue in the future field of smart medicine [87,88].
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.L.R.; Cheng, S.S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2020 Update: A Report from the American Heart Association. Circulation 2020, 141, E139–E596. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yu, H.P.; Huang, Y.P.; Jin, H.Y. ECG Signal-Enabled Automatic Diagnosis Technology of Heart Failure. J. Healthc. Eng. 2021, 2021, 5802722. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xin, H.; Zhang, J.; Fu, M.; Zhou, J.; Lian, Z. Prediction model of in-hospital mortality in intensive care unit patients with heart failure: Machine learning-based, retrospective analysis of the MIMIC-III database. BMJ Open 2021, 11, e044779. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Pasque, M.; Loh, F.; Mann, D.L.; Payne, P.R. Heart failure diagnosis, readmission, and mortality prediction using machine learning and artificial intelligence models. Curr. Epidemiol. Rep. 2020, 7, 212–219. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Cao, L.H.; Chen, R.G.; Zhao, Y.; Lv, L.K.; Xu, Z.Y.; Xu, P. Electronic healthcare records and external outcome data for hospitalized patients with heart failure. Sci. Data 2021, 8, 1–6. [Google Scholar] [CrossRef]
- Mpanya, D.; Celik, T.; Klug, E.; Ntsinjana, H. Machine learning and statistical methods for predicting mortality in heart failure. Heart Fail. Rev. 2021, 26, 545–552. [Google Scholar] [CrossRef]
- Incidence, G.B.D.D.I. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: A systematic analysis for the Global Burden of Disease Study 2016, 390. Lancet 2017, 390, E38. [Google Scholar]
- Sayed, M.; Riano, D.; Villar, J. Predicting Duration of Mechanical Ventilation in Acute Respiratory Distress Syndrome Using Supervised Machine Learning. J. Clin. Med. 2021, 10, 3824. [Google Scholar] [CrossRef]
- Marshall, J.C.; Bosco, L.; Adhikari, N.K.; Connolly, B.; Diaz, J.V.; Dorman, T.; Fowler, R.A.; Meyfroidt, G.; Nakagawa, S.; Pelosi, P. What is an intensive care unit? A report of the task force of the World Federation of Societies of Intensive and Critical Care Medicine. J. Crit. Care 2017, 37, 270–276. [Google Scholar] [CrossRef]
- Romano, M. The Role of Palliative Care in the Cardiac Intensive Care Unit. Healthcare 2019, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Haase, N.; Plovsing, R.; Christensen, S.; Poulsen, L.M.; Brochner, A.C.; Rasmussen, B.S.; Helleberg, M.; Jensen, J.U.S.; Andersen, L.P.K.; Siegel, H.; et al. Characteristics, interventions, and longer term outcomes of COVID-19 ICU patients in Denmark—A nationwide, observational study. Acta Anaesthesiol. Scand. 2020, 65, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Long, G.; Yao, L.; Sheng, Q.Z. AMRNN: Attended multi-task recurrent neural networks for dynamic illness severity prediction. World Wide Web 2019, 23, 2753–2770. [Google Scholar] [CrossRef]
- El-Rashidy, N.; El-Sappagh, S.; Abuhmed, T.; Abdelrazek, S.; El-Bakry, H.M. Intensive Care Unit Mortality Prediction: An Improved Patient-Specific Stacking Ensemble Model. IEEE Access 2020, 8, 133541–133564. [Google Scholar] [CrossRef]
- Kim, J.Y.; Yee, J.; Park, T.I.; Shin, S.Y.; Ha, M.H.; Gwak, H.S. Risk Scoring System of Mortality and Prediction Model of Hospital Stay for Critically Ill Patients Receiving Parenteral Nutrition. Healthcare 2021, 9, 853. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, L.P.; Reyfman, P.A.; Smith, A.D.R.; Zeng, Z.X.; Kho, A.; Sanchez-Pinto, L.N.; Luo, Y. Early prediction of acute kidney injury following ICU admission using a multivariate panel of physiological measurements. BMC Med. Inform. Decis. Mak. 2019, 19, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, R.; Banerjee, T.; Romine, W. Early hospital mortality prediction using vital signals. Smart Health 2018, 9, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Karunarathna, K.M. Predicting ICU death with summarized patient data. In Proceedings of the 2018 IEEE 8th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, USA, 8–10 January 2018; pp. 238–247. [Google Scholar]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonca, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Legall, J.R.; Lemeshow, S.; Saulnier, F. A new simplified acute physiology score (SAPS-II) based on a European North-American multicenter study. JAMA-J. Am. Med. Assoc. 1993, 270, 2957–2963. [Google Scholar] [CrossRef]
- Baue, A.E.; Durham, R.; Faist, E. Systemic inflammatory response syndrome (SIRS), multiple organ dysfunction syndrome (MODS), multiple organ failure (MOF): Are we winning the battle? Shock 1998, 10, 79–89. [Google Scholar] [CrossRef]
- Guo, C.H.; Lu, M.L.; Chen, J.F. An evaluation of time series summary statistics as features for clinical prediction tasks. Bmc Med. Inform. Decis. Mak. 2020, 20, 1–20. [Google Scholar] [CrossRef]
- Mitchell, T. Machine Learning; McGraw-Hill: New York, NY, USA, 1997; Volume 1. [Google Scholar]
- Adlung, L.; Cohen, Y.; Mor, U.; Elinav, E. Machine learning in clinical decision making. Med 2021, 2, 642–665. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.Y.; Joshi, H.; Tandon, P.; Freeman, R.; Reich, D.L.; Mazumdar, M.; Kohli-Seth, R.; Levin, M.A.; Timsina, P.; Kia, A. Using Machine Learning to Predict ICU Transfer in Hospitalized COVID-19 Patients. J. Clin. Med. 2020, 9, 1668. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef]
- Purushotham, S.; Meng, C.Z.; Che, Z.P.; Liu, Y. Benchmarking deep learning models on large healthcare datasets. J. Biomed. Inform. 2018, 83, 112–134. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Maugeri, A.; Favara, G.; Riela, P.M.; Gallo, G.; Mura, I.; Agodi, A.; Network, S.-U. Early Prediction of Seven-Day Mortality in Intensive Care Unit Using a Machine Learning Model: Results from the SPIN-UTI Project. J. Clin. Med. 2021, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Negassa, A.; Ahmed, S.; Zolty, R.; Patel, S.R. Prediction Model Using Machine Learning for Mortality in Patients with Heart Failure. Am. J. Cardiol. 2021, 153, 86–93. [Google Scholar] [CrossRef]
- Adler, E.; Voors, A.; Klein, L.; Macheret, F.; Sama, I.; Braun, O.; Urey, M.; Zhu, W.H.; Tadel, M.; Campagnari, C.; et al. Machine learning algorithm using 8 commonly acquired clinical variables accurately predicts mortality in heart failure. J. Am. Coll. Cardiol. 2019, 73, 689. [Google Scholar] [CrossRef]
- Jing, L.Y.; Cerna, A.E.U.; Good, C.W.; Sauers, N.M.; Schneider, G.; Hartzel, D.N.; Leader, J.B.; Kirchner, H.L.; Hu, Y.R.; Riviello, D.M.; et al. A Machine Learning Approach to Management of Heart Failure Populations. JACC-Heart Fail. 2020, 8, 578–587. [Google Scholar] [CrossRef]
- Casillas, N.; Torres, A.M.; Moret, M.; Gomez, A.; Rius-Peris, J.M.; Mateo, J. Mortality predictors in patients with COVID-19 pneumonia: A machine learning approach using eXtreme Gradient Boosting model. Intern. Emerg. Med. 2022, 17, 1929–1939. [Google Scholar] [CrossRef]
- Bi, S.W.; Chen, S.S.; Li, J.Y.; Gu, J. Machine learning-based prediction of in-hospital mortality for post cardiovascular surgery patients admitting to intensive care unit: A retrospective observational cohort study based on a large multi-center critical care database. Comput. Methods Programs Biomed. 2022, 226, e107115. [Google Scholar] [CrossRef]
- González-Nóvoa, J.A.; Busto, L.; Rodríguez-Andina, J.J.; Fariña, J.; Segura, M.; Gómez, V.; Vila, D.; Veiga, C. Using Explainable Machine Learning to Improve Intensive Care Unit Alarm Systems. Sensors 2021, 21, 7125. [Google Scholar] [CrossRef] [PubMed]
- Freund, Y.; Schapire, R.E. Experiments with a new boosting algorithm. In Proceedings of the ICML, Bari, Italy, 3–6 July 1996; pp. 148–156. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Li, J.Y.; Guo, F.C.; Sivakumar, A.; Dong, Y.J.; Krishnan, R. Transferability improvement in short-term traffic prediction using stacked LSTM network. Transp. Res. Part C Emerg. Technol. 2021, 124, e102977. [Google Scholar] [CrossRef]
- Zhai, B.; Chen, J. Development of a stacked ensemble model for forecasting and analyzing daily average PM2. 5 concentrations in Beijing, China. Sci. Total Environ. 2018, 635, 644–658. [Google Scholar] [CrossRef]
- Jia, R.; Lv, Y.; Wang, G.; Carranza, E.; Chen, Y.; Wei, C.; Zhang, Z. A stacking methodology of machine learning for 3D geological modeling with geological-geophysical datasets, Laochang Sn camp, Gejiu (China). Comput. Geosci. 2021, 151, 104754. [Google Scholar] [CrossRef]
- Zhou, T.; Jiao, H. Exploration of the stacking ensemble machine learning algorithm for cheating detection in large-scale assessment. Educ. Psychol. Meas. 2022, 1, 1–24. [Google Scholar] [CrossRef]
- Meharie, M.G.; Mengesha, W.J.; Gariy, Z.A.; Mutuku, R.N.N. Application of stacking ensemble machine learning algorithm in predicting the cost of highway construction projects. Eng. Constr. Archit. Manag. 2022, 29, 2836–2853. [Google Scholar] [CrossRef]
- Dong, Y.C.; Zhang, H.L.; Wang, C.; Zhou, X.J. Wind power forecasting based on stacking ensemble model, decomposition and intelligent optimization algorithm. Neurocomputing 2021, 462, 169–184. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.W.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.X.; Zheng, Y.L.; Zhang, R.K.; Jiang, Y.Q.; Poon, C.C.Y. Using a Multi-Task Recurrent Neural Network With Attention Mechanisms to Predict Hospital Mortality of Patients. IEEE J. Biomed. Health Inform. 2020, 24, 486–492. [Google Scholar] [CrossRef]
- Gangavarapu, T.; Jayasimha, A.; Krishnan, G.S.; Kamath, S.S. Predicting ICD-9 code groups with fuzzy similarity based supervised multi-label classification of unstructured clinical nursing notes. Knowl.-Based Syst. 2020, 190, e105321. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Zeng, X.F.; Feng, Y.L.; Chen, Q.; Liu, Z.H.; Luo, H.; Zha, L.H.; Yu, Z.X. Association of Systemic Immune-Inflammation Index With Short-Term Mortality of Congestive Heart Failure: A Retrospective Cohort Study. Front. Cardiovasc. Med. 2021, 8, 15. [Google Scholar] [CrossRef]
- Guo, W.Q.; Peng, C.N.; Liu, Q.; Zhao, L.Y.; Guo, W.Y.; Chen, X.H.; Li, L. Association between base excess and mortality in patients with congestive heart failure. ESC Heart Fail. 2021, 8, 250–258. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Lin, W.C.; Zha, L.H.; Zeng, X.F.; Zeng, X.M.; Li, G.J.; Liu, Z.H.; Yu, Z.X. Serum Anion Gap Is Associated with All-Cause Mortality among Critically Ill Patients with Congestive Heart Failure. Dis. Markers 2020, 2020, 10. [Google Scholar] [CrossRef]
- Miao, F.; Cai, Y.P.; Zhang, Y.X.; Fan, X.M.; Li, Y. Predictive Modeling of Hospital Mortality for Patients With Heart Failure by Using an Improved Random Survival Forest. IEEE Access 2018, 6, 7244–7253. [Google Scholar] [CrossRef]
- Hu, Z.Y.; Du, D.P. A new analytical framework for missing data imputation and classification with uncertainty: Missing data imputation and heart failure readmission prediction. PLoS ONE 2020, 15, e0237724. [Google Scholar] [CrossRef]
- Harutyunyan, H.; Khachatrian, H.; Kale, D.C.; Ver Steeg, G.; Galstyan, A. Multitask learning and benchmarking with clinical time series data. Sci. Data 2019, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Li, K.; Shi, Q.W.; Liu, S.R.; Xie, Y.L.; Liu, J.L. Predicting in-hospital mortality in ICU patients with sepsis using gradient boosting decision tree. Medicine 2021, 100, 5. [Google Scholar] [CrossRef]
- Maeda-Gutierrez, V.; Galvan-Tejada, C.E.; Cruz, M.; Valladares-Salgado, A.; Galvan-Tejada, J.I.; Gamboa-Rosales, H.; Garcia-Hernandez, A.; Luna-Garcia, H.; Gonzalez-Curiel, I.; Martinez-Acuna, M. Distal Symmetric Polyneuropathy Identification in Type 2 Diabetes Subjects: A Random Forest Approach. Healthcare 2021, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, S.; Fogarty, S.; Tremeer, M.; Ross, K.; Richards, B.; Bergmeir, C.; Xu, S.; Stub, D.; Smith, K.; Tacey, M.; et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med. 2018, 15, 16. [Google Scholar] [CrossRef]
- Akbari, G.; Nikkhoo, M.; Wang, L.Z.; Chen, C.P.C.; Han, D.S.; Lin, Y.H.; Chen, H.B.; Cheng, C.H. Frailty Level Classification of the Community Elderly Using Microsoft Kinect-Based Skeleton Pose: A Machine Learning Approach. Sensors 2021, 21, 4017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.C.; Li, X.L.; Zong, M.; Zhu, X.F.; Cheng, D.B. Learning k for kNN Classification. ACM Trans. Intell. Syst. Technol. 2017, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Song, J.Z.; Liu, G.X.; Jiang, J.Q.; Zhang, P.; Liang, Y.C. Prediction of Protein-ATP Binding Residues Based on Ensemble of Deep Convolutional Neural Networks and LightGBM Algorithm. Int. J. Mol. Sci. 2021, 22, 939. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.T.; Xu, J.; Ying, H.C.; Chen, X.J.; Feng, R.W.; Fang, X.L.; Gao, H.H.; Wu, J. Prediction of Extubation Failure for Intensive Care Unit Patients Using Light Gradient Boosting Machine. IEEE Access 2019, 7, 150960–150968. [Google Scholar] [CrossRef]
- Li, L.J.; Lin, Y.K.; Yu, D.X.; Liu, Z.Y.; Gao, Y.J.; Qiao, J.P. A Multi-Organ Fusion and LightGBM Based Radiomics Algorithm for High-Risk Esophageal Varices Prediction in Cirrhotic Patients. IEEE Access 2021, 9, 15041–15052. [Google Scholar] [CrossRef]
- Ali, S.; Majid, A.; Javed, S.G.; Sattar, M. Can-CSC-GBE: Developing Cost-sensitive Classifier with Gentleboost Ensemble for breast cancer classification using protein amino acids and imbalanced data. Comput. Biol. Med. 2016, 73, 38–46. [Google Scholar] [CrossRef]
- Sarmah, C.K.; Samarasinghe, S. Microarray gene expression: A study of between-platform association of Affymetrix and cDNA arrays. Comput. Biol. Med. 2011, 41, 980–986. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Choi, J.Y.; Ro, Y.M. Region based stellate features combined with variable selection using AdaBoost learning in mammographic computer-aided detection. Comput. Biol. Med. 2015, 63, 238–250. [Google Scholar] [CrossRef]
- Lee, Y.W.; Choi, J.W.; Shin, E.H. Machine learning model for predicting malaria using clinical information. Comput. Biol. Med. 2021, 129, e104151. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Pal, S. Prediction of Skin Disease with Three Different Feature Selection Techniques Using Stacking Ensemble Method. Appl. Biochem. Biotechnol. 2020, 191, 637–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, Z.W.; Chen, C.; Wei, Q.Q.; Gu, H.M.; Yu, B. DeepStack-DTIs: Predicting Drug-Target Interactions Using LightGBM Feature Selection and Deep-Stacked Ensemble Classifier. Interdiscip. Sci.-Comput. Life Sci. 2021, 14, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Yin, Y.; Wang, D.; Li, Z.; Wang, Y. A stacking-based ensemble learning method for earthquake casualty prediction. Appl. Soft Comput. 2021, 101, 107038. [Google Scholar] [CrossRef]
- Cui, S.Z.; Qiu, H.X.; Wang, S.T.; Wang, Y.Z. Two-stage stacking heterogeneous ensemble learning method for gasoline octane number loss prediction. Appl. Soft Comput. 2021, 113, e107989. [Google Scholar] [CrossRef]
- Jiang, M.Q.; Liu, J.P.; Zhang, L.; Liu, C.Y. An improved Stacking framework for stock index prediction by leveraging tree-based ensemble models and deep learning algorithms. Phys. A Stat. Mech. Its Appl. 2019, 541, e122272. [Google Scholar] [CrossRef]
- Papouskova, M.; Hajek, P. Two-stage consumer credit risk modelling using heterogeneous ensemble learning. Decis. Support Syst. 2019, 118, 33–45. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Classifying imbalanced data using SMOTE based class-specific kernelized ELM. Int. J. Mach. Learn. Cybern. 2021, 12, 1255–1280. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Teng, F.; Ma, Z.; Chen, J.; Xiao, M.; Huang, L.F. Automatic Medical Code Assignment via Deep Learning Approach for Intelligent Healthcare. IEEE J. Biomed. Health Inform. 2020, 24, 2506–2515. [Google Scholar] [CrossRef]
- Chung, I.; Lip, G.Y.H. Platelets and heart failure. Eur. Heart J. 2006, 27, 2623–2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallner, M.; Eaton, D.M.; von Lewinski, D.; Sourij, H. Revisiting the Diabetes-Heart Failure Connection. Curr. Diabetes Rep. 2018, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Barnett, O.; Horiuchi, Y.; Wettersten, N.; Murray, P.; Maisel, A. Blood urea nitrogen and biomarker trajectories in acute heart failure. Eur. J. Heart Fail. 2019, 21, 257. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2001. [Google Scholar]
- Ma, X.; Sha, J.; Wang, D.; Yu, Y.; Yang, Q.; Niu, X. Study on a prediction of P2P network loan default based on the machine learning LightGBM and XGboost algorithms according to different high dimensional data cleaning. Electron. Commer. Res. Appl. 2018, 31, 24–39. [Google Scholar] [CrossRef]
- Austin, D.E.; Lee, D.S.; Wang, C.X.; Ma, S.H.; Wang, X.S.; Porter, J.; Wang, B. Comparison of machine learning and the regression-based EHMRG model for predicting early mortality in acute heart failure. Int. J. Cardiol. 2022, 365, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.D.; Zhu, Y.; Zhu, Z.; Li, R.X.; Chen, G.Q.; Wang, Z. A machine learning-based risk stratification tool for in-hospital mortality of intensive care unit patients with heart failure. J. Transl. Med. 2022, 20, 1–9. [Google Scholar] [CrossRef]
- Sluban, B.; Lavrač, N. Relating ensemble diversity and performance: A study in class noise detection. Neurocomputing 2015, 160, 120–131. [Google Scholar] [CrossRef]
- Scherpf, M.; Grasser, F.; Malberg, H.; Zaunseder, S. Predicting sepsis with a recurrent neural network using the MIMIC III database. Comput. Biol. Med. 2019, 113, 103395. [Google Scholar] [CrossRef]
- Tootooni, M.S.; Pasupathy, K.S.; Heaton, H.A.; Clements, C.M.; Sir, M.Y. CCMapper: An adaptive NLP-based free-text chief complaint mapping algorithm. Comput. Biol. Med. 2019, 113, 13. [Google Scholar] [CrossRef]
- Ping, Y.; Chen, C.; Wu, L.; Wang, Y.; Shu, M. Automatic detection of atrial fibrillation based on CNN-LSTM and shortcut connection. Healthcare 2020, 8, 139. [Google Scholar] [CrossRef]
- Viton, F.; Elbattah, M.; Guérin, J.-L.; Dequen, G. Heatmaps for visual explainability of cnn-based predictions for multivariate time series with application to healthcare. In Proceedings of the 2020 IEEE International Conference on Healthcare Informatics (ICHI), Oldenburg, Germany, 30 November–3 December 2020; pp. 1–8. [Google Scholar]
- Laudanski, K.; Shea, G.; DiMeglio, M.; Restrepo, M.; Solomon, C. What Can COVID-19 Teach Us about Using AI in Pandemics? Healthcare 2020, 8, 527. [Google Scholar] [CrossRef] [PubMed]
- Allam, Z.; Jones, D.S. On the coronavirus (COVID-19) outbreak and the smart city network: Universal data sharing standards coupled with artificial intelligence (AI) to benefit urban health monitoring and management. Healthcare 2020, 8, 46. [Google Scholar] [CrossRef] [PubMed]




| Method | Parameters |
|---|---|
| RF | max_depth = 20, min_samples_split = 0.001, n_estimators = 20 |
| SVC | C = 1.0, kernel = ‘rbf’, degree = 3, gamma = ‘auto’, coef0 = 0.0, shrinking = True, probability = False, tol = 0.001, cache_size = 200, class_weight = None, verbose = False, max_iter = −1 |
| KNN | n_neighbors = 5, weights = ‘uniform’, algorithm = ‘auto’, leaf_size = 30, p = 2, metric = ‘minkowski’ |
| LGBM | boosting_type = ‘gbdt’, num_leaves = 31, max_depth = −1, learning_rate = 0.1, n_estimators = 100, subsample_for_bin = 200,000 |
| min_child_samples = 20, subsample = 1.0, subsample_freq = 0, colsample_bytree = 1.0, reg_alpha = 0.0, reg_lambda = 0.0 | |
| n_jobs = −1, importance_type = ‘split’ | |
| Bagging | base_estimator = None, n_estimators = 10, max_samples = 1.0, max_features = 1.0, bootstrap = True, bootstrap_features = False, oob_score = False, warm_start = False, n_jobs = None, random_state = None, verbose = 0 |
| Adaboost | base_estimator = DecistionTreeClassifer, random_state = 1, n_estimators = 50, learning_rate = 1.0, algorithm = ‘SAMME.R’ |
| Number of Survive | Number of Death | Percentage of SMOTE Increase | Class “Survived” | Class “Died” | |
|---|---|---|---|---|---|
| 3 Days | 6486 | 213 | 3000% | 6486 | 6390 |
| 30 Days | 5822 | 877 | 600% | 5822 | 5262 |
| 3 Months | 5765 | 934 | 600% | 5765 | 5604 |
| 1 Year | 5754 | 945 | 600% | 5754 | 5670 |
| Overall | Alive at ICU | Dead at ICU | |
|---|---|---|---|
| General | |||
| Number | 6699 (100%) | 5754 (85.89%) | 945 (14.11%) |
| Age (Q1–Q3) | 70.31 ± 13.04 | 69.88 ± 13.03 | 72.92 ± 12.74 |
| Gender (male) | 3694 (55.14%) | 3185 (55.35%) | 509 (53.86%) |
| Outcomes | |||
| Hospital LOS (days) (Q1–Q3) | 13.04 (5.99–16.00) | 12.78 (6.06–15.77) | 14.60 (5.27–19.09) |
| ICU LOS (days) (Q1–Q3) | 5.79 (1.93–6.23) | 5.40 (1.88–5.77) | 8.17 (2.38–10.38) |
| Admission Type | |||
| ELECTIVE | 766 (11.43%) | 721 (12.53%) | 45 (4.76%) |
| EMERGENCY | 5668 (84.61%) | 4807 (83.54%) | 861 (91.11%) |
| URGENT | 265 (3.96%) | 226 (3.93%) | 39 (4.13%) |
| Care Unit Type | |||
| CCU | 1852 (27.65%) | 1633 (28.38%) | 219 (23.17%) |
| CSRU | 1319 (19.69) | 1240 (21.55%) | 79 (8.36%) |
| MICU | 2537 (37.87%) | 2049 (35.61%) | 488 (51.64%) |
| SICU | 635 (9.48%) | 526 (9.14%) | 109 (11.34%) |
| TSICU | 356 (5.31%) | 306 (5.32%) | 50 (5.29%) |
| Insurance | |||
| Government | 96 (1.43%) | 88 (1.53%) | 8 (0.85%) |
| Medicaid | 388 (5.79%) | 352 (6.12%) | 36 (3.81%) |
| Medicare | 4748 (70.88%) | 4014 (69.76%) | 734 (77.67%) |
| Private | 1439 (21.48%) | 1277 (22.19%) | 162 (17.14%) |
| Self Pay | 28 (0.42%) | 23 (0.40%) | 5 (0.53%) |
| Variable value | |||
| Heart Rate | 85.60 ± 15.49 | 85.03 ± 15.15 | 89.04 ± 16.98 |
| Respiratory Rate | 19.51 ± 4.18 | 19.31 ± 4.01 | 20.74 ± 4.94 |
| Diastolic Blood Pressure | 57.10 ± 12.15 | 57.57 ± 12.15 | 54.35 ± 11.73 |
| Systolic Blood Pressure | 115.01 ± 19.11 | 115.70 ± 19.04 | 110.98 ± 19.01 |
| Temperature | 98.18 ± 1.42 | 98.21 ± 1.38 | 98.02 ± 1.60 |
| Oxygen Saturation | 97.00 ± 2.24 | 97.09 ± 2.02 | 96.51 ± 3.25 |
| Fractional Inspired Oxygen | 15.55 ± 26.57 | 16.12 ± 26.69 | 12.73 ± 25.81 |
| Blood Urea Nitrogen | 33.53 ± 24.12 | 31.73 ± 22.70 | 44.50 ± 29.12 |
| Creatinine | 1.76 ± 2.06 | 1.72 ± 2.13 | 1.99 ± 1.57 |
| Mean Blood Pressure | 76.97 ± 12.45 | 76.98 ± 11.50 | 76.93 ± 17.14 |
| Glucose | 146.11 ± 46.21 | 144.65 ± 45.09 | 154.76 ± 51.53 |
| White Blood Cell | 12.63 ± 11.94 | 12.21 ± 7.27 | 15.15 ± 26.12 |
| Red Blood Cell | 3.54 ± 0.52 | 3.54 ± 0.52 | 3.50 ± 0.55 |
| Prothrombin Time | 16.61 ± 6.74 | 16.43 ± 6.63 | 17.66 ± 7.24 |
| International Normalized Ratio | 1.65 ± 1.00 | 1.61 ± 0.94 | 1.87 ± 1.28 |
| Platelets | 216.09 ± 95.72 | 217.45 ± 93.55 | 207.80 ± 107.65 |
| GCS eye | 3.32 ± 0.86 | 3.38 ± 0.80 | 2.90 ± 1.05 |
| GCS motor | 5.33 ± 1.11 | 5.41 ± 1.01 | 4.88 ± 1.49 |
| GCS verbal | 3.44 ± 1.67 | 3.54 ± 1.64 | 2.80 ± 1.74 |
| RF | SVC | KNN | LGBM | Bagging | Adaboost | Stacking | |
|---|---|---|---|---|---|---|---|
| 3 Days | 0.7598 ± 0.0092 | 0.7249 ± 0.0176 | 0.7490 ± 0.0220 | 0.7868 ± 0.0084 | 0.7534 ± 0.0151 | 0.7230 ± 0.0076 | 0.8255 ± 0.0201 |
| 30 Days | 0.7472 ± 0.0120 | 0.7179 ± 0.0086 | 0.7476 ± 0.0049 | 0.7724 ± 0.0086 | 0.7442 ± 0.0104 | 0.7168 ± 0.0078 | 0.8052 ± 0.0049 |
| 3 Months | 0.7433 ± 0.0121 | 0.7002 ± 0.0145 | 0.7313 ± 0.0110 | 0.7596 ± 0.0078 | 0.7338 ± 0.0120 | 0.7005 ± 0.0121 | 0.7830 ± 0.0155 |
| 1 Year | 0.6998 ± 0.0097 | 0.6671 ± 0.0081 | 0.6958 ± 0.0125 | 0.7269 ± 0.0062 | 0.7014 ± 0.0106 | 0.6706 ± 0.0048 | 0.7532 ± 0.0064 |
| Method | Precision | Recall | F-Score | Accuracy | |
|---|---|---|---|---|---|
| 3 Days | RF | 0.9167 ± 0.0328 | 0.3429 ± 0.0175 | 0.4989 ± 0.0210 | 0.9343 ± 0.0097 |
| SVC | 0.8991 ± 0.0382 | 0.1920 ± 0.0352 | 0.3105 ± 0.0487 | 0.9208 ± 0.0114 | |
| KNN | 0.6635 ± 0.0207 | 0.5198 ± 0.0415 | 0.5824 ± 0.0328 | 0.9288 ± 0.0125 | |
| LGBM | 0.8763 ± 0.0532 | 0.5821 ± 0.0163 | 0.6989 ± 0.0223 | 0.9425 ± 0.0050 | |
| Bagging | 0.8176 ± 0.0616 | 0.3959 ± 0.0309 | 0.5322 ± 0.0324 | 0.9343 ± 0.0055 | |
| AdaBoost | 0.5937 ± 0.0633 | 0.3082 ± 0.0146 | 0.4044 ± 0.0202 | 0.9139 ± 0.0078 | |
| Stacking | 0.8030 ± 0.0108 | 0.6682 ± 0.0402 | 0.7286 ± 0.0223 | 0.9525 ± 0.0081 | |
| 30 Days | RF | 0.7857 ± 0.0217 | 0.5455 ± 0.0254 | 0.6435 ± 0.0201 | 0.8457 ± 0.0056 |
| SVC | 0.7389 ± 0.0269 | 0.4962 ± 0.0173 | 0.5934 ± 0.0161 | 0.8262 ± 0.0050 | |
| KNN | 0.6399 ± 0.0242 | 0.6145 ± 0.0181 | 0.6264 ± 0.0105 | 0.8126 ± 0.0060 | |
| LGBM | 0.7704 ± 0.0104 | 0.6069 ± 0.0175 | 0.6789 ± 0.0144 | 0.8533 ± 0.0042 | |
| Bagging | 0.7526 ± 0.0246 | 0.5508 ± 0.0248 | 0.6355 ± 0.0171 | 0.8387 ± 0.0037 | |
| AdaBoost | 0.6827 ± 0.0380 | 0.5170 ± 0.0249 | 0.5871 ± 0.0136 | 0.8142 ± 0.0057 | |
| Stacking | 0.7831 ± 0.0171 | 0.6747 ± 0.0125 | 0.7247 ± 0.0086 | 0.8690 ± 0.0032 | |
| 3 Months | RF | 0.7878 ± 0.0102 | 0.5372 ± 0.0262 | 0.6385 ± 0.0200 | 0.8429 ± 0.0052 |
| SVC | 0.7577 ± 0.0207 | 0.4505 ± 0.0284 | 0.5647 ± 0.0263 | 0.8206 ± 0.0085 | |
| KNN | 0.6507 ± 0.0149 | 0.5692 ± 0.0201 | 0.6072 ± 0.0176 | 0.8095 ± 0.0074 | |
| LGBM | 0.7712 ± 0.0118 | 0.5792 ± 0.0188 | 0.6613 ± 0.0119 | 0.8466 ± 0.0037 | |
| Bagging | 0.7476 ± 0.0226 | 0.5304 ± 0.0270 | 0.6200 ± 0.0201 | 0.8320 ± 0.0055 | |
| AdaBoost | 0.6844 ± 0.0057 | 0.4780 ± 0.0264 | 0.5625 ± 0.0195 | 0.8079 ± 0.0074 | |
| Stacking | 0.7720 ± 0.0114 | 0.6311 ± 0.0350 | 0.6939 ± 0.0221 | 0.8564 ± 0.0059 | |
| 1 Year | RF | 0.7600 ± 0.0175 | 0.4412 ± 0.0223 | 0.5577 ± 0.0156 | 0.8397 ± 0.0076 |
| SVC | 0.7490 ± 0.0383 | 0.3722 ± 0.0221 | 0.4961 ± 0.0154 | 0.8267 ± 0.0103 | |
| KNN | 0.6252 ± 0.0196 | 0.4767 ± 0.0209 | 0.5408 ± 0.0198 | 0.8144 ± 0.0108 | |
| LGBM | 0.7405 ± 0.0231 | 0.5069 ± 0.0071 | 0.6017 ± 0.0107 | 0.8460 ± 0.0092 | |
| Bagging | 0.7114 ± 0.0172 | 0.4583 ± 0.0268 | 0.5569 ± 0.0192 | 0.8332 ± 0.0044 | |
| AdaBoost | 0.6558 ± 0.0149 | 0.4046 ± 0.0136 | 0.5001 ± 0.0090 | 0.8147 ± 0.0058 | |
| Stacking | 0.7428 ± 0.0229 | 0.5651 ± 0.0152 | 0.6414 ± 0.0093 | 0.8551 ± 0.0085 |
| Variable Importance | 3 Days | 30 Days | 3 Months | 1 Year |
|---|---|---|---|---|
| 1 | Platelets | Platelets | Platelets | Glucose |
| 2 | Glucose | Glucose | Glucose | Platelets |
| 3 | Blood Urea Nitrogen | Blood Urea Nitrogen | Blood Urea Nitrogen | Heart Rate |
| 4 | Age | Age | Diastolic Blood Pressure | Age |
| 5 | Systolic Blood Pressure | Heart Rate | Age | Blood Urea Nitrogen |
| 6 | Heart Rate | Diastolic Blood Pressure | Heart Rate | Respiratory Rate |
| 7 | White Blood Cell | Respiratory Rate | Mean Blood Pressure | Systolic Blood Pressure |
| 8 | Mean Blood Pressure | Systolic Blood Pressure | Respiratory Rate | Diastolic Blood Pressure |
| 9 | Diastolic Blood Pressure | Prothrombin Time | Systolic Blood Pressure | White Blood Cell |
| 10 | Prothrombin Time | Mean Blood Pressure | Prothrombin Time | Mean Blood Pressure |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, C.-C.; Wu, C.-M.; Chien, T.-N.; Kao, L.-J.; Li, C.; Jiang, H.-L. Applying an Improved Stacking Ensemble Model to Predict the Mortality of ICU Patients with Heart Failure. J. Clin. Med. 2022, 11, 6460. https://doi.org/10.3390/jcm11216460
Chiu C-C, Wu C-M, Chien T-N, Kao L-J, Li C, Jiang H-L. Applying an Improved Stacking Ensemble Model to Predict the Mortality of ICU Patients with Heart Failure. Journal of Clinical Medicine. 2022; 11(21):6460. https://doi.org/10.3390/jcm11216460
Chicago/Turabian StyleChiu, Chih-Chou, Chung-Min Wu, Te-Nien Chien, Ling-Jing Kao, Chengcheng Li, and Han-Ling Jiang. 2022. "Applying an Improved Stacking Ensemble Model to Predict the Mortality of ICU Patients with Heart Failure" Journal of Clinical Medicine 11, no. 21: 6460. https://doi.org/10.3390/jcm11216460
APA StyleChiu, C.-C., Wu, C.-M., Chien, T.-N., Kao, L.-J., Li, C., & Jiang, H.-L. (2022). Applying an Improved Stacking Ensemble Model to Predict the Mortality of ICU Patients with Heart Failure. Journal of Clinical Medicine, 11(21), 6460. https://doi.org/10.3390/jcm11216460






