Screening and Identification of Key Biomarkers in Metastatic Uveal Melanoma: Evidence from a Bioinformatic Analysis
Abstract
:1. Introduction
2. Methods
2.1. Microarray Data
2.2. Identification of DEGs
2.3. Pathway and Process Enrichment Analysis of DEGs
2.4. PPI Network Construction and Hub Gene Identification
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics
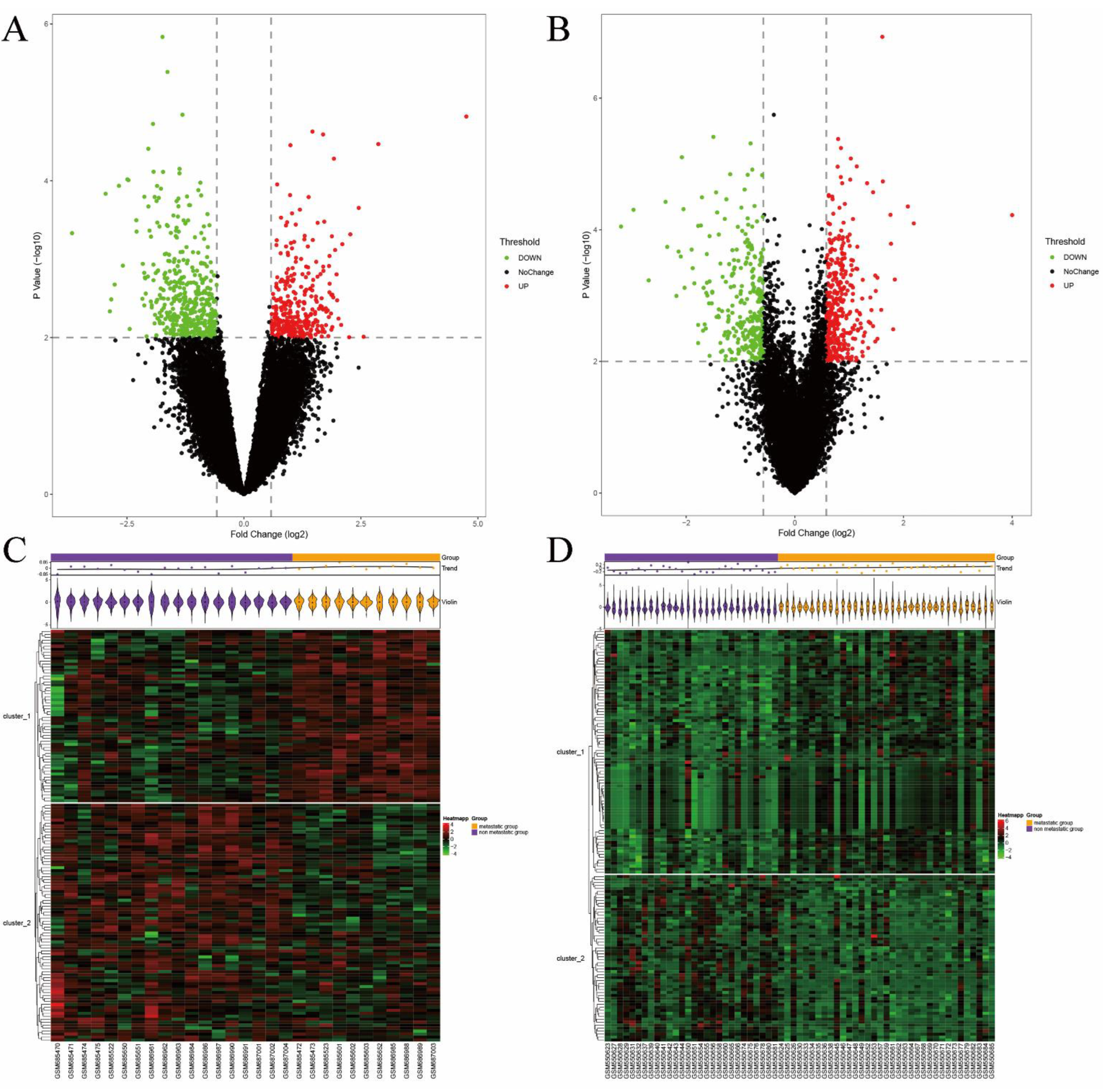
3.2. Identification of DEGs
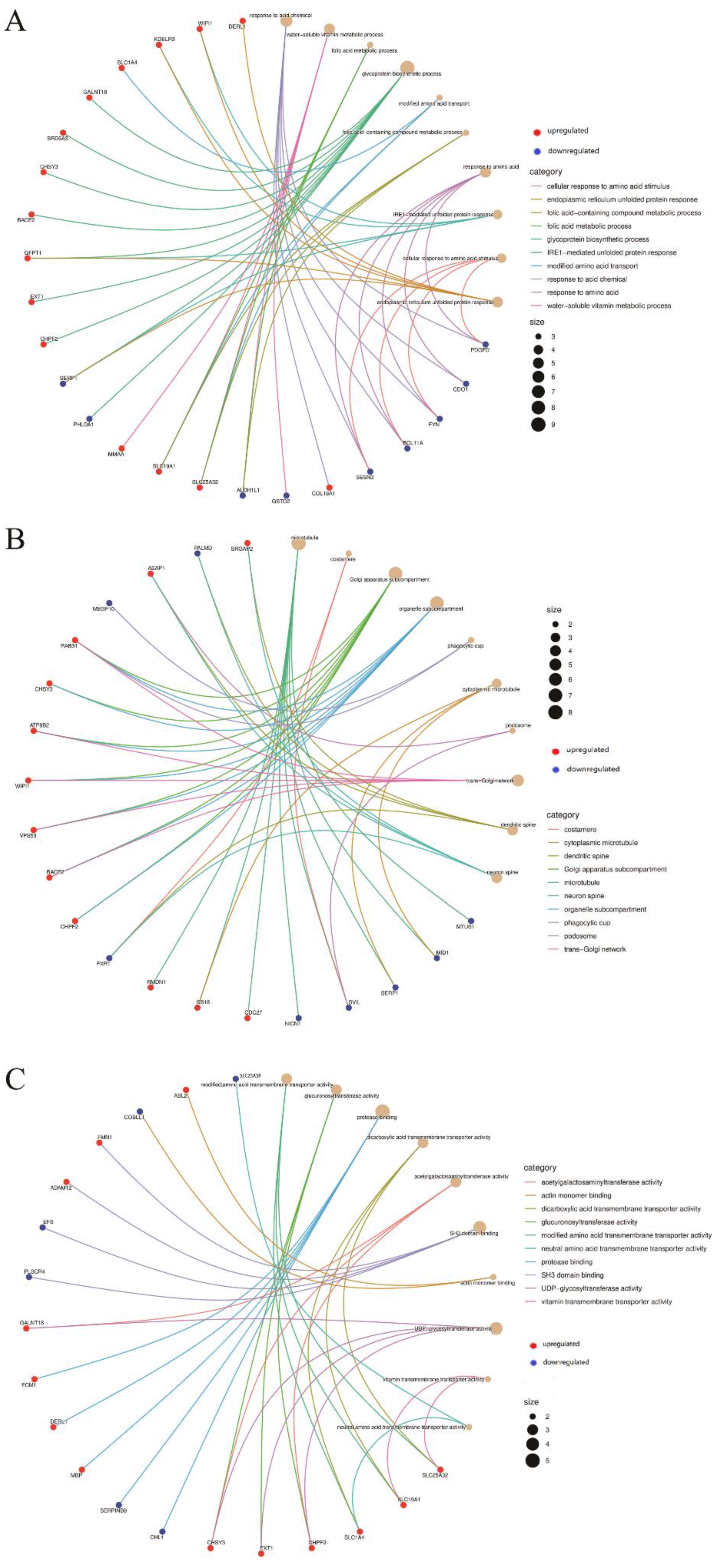
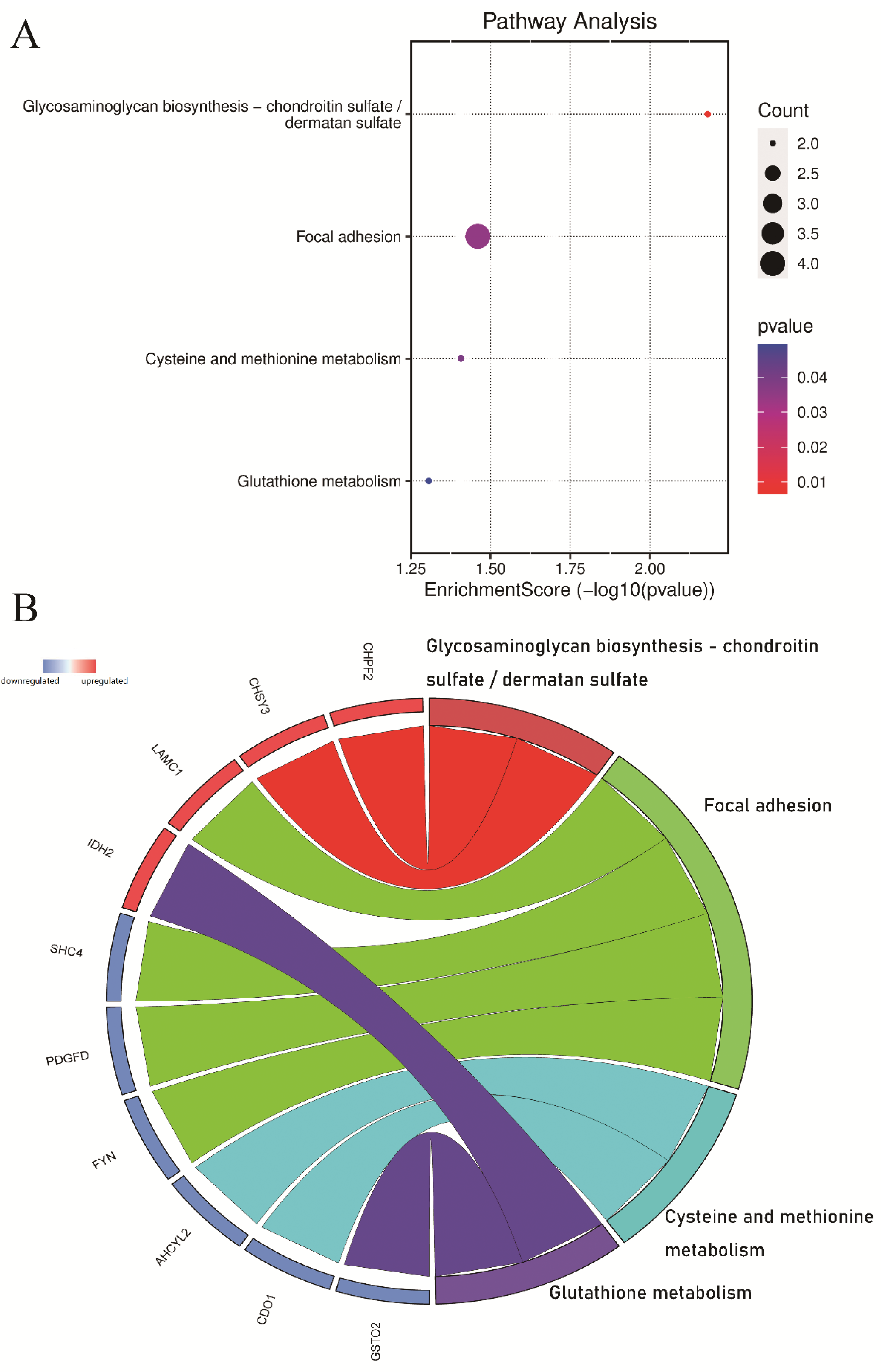
3.3. Pathway and Process Enrichment Analysis of DEGs
3.4. PPI Network Construction and Hub Gene Identification
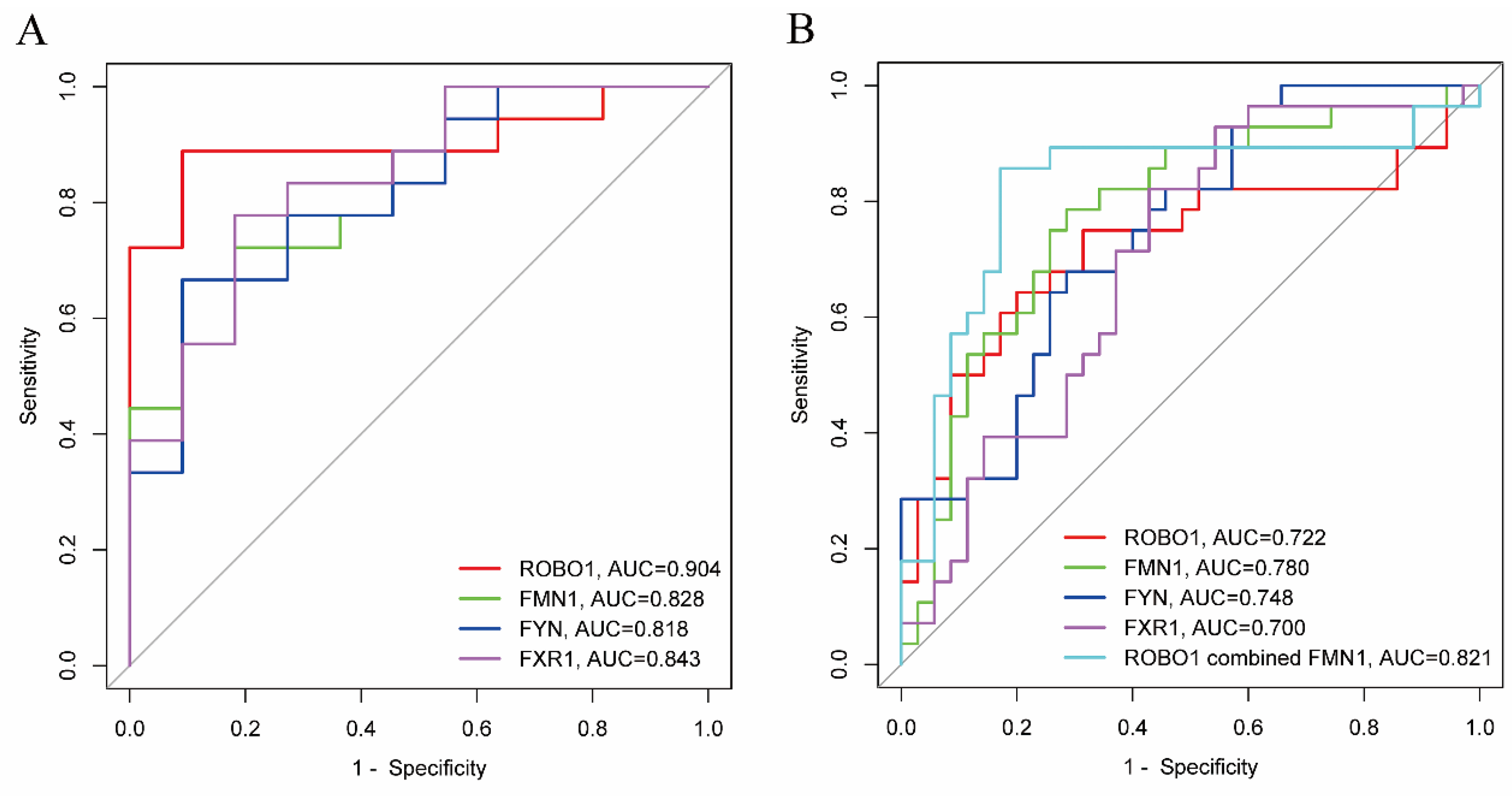
3.5. Biomarker Analysis of the Hub Genes
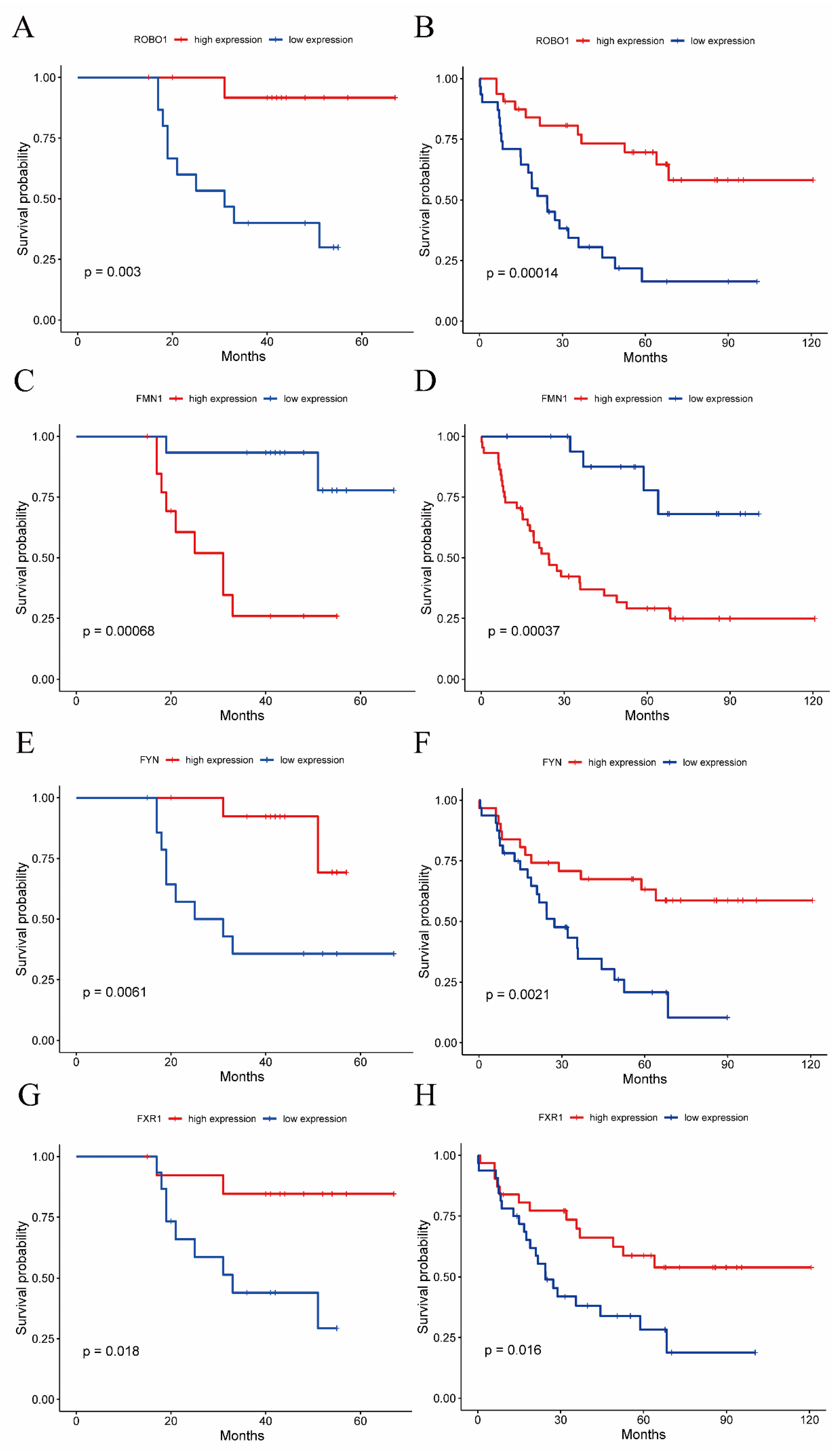
3.6. Metastasis-Free Survival Analysis of the Hub Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gragoudas, E.S.; Egan, K.M.; Seddon, J.M.; Glynn, R.J.; Walsh, S.M.; Finn, S.M.; Munzenrider, J.E.; Spar, M.D. Survival of patients with metastases from uveal melanoma. Ophthalmology 1991, 98, 383–389; discussion 390. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashyap, S.; Meel, R.; Singh, L.; Singh, M. Uveal melanoma. Semin. Diagn. Pathol. 2016, 33, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Chen, L.; Zhang, J. Expression analysis of genes and pathways associated with liver metastases of the uveal melanoma. BMC Med. Genet. 2014, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Gangemi, R.; Mirisola, V.; Barisione, G.; Fabbi, M.; Brizzolara, A.; Lanza, F.; Mosci, C.; Salvi, S.; Gualco, M.; Truini, M.; et al. Mda-9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS ONE 2012, 7, e29989. [Google Scholar] [CrossRef]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B.; et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006, 34, D322–D326. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [Green Version]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef]
- Isager, P.; Ehlers, N.; Overgaard, J. Prognostic factors for survival after enucleation for choroidal and ciliary body melanomas. Acta Ophthalmol. Scand. 2004, 82, 517–525. [Google Scholar] [CrossRef]
- Russo, A.; Avitabile, T.; Reibaldi, M.; Bonfiglio, V.; Pignatelli, F.; Fallico, M.; Caltabiano, R.; Broggi, G.; Russo, D.; Varricchio, S.; et al. Iris Melanoma: Management and Prognosis. Appl. Sci. 2020, 10, 8766. [Google Scholar] [CrossRef]
- Liu-Smith, F.; Farhat, A.M.; Arce, A.; Ziogas, A.; Taylor, T.; Wang, Z.; Yourk, V.; Liu, J.; Wu, J.; McEligot, A.J.; et al. Sex differences in the association of cutaneous melanoma incidence rates and geographic ultraviolet light exposure. J. Am. Acad. Dermatol. 2017, 76, 499–505.e3. [Google Scholar] [CrossRef]
- Kim, Y.I. Will mandatory folic acid fortification prevent or promote cancer? Am. J. Clin. Nutr. 2004, 80, 1123–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silsirivanit, A. Glycosylation markers in cancer. Adv. Clin. Chem. 2019, 89, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Kiewe, P.; Bechrakis, N.E.; Schmittel, A.; Ruf, P.; Lindhofer, H.; Thiel, E.; Nagorsen, D. Increased chondroitin sulphate proteoglycan expression (B5 immunoreactivity) in metastases of uveal melanoma. Ann. Oncol. 2006, 17, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Mackenzie, P.I.; McKinnon, R.A.; Meech, R. Genetic polymorphisms of human UDP-glucuronosyltransferase (UGT) genes and cancer risk. Drug Metab. Rev. 2016, 48, 47–69. [Google Scholar] [CrossRef]
- Hussain, M.R.; Hoessli, D.C.; Fang, M. N-acetylgalactosaminyltransferases in cancer. Oncotarget 2016, 7, 54067–54081. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; Shao, Y.; Zhao, X.; Hong, C.S.; Wang, F.; Lu, X.; Li, J.; Ye, G.; Yan, M.; Zhuang, Z.; et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol. Cell. Proteom. MCP 2016, 15, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxidative Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.Z.; Ibrahim, L.A.; Kim, Y.J.; Gibson, D.A.; Leung, H.C.; Yuan, W.; Zhang, K.K.; Tao, H.W.; Ma, L.; Zhang, L.I. Slit/Robo signaling mediates spatial positioning of spiral ganglion neurons during development of cochlear innervation. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 12242–12254. [Google Scholar] [CrossRef] [Green Version]
- Whitford, K.L.; Marillat, V.; Stein, E.; Goodman, C.S.; Tessier-Lavigne, M.; Chédotal, A.; Ghosh, A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron 2002, 33, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Blockus, H.; Chédotal, A. The multifaceted roles of Slits and Robos in cortical circuits: From proliferation to axon guidance and neurological diseases. Curr. Opin. Neurobiol. 2014, 27, 82–88. [Google Scholar] [CrossRef]
- Blockus, H.; Chédotal, A. Slit-Robo signaling. Development 2016, 143, 3037–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Funahashi, S.; Yamauchi, N.; Shibahara, J.; Midorikawa, Y.; Kawai, S.; Kinoshita, Y.; Watanabe, A.; Hippo, Y.; Ohtomo, T.; et al. Identification of ROBO1 as a novel hepatocellular carcinoma antigen and a potential therapeutic and diagnostic target. Clin. Cancer Res. 2006, 12, 3257–3264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallol, A.; Krex, D.; Hesson, L.; Eng, C.; Maher, E.R.; Latif, F. Frequent epigenetic inactivation of the SLIT2 gene in gliomas. Oncogene 2003, 22, 4611–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallol, A.; Da Silva, N.F.; Viacava, P.; Minna, J.D.; Bieche, I.; Maher, E.R.; Latif, F. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002, 62, 5874–5880. [Google Scholar] [PubMed]
- Dallol, A.; Morton, D.; Maher, E.R.; Latif, F. SLIT2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Cancer Res. 2003, 63, 1054–1058. [Google Scholar]
- Mehlen, P.; Delloye-Bourgeois, C.; Chédotal, A. Novel roles for Slits and netrins: Axon guidance cues as anticancer targets? Nat. Rev. Cancer 2011, 11, 188–197. [Google Scholar] [CrossRef]
- Narayan, G.; Goparaju, C.; Arias-Pulido, H.; Kaufmann, A.M.; Schneider, A.; Dürst, M.; Mansukhani, M.; Pothuri, B.; Murty, V.V. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol. Cancer 2006, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhou, F.L.; Li, W.P.; Wang, J.; Wang, L.J. Slit2-Robo1 signaling promotes the adhesion, invasion and migration of tongue carcinoma cells via upregulating matrix metalloproteinases 2 and 9, and downregulating E-cadherin. Mol. Med. Rep. 2016, 14, 1901–1906. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Xiao, Y.; Ding, B.B.; Zhang, N.; Yuan, X.; Gui, L.; Qian, K.-X.; Duan, S.; Chen, Z.; Rao, Y.; et al. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell 2003, 4, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, S.; Mitsui, K.; Kawamura, T.; Iwanari, H.; Daigo, K.; Horiuchi, K.; Minami, T.; Kodama, T.; Hamakubo, T. Suppression of Slit2/Robo1 mediated HUVEC migration by Robo4. Biochem. Biophys. Res. Commun. 2016, 469, 797–802. [Google Scholar] [CrossRef]
- Prasad, A.; Fernandis, A.Z.; Rao, Y.; Ganju, R.K. Slit protein-mediated inhibition of CXCR4-induced chemotactic and chemoinvasive signaling pathways in breast cancer cells. J. Biol. Chem. 2004, 279, 9115–9124. [Google Scholar] [CrossRef] [PubMed]
- Stella, M.C.; Trusolino, L.; Comoglio, P.M. The Slit/Robo system suppresses hepatocyte growth factor-dependent invasion and morphogenesis. Mol. Biol. Cell 2009, 20, 642–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, M.J.; Blaine, S.; Van Putten, V.; Saavedra, M.; Nemenoff, R.A. Lung Krüppel-like factor (LKLF) is a transcriptional activator of the cytosolic phospholipase A2 alpha promoter. Biochem. J. 2005, 387, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson-Grusby, L.; Kuo, A.; Leder, P. A variant limb deformity transcript expressed in the embryonic mouse limb defines a novel formin. Genes Dev. 1992, 6, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Woychik, R.P.; Maas, R.L.; Zeller, R.; Vogt, T.F.; Leder, P. ‘Formins’: Proteins deduced from the alternative transcripts of the limb deformity gene. Nature 1990, 346, 850–853. [Google Scholar] [CrossRef]
- Kobielak, A.; Pasolli, H.A.; Fuchs, E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004, 6, 21–30. [Google Scholar] [CrossRef]
- Dettenhofer, M.; Zhou, F.; Leder, P. Formin 1-isoform IV deficient cells exhibit defects in cell spreading and focal adhesion formation. PLoS ONE 2008, 3, e2497. [Google Scholar] [CrossRef] [Green Version]
- Simon-Areces, J.; Dopazo, A.; Dettenhofer, M.; Rodriguez-Tebar, A.; Garcia-Segura, L.M.; Arevalo, M.A. Formin1 mediates the induction of dendritogenesis and synaptogenesis by neurogenin3 in mouse hippocampal neurons. PLoS ONE 2011, 6, e21825. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.D.; Jensen, A.R.; Salgia, R.; Posadas, E.M. Fyn: A novel molecular target in cancer. Cancer 2010, 116, 1629–1637. [Google Scholar] [CrossRef]
- Semba, K.; Nishizawa, M.; Miyajima, N.; Yoshida, M.C.; Sukegawa, J.; Yamanashi, Y.; Sasaki, M.; Yamamoto, T.; Toyoshima, K. yes-related protooncogene, syn, belongs to the protein-tyrosine kinase family. Proc. Natl. Acad. Sci. USA 1986, 83, 5459–5463. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.H.; Yoo, K.C.; An, Y.; Lee, H.J.; Lee, M.; Uddin, N.; Kim, M.-J.; Kim, I.-G.; Suh, Y.; Lee, S.-J. FYN promotes mesenchymal phenotypes of basal type breast cancer cells through STAT5/NOTCH2 signaling node. Oncogene 2018, 37, 1857–1868. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, G.; Hao, Y.; Hammell, M.; Wilkinson, J.E.; Tonks, N.K. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev. 2017, 31, 1939–1957. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Li, Z.; Tian, F.; Li, X.; Yang, J. Fyn/heterogeneous nuclear ribonucleoprotein E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin β1. Int. J. Oncol. 2017, 51, 169–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Qi, Q.; Chan, C.B.; Zhou, W.; Chen, J.; Luo, H.R.; Appin, C.; Brat, D.J.; Ye, K. Fyn-phosphorylated PIKE-A binds and inhibits AMPK signaling, blocking its tumor suppressive activity. Cell Death Differ. 2016, 23, 52–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gujral, T.S.; Chan, M.; Peshkin, L.; Sorger, P.K.; Kirschner, M.W.; MacBeath, G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014, 159, 844–856. [Google Scholar] [CrossRef] [Green Version]
- Malho, P.; Dunn, K.; Donaldson, D.; Dubielzig, R.R.; Birand, Z.; Starkey, M. Investigation of prognostic indicators for human uveal melanoma as biomarkers of canine uveal melanoma metastasis. J. Small Anim. Pract. 2013, 54, 584–593. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Tuscan, M.D.; Harbour, J.W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J. Mol. Diagn. JMD 2010, 12, 461–468. [Google Scholar] [CrossRef]
- Comtesse, N.; Keller, A.; Diesinger, I.; Bauer, C.; Kayser, K.; Huwer, H.; Lenhof, H.-P.; Meese, E. Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. Int. J. Cancer 2007, 120, 2538–2544. [Google Scholar] [CrossRef]
- Qian, J.; Hassanein, M.; Hoeksema, M.D.; Harris, B.K.; Zou, Y.; Chen, H.; Lu, P.; Eisenberg, R.; Wang, J.; Espinosa, A.; et al. The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 3469–3474. [Google Scholar] [CrossRef] [Green Version]
- McClure, J.J.; Palanisamy, V. Muscle-Specific FXR1 Isoforms in Squamous Cell Cancer. Trends Cancer 2019, 5, 82–84. [Google Scholar] [CrossRef]
- Cao, S.; Zheng, J.; Liu, X.; Liu, Y.; Ruan, X.; Ma, J.; Liu, L.; Wang, D.; Yang, C.; Cai, H.; et al. FXR1 promotes the malignant biological behavior of glioma cells via stabilizing MIR17HG. J. Exp. Clin. Cancer Res. CR 2019, 38, 37. [Google Scholar] [CrossRef]
- Jin, X.; Zhai, B.; Fang, T.; Guo, X.; Xu, L. FXR1 is elevated in colorectal cancer and acts as an oncogene. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 2683–2690. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Gao, R.; Yu, C.; Chen, L.; Feng, Y. The RNA-binding protein FXR1 modulates prostate cancer progression by regulating FBXO4. Funct. Integr. Genom. 2019, 19, 487–496. [Google Scholar] [CrossRef] [PubMed]

| GSE27831 | GSE22138 | ||||||
|---|---|---|---|---|---|---|---|
| Variables | Metastatic Group (n = 11) | Nonmetastatic Group (n = 18) | p† | Metastatic Group (n = 35) | Nonmetastatic Group (n = 28) | p† | |
| Age, y | 66.7 ± 12.3 | 65.6 ± 13.4 | 0.823 t | 62.5 ± 9.6 | 59.1 ± 15.0 | 0.306 t | |
| Male gender (%) | 7 (63.6) | 10 (55.6) | 0.717 F | 22 (62.9) | 17 (60.7) | 0.862 P | |
| Tumor location | anterior | 2 (18.2) | 2 (12.5) | 0.653 F | 2 (5.9) | 1 (4.2) | 0.305 F |
| middle | 5 (45.5) | 10 (62.5) | 22 (64.7) | 20 (83.3) | |||
| posterior | 4 (36.4) | 4 (25.0) | 6 (17.6) | 3 (12.5) | |||
| 2 or 3 locations | 4 (11.8) | 0 (0) | |||||
| Tumor diameter (mm) | 13.0 ± 4.0 | 13.0 ± 6.0 | 0. 912 U | 15.2 ± 3.7 | 15.6 ± 3.9 | 0.921 t | |
| Tumor thickness (mm) | 9.9 ± 4.1 | 7.8 ± 3.4 | 0.236 t | 11.9 ± 1.9 | 11.3 ± 2.1 | 0.306 t | |
| Monosomy of chromosome 3 (%) | 10 (90.9) | 8 (44.4) | 0.019 P | 12 (86.2) | 12 (46.2) | 0.002 P | |
| Extrascleral extension (%) | 4 (36.4) | 10 (48.3) | 0.450 F | 5 (17.2) | 0 (0) | 0.056 F | |
| Tumor cell type | spindle | 1 (9.1) | 8 (50.0) | 0.117 F | 0 (0) | 0 (0) | 0.112 p |
| epithelioid | 3 (27.3) | 3 (18.8) | 15 (57.7) | 6 (33.3) | |||
| mixed | 7 (63.6) | 5 (31.3) | 11 (42.3) | 12 (66.7) | |||
| Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | ||
| GSE27831 | ROBO1 | 1.002 (1.001–1.004) | 0.010 | 1.002 (1.001–1.004) | 0.010 |
| FMN1 | 0.997 (0.995–0.999) | 0.013 | 0.998 (0.995–1.001) | 1.197 | |
| FYN | 1.004 (1.000–1.008) | 0.030 | 1.003 (0.995–1.011) | 0.424 | |
| FXR1 | 1.006 (1.001–1.010) | 0.010 | 1.004 (0.999–1.009) | 0.134 | |
| GSE22138 | ROBO1 | 1.583 (1.173–2.136) | 0.003 | 1.456 (1.054–2.011) | 0.023 |
| FMN1 | 0.432 (0.260–0.719) | 0.001 | 0.577 (0.336–0.992) | 0.047 | |
| FYN | 2.491 (1.414–4.387) | 0.002 | 1.804 (0.929–3.500) | 0.081 | |
| FXR1 | 2.009 (1.166–3.461) | 0.012 | 1.078 (0.543–2.139) | 0.830 | |
| GSE27831 | GSE22138 | |||||
|---|---|---|---|---|---|---|
| Genes | 95% CI | AUC | p | AUC | p | |
| ROBO1 | 0.788–1.000 | 0.904 | <0.001 | 0.586–0.859 | 0.722 | 0.003 |
| FMN1 | 0.678–0.978 | 0.828 | 0.003 | 0.662–0.897 | 0.780 | <0.001 |
| FYN | 0.661–0.975 | 0.818 | 0.005 | 0.629–0.867 | 0.748 | 0.001 |
| FXR1 | 0.697–0.990 | 0.843 | 0.002 | 0.570–0.830 | 0.700 | 0.007 |
| ROBO1 combined FMN1 | 0.703–0.939 | 0.821 | <0.001 | |||
| Low-Expression Group (Months) | High-Expression Group (Months) | ||
|---|---|---|---|
| GSE27831 | ROBO1 | 31.0 | 41.5 |
| FMN1 | 44.0 | 23.0 | |
| FYN | 25.0 | 41.5 | |
| FXR1 | 31.0 | 43.5 | |
| GSE22138 | ROBO1 | 24.4 | 57.9 |
| FMN1 | 55.8 | 23.1 | |
| FYN | 24.5 | 58.7 | |
| FXR1 | 24.5 | 55.8 |
| Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| GSE27831 | ROBO1 | 0.999 (0.997–1.000) | 0.009 | 0.999 (0.997–1.000) | 0.009 |
| FMN1 | 1.002 (1.001–1.002) | 0.002 | 1.000 (0.999–1.002) | 0.455 | |
| FYN | 0.996 (0.993–0.999) | 0.021 | 0.997 (0.993–1.001) | 0.125 | |
| FXR1 | 0.996 (0.993–0.999) | 0.009 | 1.004 (0.995–1.002) | 0.534 | |
| GSE22138 | ROBO1 | 0.768 (0.644–0.915) | 0.003 | 0.768 (0.634–0.931) | 0.007 |
| FMN1 | 1.839 (1.346–2.512) | <0.001 | 1.832 (1.316–2.552) | <0.001 | |
| FYN | 0.605 (0.461–0.794) | <0.001 | 0.788 (0.570–1.090) | 0.150 | |
| FXR1 | 0.688 (0.511–0.928) | 0.014 | 0.960 (0.645–1.430) | 0.842 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Wang, Z.; Yang, J.; Chen, Y.; Min, H. Screening and Identification of Key Biomarkers in Metastatic Uveal Melanoma: Evidence from a Bioinformatic Analysis. J. Clin. Med. 2022, 11, 7224. https://doi.org/10.3390/jcm11237224
Wang T, Wang Z, Yang J, Chen Y, Min H. Screening and Identification of Key Biomarkers in Metastatic Uveal Melanoma: Evidence from a Bioinformatic Analysis. Journal of Clinical Medicine. 2022; 11(23):7224. https://doi.org/10.3390/jcm11237224
Chicago/Turabian StyleWang, Tan, Zixing Wang, Jingyuan Yang, Youxin Chen, and Hanyi Min. 2022. "Screening and Identification of Key Biomarkers in Metastatic Uveal Melanoma: Evidence from a Bioinformatic Analysis" Journal of Clinical Medicine 11, no. 23: 7224. https://doi.org/10.3390/jcm11237224






